Significance
The accurate removal of introns by pre-mRNA splicing is a critical step in proper gene expression. Most eukaryotic genomes, from plant to human, contain a tiny subset of “minor class” introns with unique sequence elements that require their own splicing machinery. The significance of this second splicing pathway has intrigued RNA biologists for two decades, but its biological relevance was recently underscored when defects in the process were firmly linked to human disease. Here, we use a novel zebrafish mutant with defective minor class splicing to investigate how this pathway shapes the transcriptome during vertebrate development. We link its pleiotropic phenotype to widespread changes in gene expression that disrupt essential cellular pathways, including mRNA processing.
Abstract
Minor class or U12-type splicing is a highly conserved process required to remove a minute fraction of introns from human pre-mRNAs. Defects in this splicing pathway have recently been linked to human disease, including a severe developmental disorder encompassing brain and skeletal abnormalities known as Taybi-Linder syndrome or microcephalic osteodysplastic primordial dwarfism 1, and a hereditary intestinal polyposis condition, Peutz-Jeghers syndrome. Although a key mechanism for regulating gene expression, the impact of