Significance
The ability of printing single-cell arrays with high precision and efficiency, single-cell resolution, multiple cell types, and maintenance of cell viability and function is essential for cell function and heterogeneity measurement. It is still hard for current methods to completely satisfy the above requirements. We report a unique live-cell printing technique, Block-Cell-Printing, that allows for convenient, precise, multiplexed, and high-throughput printing of functional single-cell arrays. Block-Cell-Printing has a minimum turnaround time of 0.5 h, a maximum resolution of 5 µm, and close to 100% cell viability. This method has been applied to study cell communications in heterotypic cell pairs with controlled morphology, characterize cells’ abilities to extend their membranes, and print primary neurons.
Keywords: cell array, cell communication, protrusion profiling, neuron patterning
Abstract
A unique live-cell printing technique, termed “Block-Cell-Printing” (BloC-Printing), allows for convenient, precise, multiplexed, and high-throughput printing of functional single-cell arrays. Adapted from woodblock printing techniques, the approach employs microfluidic arrays of hook-shaped traps to hold cells at designated positions and directly transfer the anchored cells onto various substrates. BloC-Printing has a minimum turnaround time of 0.5 h, a maximum resolution of 5 µm, close to 100% cell viability, the ability to handle multiple cell types, and efficiently construct protrusion-connected single-cell arrays. The approach enables the large-scale formation of heterotypic cell pairs with controlled morphology and allows for material transport through gap junction intercellular communication. When six types of breast cancer cells are allowed to extend membrane protrusions in the BloC-Printing device for 3 h, multiple biophysical characteristics of cells—including the protrusion percentage, extension rate, and cell length—are easily quantified and found to correlate well with their migration levels. In light of this discovery, BloC-Printing may serve as a rapid and high-throughput cell protrusion characterization tool to measure the invasion and migration capability of cancer cells. Furthermore, primary neurons are also compatible with BloC-Printing.
Current high-throughput screening of cell function and heterogeneity and in vitro cell–cell communication studies requires routine generation of large-scale single-cell arrays with high precision and efficiency, single-cell resolution, multiple cell types, and maintenance of cell viability and function (1, 2). Several approaches have been designed for this purpose, e.g., inkjet cell printing (3–6), surface engineering (7–15), and physical constraints (16–23). However, finding a method that completely satisfies the above requirements remains a challenge. Potentially useful and convenient tools may be available by adapting traditional printing tools to cell printing. In particular, woodblock printing is an efficient and convenient technology that revolutionized the printing world more than 1,800 y ago and was extended to microcontact molecular printing ∼20 y ago (24). However, application of the block-printing concept to cells has never been achieved. The main challenges are (i) inking cells to their molds with precision and maintaining viability, (ii) evenly and gently applying and transferring the cells to a substrate and successfully lifting off the mold without detaching the cells, and (iii) maintaining cell functions after printing.
We report here the development and testing of a technology called “Block-Cell-Printing” (BloC-Printing), which involves directly inking cells to a predesigned mold and then transferring the cells to a substrate. We overcome the challenges described above by flow patterning, instead of pressing, the ink objects, as in woodblock or microcontact printing. By performing various validation experiments, we prove that BloC-Printing can achieve a maximum spatial resolution of 5 µm, has the ability to simultaneously handle multiple cell types, results in close to 100% cell viability, and requires a minimum turnaround time of ∼0.5 h. The Block-Cell-Mold (BloC-Mold) is reusable for hundreds of printings. This approach does not require sophisticated equipment or large sample volumes. It is also straightforward and convenient, and it permits the subsequent culture of cells and image analysis under standard conditions.
Results and Discussion
Design and Operation of BloC-Printing.
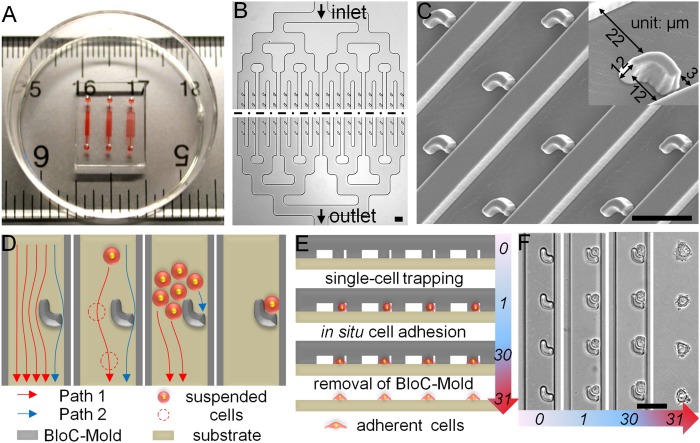
In a typical BloC-Printing process, the BloC-Mold, designed using AutoCAD (Autodesk) and fabricated by photolithography and polydimethylsiloxane (PDMS) molding techniques, was laid onto a Petri dish, glass slide, or other type of substrate, without thermal or oxygen plasma treatment. This formed an assembled BloC-Printing device with a network of microfluidic channels. A typical assembled BloC-Printing device is shown in Fig. 1A. After removal of air by vacuum pressure, cell culture medium is drawn into the channels by the application of negative pressure at the outlet (Fig. 1B, Lower). Then, the suspended cells are introduced into the BloC-Printing device from the inlet (Fig. 1B, Upper) and the flow of cells is driven by 1 psi vacuum pressure applied to the outlet. The flow force was carefully distributed to create a uniform flow of cells throughout the entire device. Single-cell traps were located along the sides of the flow channels with 3-µm gaps (Fig. 1C and SI Appendix, Fig. S1). The initial printings were performed with SK-BR-3 breast cancer cells (American Type Culture Collection, ATCC).
Fig. 1.
Design and operation of BloC-Printing technique. (A) A typical BloC-Printing device consists of a PDMS BloC-Mold and a commercially available PS Petri dish. The device is displayed on a ruler to show scale, and red dye has been injected to aid visualization of the three distinct channel networks with trap spacings of 30, 50, and 90 µm, from left to right (see also SI Appendix, Fig. S9A). (B) The BloC-Mold features symmetrical microfluidic channel networks and microarrays of traps. The black dashed line represents a large extended region between the input and output sides of the chip. (C) Scanning electron micrograph of the trap microarray, taken at a 20° tilt-angle. A magnified, single trap is shown (Inset). (D) Schematic diagram of cell flow paths. Cross-sectional schematics (E) and corresponding bright-field micrographs (F) showing the entire BloC-Printing process: (i) single-cell trapping by the traps, (ii) in situ cell adhesion, and (iii) removal of the BloC-Mold. The numbers in E and F represent time in minutes. (Scale bars: 50 µm.)
There are two potential flow paths around a trap structure (Fig. 1D, Left). The wide side consists of a 22-µm gap and the narrow side consists of a 3-µm gap, and these are labeled as paths 1 and 2, respectively. The fluid resistance ratio of paths 1 to 2 is ∼1:41, according to a theoretical calculation (SI Appendix, Fig. S2A). Therefore, at low cell densities (<104 cells per mL), almost all cells will flow through the wide side of the trap area (Fig. 1D, Center Left), because of the Zweifach–Fung bifurcation law. However, at high densities (>106 cells per mL), the wide side may be temporarily blocked by a group of cells such that an individual cell may be forced into the narrow channel and trapped (Fig. 1D, Center Right and Movies S1–S4). Because of the flexibility of the cells, the flow force will immediately clear such temporary blockages and keep an almost continuous cell flow (Fig. 1D, Right and SI Appendix, Fig. S2B). This crowding and trapping process takes place in the millisecond range and through the entire channel. The trapping efficiency has been carefully optimized by adjusting the trap size and shape, the fluid resistance ratio between paths 1 and 2, cell density, and the flow rate.
Again, because of the inherent cell flexibility, cells in the size range of 8 to 20 µm may all be effectively trapped by the 12 × 10-µm trap (SI Appendix, Fig. S2C). Additionally, precise cell positioning on the traps can be achieved by a slight increment in the vacuum flow pressure (SI Appendix, Fig. S2D). The flow of cells is not stopped until all traps capture their cells. In very rare circumstances, when a trap has captured more than one cell, the continuous flow of medium is able to remove any additional cells (Movie S2). The single-cell trapping efficiency in BloC-Printing is able to reach 100% (Fig. 2A and SI Appendix, Fig. S3).
Fig. 2.
Various cell arrays generated by BloC-Printing. (A) A bright-field image displays single-cell trapping efficiency in a 6 × 9 cell array. (B) Phase-contrast image of a printed 16 × 44 cell array. Bright field (C) and corresponding fluorescence image (D) of a 3 × 8 cell array. The cell microarray was stained with calcein AM (green) to show live cells and EthD-1 (red) to show dead cells (no dead cells appear in this array) immediately after BloC-Printing, to evaluate cell viability during the procedure. (E) Phase-contrast images of single-cell arrays with intercellular spacing of 30, 50, and 90 µm from left to right. (F) Phase-contrast images of cell pairs with intercellular spacing of 5, 10, and 20 µm from left to right, respectively. (G) Phase-contrast images of various single-cell arrays including a concentric square, a spiral square, capital letters “TMH” (abbreviation for “The Methodist Hospital”), an hourglass, a smiley face, and a ribbon (a bright-field image is also shown for the ribbon). SK-BR-3 cells were used in all images. (Scale bars: 50 µm.)
The next BloC-Printing step was to transfer the cells to a Petri dish or other substrates (Fig. 1 E and F). In this step, cells were allowed to adhere to the substrate by incubating the device for ∼0.5–1 h, depending on the adhesive capability of the cells to the substrates. SUM 159 cells and 3T3 fibroblasts were separately tested on both the polystyrene (PS) and glass substrates. Results revealed that both cell types could spread into cell flow channels within 1 h (SI Appendix, Figs. S4 and S5). After the cells were gently attached to the substrate, the BloC-Mold was then removed from the substrate, leaving behind the patterned array of cells on the substrate. Because the PDMS BloC-Mold is less adhesive to cells than the substrate, the transfer efficiency could be greater than 98%, shown as an orderly and high-density (more than 3 × 104 cells per cm2) live single-cell array (Fig. 2B and SI Appendix, Fig. S6).
Cell Viability Assays.
Cell viability was validated by calcein acetoxymethyl ester (AM) and ethdium homodimer-1 (EthD-1) staining (Fig. 2 C and D), whereby green fluorescence indicates live cells and red indicates dead ones. Only green fluorescence was observed. Cells were not damaged during the BloC-Printing procedures because (i) the flow rate was very gentle (less than 100 µm/s) with a short flow time (less than 2 min), (ii) the PDMS traps were fabricated from flexible elastomeric materials designed without sharp edges, and (iii) the gas permeability of the PDMS material allowed the cells to “breathe” during the cell adhesion process. When the medium was refreshed via a specially designed gravity-induced flow, MDA-MB-231/green fluorescent protein (GFP) cells inside the BloC-Printing device were able to grow and migrate, demonstrating normal morphology, for more than 48 h (SI Appendix, Fig. S7). After removal of the BloC-Mold, the printed SK-BR-3 cells were also able to divide and propagate on the PS cell culture dish for 5 d, indicating close to 100% cell viability (SI Appendix, Fig. S8).
Various Single-Cell Arrays Generated by BloC-Printing.
Through precise placement of traps in the BloC-Mold, the spatial resolution of the printed cell array was approximately equivalent to the cell sizes, with an edge-to-edge distance of less than 30 µm vertically and 5 µm horizontally (Fig. 2 E and F). Fig. 2E and SI Appendix, Fig. S9 show the controlled edge-to-edge cell spaces of 30, 50, and 90 μm and printing efficiency of more than 96%. The corresponding precision of the cell positions was within ±2.5 µm both horizontally and vertically (SI Appendix, Fig. S10). Because of cell spreading and the cell membrane extension on the substrate, actual distances were slightly shorter in the finished arrays. Therefore, this method will be very useful to precisely generate density-controlled cell patterns (25).
Control of cell-pair distance is remarkably simple with BloC-Printing, providing a potential approach for studying cell–cell interaction (26, 27) and cell fusion (28). By designing trap pairs with edge-to-edge spacing from 5 to 20 μm (SI Appendix, Fig. S11A), corresponding cell pairs with more than 94% printing efficiency were obtained (Fig. 2F and SI Appendix, Fig. S11B). The fluctuation in cell spacing was within ±3 μm horizontally (SI Appendix, Fig. S12). Therefore, sophisticated and high-resolution single-cell arrays could also be made in various shapes, including a concentric square, a spiral square, shapes of the capital letters “TMH,” an hourglass, a smiley face, and a ribbon (Fig. 2G and SI Appendix, Fig. S13). Moreover, the BloC-Printing approach also allows for flexibility in printing substrates. In addition to the PS Petri dish surface, direct printing on ultrathin glass (0.085–0.13 mm thickness) and elastic polyethylene napthalate (PEN) membranes (29) has also been achieved (SI Appendix, Fig. S14).
BloC-Printing has resolution limits of 5 µm horizontally and 30 µm vertically. In the cell pairing design, cell pairs are isolated with a PDMS wall. The wall thickness determines the final printed gap size. To maintain the strength of the PDMS wall, the wall must be at 5 µm thick. For the regularly spaced arrays, sufficient space needs to be reserved between two adjacent hooks to allow cells to flow inside the hooks, which results in a resolution limit of around 30 µm. Because we measured cell spacing by edge-to-edge distance according to standards in the literature (15, 20) rather than center-to-center, the cell spreading will decrease the final average spacing. Although we predesigned the cell array as 30, 50, and 90 µm edge-to-edge spacings (SI Appendix, Fig. S9) with an estimated cell size at 12 µm, the experimental results showed slightly larger cell spreading than the estimation and exhibited 27.1, 47.9, and 89.9 µm measured ones (SI Appendix, Fig. S10). Herein cells spread more for high-density arrays. Such a variation is most likely because high cell density promotes cell spreading, in light of the high growth factor concentration generated from neighbor cells.
Multiplexed Single-Cell Arrays by BloC-Printing.
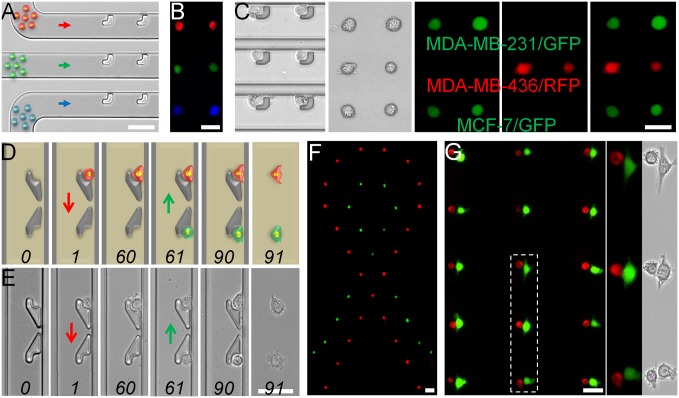
The BloC-Printing approach allows for printing of multiple cell types in both the vertical and horizontal direction. A parallel arrangement of cell flow channels allowed for multiple cell types to be simultaneously anchored to the BloC-Printing device (Fig. 3 A–C). All cells flow in the same direction, and each channel only allows one type of cell to flow through. In a proof-of-concept experiment, SK-BR-3 cells, labeled with red, green, and blue CellTracker fluorescent dye, were applied to three parallel channels and imaged with a multichannel fluorescence microscope (Fig. 3B). The same approach can also be applied to different cell types by slightly adjusting the trap size (SI Appendix, Fig. S15). When MDA-MB-231/GFP, MDA-MB-436/red fluorescent protein (RFP), and MCF-7/GFP cells were simultaneously flowed through the BloC-Printing device and transferred to a Petri dish, an array of the three types of cells was obtained and validated in bright-field and fluorescence images (Fig. 3C). Using this method, the total time for cell printing was about 0.5 h.
Fig. 3.
Multiplexed cell arrays generated by BloC-Printing. (A) The BloC-Mold for patterning three types of cells. Red, green, and blue arrows represent the direction of flow of the three types of cells by dye color. (B) Patterning of a 3 × 2 single-cell microarray with red, green, and blue CellTracker-labeled SK-BR-3 cells. (C) Patterning of a 3 × 2 single-cell microarray with MDA-MB-231/GFP, MDA-MB-436/RFP, and MCF-7/GFP cells. Schematic (D) and corresponding micrographs (E) showing the whole process of patterning with two types of cells differentially labeled with green or red dyes. The numbers and arrows in D and E, respectively, represent time and direction of flow. Patterning of a ribbon (F) and cell pairing (G) with red and green CellTracker-labeled cells. The two right-hand panels in G are enlarged views of three cell pairs within the dotted box. (Scale bars: 50 µm.)
As an alternative, arrays of two types of cells could also be printed by placing sets of two long-tail traps facing opposite directions and with each trap aligned in the direction of flow of one cell type but not the other (Fig. 3 D and E, SI Appendix, Fig. S16A, and Movies S3 and S4). In this experiment, two strategies were used to avoid washing cells away. First, cell adhesion was ensured for the first set of cells by observing the cell spreading, which could be established in less than 1 h. The spread cells would encounter lower shear force from fluid and have stronger adherence to the substrate. Second, a reduced flow rate was used when loading the second set of cells. As a result, more than 80% of the first set of cells was retained. This technique allowed for sophisticated two-cell patterns to be made, such as a dual-cell ribbon (Fig. 3F and SI Appendix, Fig. S16B) and a heterotypic cell-pair array (Fig. 3G and SI Appendix, Fig. S16C). Using this method, the total time for cell printing was about 1.5 h.
Protrusion-Connected Single-Cell Arrays.
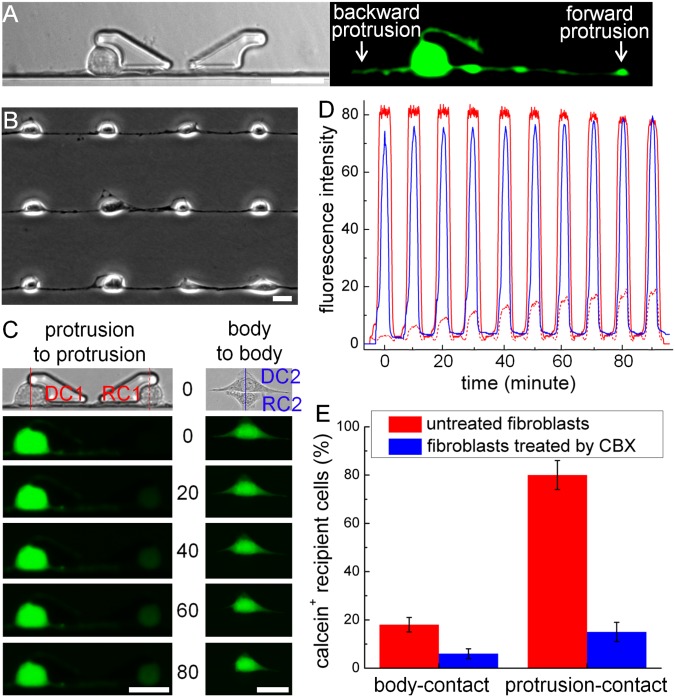
BloC-Printing can also be used to efficiently construct protrusion-connected single-cell arrays. When cells were trapped and allowed to spread for more than 2 h, they would typically generate two protrusions along the wall of the flow channel in both forward and backward directions (Fig. 4A). Cell spreading in random directions was rare in the BloC-Printing device (Fig. 4B). Such protrusion-connected single-cell arrays were successfully obtained with fibroblasts [National Institutes of Health (NIH 3T3)] and cancer cells (HeLa) on PS substrates and PEN membranes (Fig. 4B and SI Appendix, Fig. S17).
Fig. 4.
Dye transfer via GJIC in cell pairs with controllable morphology. (A) A fibroblast generates forward and backward protrusions along the channel wall after 3 h of trapping. (B) Protrusion-connected NIH 3T3 fibroblast cell arrays, cultured in the BloC-Printing device for 3 h, followed by removal of the BloC-Mold. (C) Comparison of calcein transport between protrusion-to-protrusion (Left) and body-to-body (Right) cell pairs. (D) Fluorescence intensity tracking of the DCs and RCs in C for 90 min. Signal intensities are read from the lines shown in bright-field images in C (Top). (E) Statistical data demonstrates the dye transfer efficiency in untreated versus GJIC-inhibited cells. The efficiency is expressed as the ratio of RCs that successfully received dye to the total number of RCs, after 3 h of cell trapping. The error bars represent the standard deviations of three independent experiments. (Scale bars: 20 μm.)
Gap Junction Intercellular Communication in Cell Pairs with Controlled Morphology.
To demonstrate the functionality and application of protrusion-connected single-cell arrays formed by BloC-Printing, we developed a challenging cell–cell communication model that had never been artificially created on a large scale. In this experiment, we formed heterotypic cell pairs with controlled morphology to study material transport through gap junction intercellular communication (GJIC) (30). Studying GJIC at the single-cell level is significant and challenging and has traditionally been carried out using techniques such as microinjection (31), dual whole-cell patch clamp (32), gap–fluorescence recovery after photobleaching (33), and local activation of a molecular fluorescent probe (34). However, such studies are impeded by the uncontrollability of gap junction formation and conflicting aims of reducing invasiveness while maintaining high throughput. The BloC-Printing method provides both noninvasive and high-throughput formation of cell pairs for studying GJIC in the form of controllable cell-to-cell contacts.
NIH 3T3 fibroblasts were chosen for the initial GJIC study. Fibroblasts labeled with calcein AM [the donor cells (DCs)] were patterned from top to bottom, and nonlabeled cells [the recipient cells (RCs)] were patterned in the opposite direction (SI Appendix, Fig. S18). Thus, the DCs and RCs were brought together as close neighbors in the cell array. When the forward protrusion from a DC physically contacted the backward protrusion from a neighboring RC, dye (calcein) transfer, a popular method for evaluating GJIC, was monitored and observed (SI Appendix, Fig. S19A). The dye transfer rate was slow at the beginning due to the limited number of gap junctions (35). With the generation of more gap junctions as the experiment continued, the dye transfer rate increased significantly. After 3 h of dye transfer, a balance was reached between the DCs and the RCs (SI Appendix, Fig. S19B).
In addition to the forward protrusion, the backward protrusion, although shorter in length, could also transfer calcein efficiently (SI Appendix, Fig. S19C), indicating the potential to study GJIC in protrusion-connected single-cell arrays. Two different arrangements of cell pairing (protrusion-to-protrusion and body-to-body contacts) were created between DCs and RCs, using the multiplexed BloC-Printing approach. Dye transfer experiments showed that cells engaged in GJIC more readily through protrusion contact than through body contact (Fig. 4 C–E). Dye transfer was inhibited by the GJIC blocker carbenoxolone, indicating that transfer of dye was indeed occurring through connected protrusions (Fig. 4E and SI Appendix, Fig. S20). This discovery will be useful for future study of cell morphology- and protrusion-related GJIC (36).
Characterization of Cells’ Capability to Extend the Membrane.
The ability of cells to generate membrane protrusions plays an important role in numerous biological activities, particularly in cell migration and invasion (37–40), which are mainly mediated by protrusions in the form of filopodia and lamellipodia (41). BloC-Printing provides a rapid and high-throughput method to characterize cell protrusions, including protrusion percentage and extension rate, and cell length, which are challenging to achieve using existing methods. A BloC-Mold containing a hook array with a longitudinal spacing of 200-μm was specifically designed to observe the extension of cell length (SI Appendix, Fig. S21A). Individual cells with long, thin protrusions were clearly visualized after on-chip culture (Fig. 5A and SI Appendix, Fig. S21B). Measurement after 3 h of culture was found to best represent the results as (i) extension of cell protrusions had almost stopped and (ii) cells began to move away from the trap with longer time in culture (SI Appendix, Fig. S21C). In such experiments, BloC-Molds could also be removed leaving the cell protrusions printed in the Petri dishes, if needed (Fig. 5B). We used this method to characterize protrusions of six types of breast cancer cells, including MDA-MB-231, MDA-MB-436, SUM 159, SUM 149, SK-BR-3, and MCF-7 (Fig. 5C).
Fig. 5.
Cells’ capability to extend membranes in the BloC-Printing device. (A) The morphology of MDA-MB-231/GFP cells after culture in the BloC device for 3 h. The hook, cell body, and cell membrane protrusions are indicated. (B) The morphology of the printed MDA-MB-436 cells on a Petri dish after removal of the BloC-Mold. (C) Representative images of six types of breast cancer cells after 3 h of culture in the BloC-Printing device. (D) Percentages of cells of the six selected cell lines that have extended their membranes. (E) The average extended cell length for the indicated cell lines. The red dashed line indicates average cell length before extension of cell protrusions. (F) The protrusion-generation rate calculated as the protrusion length divided by the cell extension time. More than 300 cells were counted following 3 h of BloC-Printing device culture. The averages in E and F were calculated for elongated and nonelongated cells combined. The error bars represent the SDs of three independent experiments. (Scale bars: 25 μm.)
For all six cell lines, the percentage of cells that elongated, the average cell length, and the average cell-extension rate were calculated and plotted in Fig. 5 D–F and SI Appendix, Fig. S22. It is not surprising that the percentages of cells that elongated (Fig. 5D) for the six cell lines correlate with their reported tumorigenicity (42), with invasiveness increasing from MCF-7 to SK-BR-3, SUM149, SUM159, MDA-MB-436, and MDA-MB-231. The same trend applied to the average cell length (Fig. 5E) and extension rate (Fig. 5F), when the averages were calculated for both elongated and nonelongated cells. In general, compared with luminal-like cancer cells, basal-like cancer cells, especially MDA-MB-231 and MDA-MB-436, had greater membrane elongation abilities, indicating their stronger migratory abilities (41). There was a slight change in the trend when the averages were calculated for only elongated cells, with MDA-MB-436 having the longest average length of protrusion (SI Appendix, Fig. S22); this seems reasonable, as cells are quite heterogeneous, and quantitation of cell invasiveness still remains a challenge given the complexity of the live-cell system.
BloC-Printing of Individual Primary Cortical Neurons.
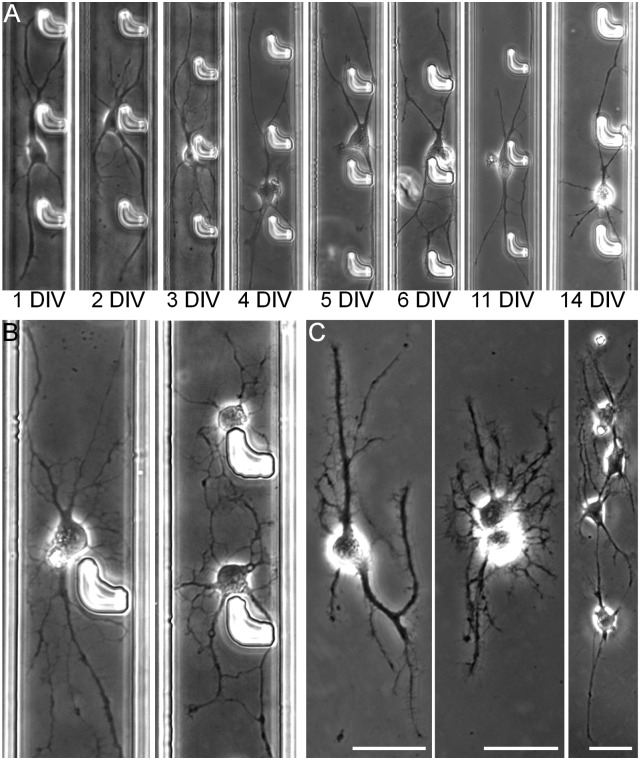
In addition to efficient printing of cancer and fibroblast cell lines, BloC-Printing can also be used for controllable printing of individual primary neurons. Positioning and addressing individual neurons are desirable for neuronal imaging and studies of signal transduction. Current methods are often limited by the difficulty of long term in vitro culture of individual neurons or the requirement of coculture with glial cells (43). Microfluidic devices have been described for culture of individual neurons for up to 11 d in vitro (DIV), without the use of any coculture or feeder layers (44). Such devices are still difficult to adapt to cell culture Petri dishes or substrates for measurement of neuronal activity because the neurons are retained in the PDMS device, and the PDMS material also requires complicated treatment. Herein, BloC-Printing was introduced to overcome such limitations. First, by heating the BloC-Mold at 110 °C for 60 min and then exposing it to UV light for 12 h, one can sterilize and completely cross-link the PDMS. Such a step does not require days of solvent exchange treatment for PDMS, as with earlier studies (44). Second, stopped-flow incubation was adapted to the BloC-Printing of neurons to minimize outside interference and maintain localized concentration of secretions (43). As a result, individual primary rat cortical neurons were successfully cultured for up to 14 DIV in the BloC-Mold (Fig. 6A). The neurons showed normal morphology and clear neurite outgrowth. The confined cell-spreading channel also increased the possibility of autapse formation (6 and 11 DIV) (45). By controlling the number and spacing of hooks (SI Appendix, Fig. S23), single and paired neurons with highly branched dendrites could be obtained at 7 DIV (Fig. 6B). Because neurons adhere to the selected substrates, the fine axons and dendrites could be successfully printed to these substrates via BloC-Printing (Fig. 6C), facilitating future analyses, such as measuring electrical signals via patch-clamp technique.
Fig. 6.
BloC-Printing of individual primary cortical neurons. (A) Morphology of individual neurons from 1 to 14 DIV in the BloC-Printing device. Autapses are observed at 6 and 11 DIV. (B) Single and paired neurons in the BloC-Printing device at 7 DIV. (C) Individual neurons at 9 DIV on the BloC-Printing substrate after the removal of BloC-Mold. The width of channel is 42 μm. (Scale bars: 25 μm.)
Conclusions
In conclusion, a unique live single-cell printing method, BloC-Printing, has been introduced. The approach allows for convenient and highly efficient formation of multiplexed single-cell arrays with precise, adjustable cell spacing, sophisticated single-cell patterning, coculture of heterotypic cell pairs, and an elongated cell array. The BloC-Mold can be reused hundreds of times without loss of precision, and single cells can be directly printed on to commonly used materials, including PS and glass cell-culture dishes. This method has been applied to the study of GJIC in heterotypic cell pairs with controlled morphology, rapidly characterizing cells’ ability to extend membranes, and for controlled printing of individual primary neurons. In the future, BloC-Printing may be combined with well-established molecular printing technology (16, 22, 46) to obtain multiplexed single-cell arrays for high-throughput drug screening (47).
Materials and Methods
Design and Fabrication of the BloC-Mold.
All designs were drawn with AutoCAD software and printed out as glass photomasks (Photo Sciences Inc.). PDMS BloC-Molds were fabricated by standard photolithography and elastomer molding. We used SPR 220–7 positive photoresist (MicroChem Corp.) to fabricate 12-μm thick channels and SU-8 3025 negative photoresist (MicroChem Corp.) to fabricate 17-μm thick channels. The SPR 220–7 photoresist was spin-coated onto a 4-inch silicon wafer (Silicon Quest International Inc.) at 1,500 rpm (Laurell Technologies Corp., Model: WS-400B-6NPP/LITE/AS) for 40 s to form a layer ∼12 μm thick. After baking at 75 °C for 3 min and then at 115 °C for 5 min, the wafer was cooled, exposed to UV light for 7 s, and developed. The SU-8 3025 photoresist was spin-coated onto a 4-inch silicon wafer at 4,000 rpm for 60 s to form a layer ∼17 μm in thickness. After soft baking at 65 °C for 2 min and then at 95 °C for 10 min, the wafer was cooled and exposed to UV light for 6 s. It was then heated for postexposure baking at 65 °C for 1 min and then at 95 °C for 3 min. After cooling down, the wafer was developed and heated for hard baking at 135 °C for 20 min. Finally PDMS (10A:1B; Dow Corning Corp.) was poured onto the photoresist mold and heated at 75 °C for 30 min. After curing, the PDMS was peeled off, cut to the appropriate size, and then punched to form the BloC-Printing device.
Cell Culture and Staining.
The cell line SK-BR-3 (ATCC) was cultured in RPMI medium 1640 supplemented with 10% (vol/vol) FBS and 1% penicillin-streptomycin. The cell lines MDA-MB-231/GFP, MDA-MB-436/RFP, and MCF-7/GFP (Cell Biolabs); SUM 159 (Asterand); and HeLa cells and NIH 3T3 fibroblasts (ATCC) were cultured in Dulbecco’s modified Eagle medium supplemented with 10% (vol/vol) FBS and 1% penicillin–streptomycin. The cell line SUM 149 (Asterand) was grown in Ham’s F-12 medium (Life Technologies Corp.) supplemented with 5% (vol/vol) FBS, 1% penicillin–streptomycin, 5 µg/mL insulin, and 1 µg/mL hydrocortisone. All cells were grown in a humidified atmosphere of 5% (vol/vol) CO2 at 37 °C. In the cell viability test (Fig. 2D), SK-BR-3 cells were stained with calcein AM and EthD-1 (Life Technologies Corp.), in accordance with the manufacturer’s instructions. In multiplexed BloC-Printing (Fig. 3), SK-BR-3 cells were stained with CellTracker Red CMTPX, CellTracker Green CMFDA, and CellTracker Blue CMAC (Life Technologies Corp.), following the manufacturer’s instructions. In the dye transfer study (Fig. 4), fibroblasts were stained with 4 μM calcein AM for 15 min at 37 °C.
Preparation, Printing, and Culture of Primary Rat Cortical Neurons.
Animal tissues were obtained following the protocol approved by Houston Methodist Hospital Institutional Animal Care and Use Committee. Embryonic day 18 (E18) cortical rat neurons were prepared following established procedures (48). Briefly, cortical neurons were dissected from E18 Sprague–Dawley rats, and dissociated with trypsin to single cells. The dissociated cells were cultured within 4 h after dissection, and cell viability was determined to be greater than 95%. Cells were cultured in a medium consisting of neural basal media, B-27 supplement, and Glutamax (Life Technologies). To sterilize and reduce amounts of the un–cross-linked oligomer, which adversely affects neuron viability, before use, the BloC-Molds were heated at 110 °C for 60 min and then exposed to UV light (UV output: 13.9 W) overnight. The PS culture dish was first coated with 100 µg/mL poly-d-lysine (Sigma) for 1 h. The poly-d-lysine solution was then aspirated, and the dish was rinsed once with double-distilled water. When the culture dish was dry, a degassed BloC-Mold was laid on the dish surface to form sealed channels. Cell-free medium was loaded into channels before cell loading. After individual neurons were trapped by the hooks, the medium was refreshed every 12 h.
BloC-Printing Cell Pairs for the Formation of Protrusions.
Cell pairs with a protrusion-to-protrusion morphology were obtained using the strategy of sequential cell anchoring. Cells without fluorescent labels (RCs) were first loaded and anchored. After adhesion of the RCs to the substrate, cells labeled with calcein (DCs) were loaded and anchored. After culture, both classes of cells generated protrusions along the wall of the channel that contacted each other. Cell pairs showing body-to-body contact were obtained by simultaneously loading RCs and DCs and anchoring them in trap pairs with 5 μm of spacing between traps. After cells had adhered for 2 h, they were polarized in the same direction. The BloC-Mold was then removed to allow anchored cells to spread and contact each other in a body-to-body arrangement.
Image Acquisition and Analysis.
Bright-field, phase-contrast, and fluorescence images were obtained with an AMG EVOS fl digital inverted fluorescence microscope, an Olympus IX81 inverted fluorescence microscope, and a Leica gated stimulated emission depletion super resolution microscope. Movies were filmed on the Olympus IX81 microscope.
Supplementary Material
Acknowledgments
We thank the Houston Methodist Research Institute SEM core facility for assistance with instrumentation and Dr. Dongfang Liu and Dr. Peilin Zheng (Baylor College of Medicine) for help in obtaining the high-resolution fluorescence images shown in SI Appendix, Fig. S19B. We also acknowledge funding support from the Cancer Prevention and Research Institute of Texas (CPRIT-R1007), NIH Grants NIH-CA180083 and NIH-DA035868, the Emily Herman Research Fund, the Department of Defense (W81XWH-11-02-0168), the Alliance of Nanohealth, and the Golfers Against Cancer Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313661111/-/DCSupplemental.
References
- 1.Yarmush ML, King KR. Living-cell microarrays. Annu Rev Biomed Eng. 2009;11:235–257. doi: 10.1146/annurev.bioeng.10.061807.160502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Carlo D, Lee LP. Dynamic single-cell analysis for quantitative biology. Anal Chem. 2006;78(23):7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 3.Yusof A, et al. Inkjet-like printing of single-cells. Lab Chip. 2011;11(14):2447–2454. doi: 10.1039/c1lc20176j. [DOI] [PubMed] [Google Scholar]
- 4.Calvert P. Materials science. Printing cells. Science. 2007;318(5848):208–209. doi: 10.1126/science.1144212. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura M, et al. Biocompatible inkjet printing technique for designed seeding of individual living cells. Tissue Eng. 2005;11(11-12):1658–1666. doi: 10.1089/ten.2005.11.1658. [DOI] [PubMed] [Google Scholar]
- 6.Xu T, Jin J, Gregory C, Hickman JJ, Boland T. Inkjet printing of viable mammalian cells. Biomaterials. 2005;26(1):93–99. doi: 10.1016/j.biomaterials.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Tseng P, Judy JW, Di Carlo D. Magnetic nanoparticle-mediated massively parallel mechanical modulation of single-cell behavior. Nat Methods. 2012;9(11):1113–1119. doi: 10.1038/nmeth.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giam LR, et al. Scanning probe-enabled nanocombinatorics define the relationship between fibronectin feature size and stem cell fate. Proc Natl Acad Sci USA. 2012;109(12):4377–4382. doi: 10.1073/pnas.1201086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermesh U, et al. High-density, multiplexed patterning of cells at single-cell resolution for tissue engineering and other applications. Angew Chem Int Ed Engl. 2011;50(32):7378–7380. doi: 10.1002/anie.201102249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azioune A, Storch M, Bornens M, Théry M, Piel M. Simple and rapid process for single cell micro-patterning. Lab Chip. 2009;9(11):1640–1642. doi: 10.1039/b821581m. [DOI] [PubMed] [Google Scholar]
- 11.Tan CP, et al. Parylene peel-off arrays to probe the role of cell-cell interactions in tumour angiogenesis. Integr Biol (Camb) 2009;1(10):587–594. doi: 10.1039/b908036h. [DOI] [PubMed] [Google Scholar]
- 12.Falconnet D, Csucs G, Grandin HM, Textor M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials. 2006;27(16):3044–3063. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Suh KY, Seong J, Khademhosseini A, Laibinis PE, Langer R. A simple soft lithographic route to fabrication of poly(ethylene glycol) microstructures for protein and cell patterning. Biomaterials. 2004;25(3):557–563. doi: 10.1016/s0142-9612(03)00543-x. [DOI] [PubMed] [Google Scholar]
- 14.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295(5560):1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 15.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 16.Lin L, Chu YS, Thiery JP, Lim CT, Rodriguez I. Microfluidic cell trap array for controlled positioning of single cells on adhesive micropatterns. Lab Chip. 2013;13(4):714–721. doi: 10.1039/c2lc41070b. [DOI] [PubMed] [Google Scholar]
- 17.Chung K, Rivet CA, Kemp ML, Lu H. Imaging single-cell signaling dynamics with a deterministic high-density single-cell trap array. Anal Chem. 2011;83(18):7044–7052. doi: 10.1021/ac2011153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci USA. 2010;107(22):10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright D, et al. Reusable, reversibly sealable parylene membranes for cell and protein patterning. J Biomed Mater Res A. 2008;85(2):530–538. doi: 10.1002/jbm.a.31281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenthal A, Macdonald A, Voldman J. Cell patterning chip for controlling the stem cell microenvironment. Biomaterials. 2007;28(21):3208–3216. doi: 10.1016/j.biomaterials.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem. 2005;77(17):5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 22.Ostuni E, Kane R, Chen CS, Ingber DE, Whitesides GM. Patterning mammalian cells using elastomeric membranes. Langmuir. 2000;16(20):7811–7819. [Google Scholar]
- 23.Folch A, Jo BH, Hurtado O, Beebe DJ, Toner M. Microfabricated elastomeric stencils for micropatterning cell cultures. J Biomed Mater Res. 2000;52(2):346–353. doi: 10.1002/1097-4636(200011)52:2<346::aid-jbm14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 24.Kumar A, Whitesides GM. Features of gold having micrometer to centimeter dimensions can be formed through a combination of stamping with an elastomeric stamp and an alkanethiol “ink” followed by chemical etching. Appl Phys Lett. 1993;63(14):2002–2004. [Google Scholar]
- 25.Liu WY, Zhang Y, Thomopoulos S, Xia YN. Generation of controllable gradients in cell density. Angew Chem Int Ed Engl. 2013;52(1):429–432. doi: 10.1002/anie.201206060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Théry M. Micropatterning as a tool to decipher cell morphogenesis and functions. J Cell Sci. 2010;123(Pt 24):4201–4213. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]
- 27.Nelson CM, Chen CS. Cell-cell signaling by direct contact increases cell proliferation via a PI3K-dependent signal. FEBS Lett. 2002;514(2-3):238–242. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- 28.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Microfluidic control of cell pairing and fusion. Nat Methods. 2009;6(2):147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Espina V, et al. Laser-capture microdissection. Nat Protoc. 2006;1(2):586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 30.Abbaci M, Barberi-Heyob M, Blondel W, Guillemin F, Didelon J. Advantages and limitations of commonly used methods to assay the molecular permeability of gap junctional intercellular communication. Biotechniques. 2008;45(1):33–52, 56–62. doi: 10.2144/000112810. [DOI] [PubMed] [Google Scholar]
- 31.Kanno Y, Loewenstein WR. Intercellular diffusion. Science. 1964;143(3609):959–960. doi: 10.1126/science.143.3609.959. [DOI] [PubMed] [Google Scholar]
- 32.Neyton J, Trautmann A. Single-channel currents of an intercellular junction. Nature. 1985;317(6035):331–335. doi: 10.1038/317331a0. [DOI] [PubMed] [Google Scholar]
- 33.Wade MH, Trosko JE, Schindler MA. A fluorescence photobleaching assay of gap junction-mediated communication between human cells. Science. 1986;232(4749):525–528. doi: 10.1126/science.3961495. [DOI] [PubMed] [Google Scholar]
- 34.Dakin K, Zhao YR, Li WH. LAMP, a new imaging assay of gap junctional communication unveils that Ca2+ influx inhibits cell coupling. Nat Methods. 2005;2(1):55–62. doi: 10.1038/nmeth730. [DOI] [PubMed] [Google Scholar]
- 35.Lee PJ, Hung PJ, Shaw R, Jan L, Lee LP. Microfluidic application-specific integrated device for monitoring direct cell-cell communication via gap junctions between individual cell pairs. Appl Phys Lett. 2005;86:223902. [Google Scholar]
- 36.Tseng QZ, et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci USA. 2012;109(5):1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer AS, et al. 2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen. J Cell Biol. 2012;197(6):721–729. doi: 10.1083/jcb.201201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Short B. Protrusion provides a prognosis. J Cell Biol. 2012;197(6):692. [Google Scholar]
- 39.Ridley AJ. Life at the leading edge. Cell. 2011;145(7):1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 40.DeMali KA, Burridge K. Coupling membrane protrusion and cell adhesion. J Cell Sci. 2003;116(Pt 12):2389–2397. doi: 10.1242/jcs.00605. [DOI] [PubMed] [Google Scholar]
- 41.Mattila PK, Lappalainen P. Filopodia: Molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9(6):446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 42.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1(5):2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 44.Millet LJ, Stewart ME, Sweedler JV, Nuzzo RG, Gillette MU. Microfluidic devices for culturing primary mammalian neurons at low densities. Lab Chip. 2007;7(8):987–994. doi: 10.1039/b705266a. [DOI] [PubMed] [Google Scholar]
- 45.Fan Y, Xu F, Huang G, Lu TJ, Xing W. Single neuron capture and axonal development in three-dimensional microscale hydrogels. Lab Chip. 2012;12(22):4724–4731. doi: 10.1039/c2lc40312a. [DOI] [PubMed] [Google Scholar]
- 46.Braunschweig AB, Huo F, Mirkin CA. Molecular printing. Nat Chem. 2009;1(5):353–358. doi: 10.1038/nchem.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandes TG, Diogo MM, Clark DS, Dordick JS, Cabral JMS. High-throughput cellular microarray platforms: Applications in drug discovery, toxicology and stem cell research. Trends Biotechnol. 2009;27(6):342–349. doi: 10.1016/j.tibtech.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris J, et al. Preparing e18 cortical rat neurons for compartmentalization in a microfluidic device. J Vis Exp. 2007;(8):305. doi: 10.3791/305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.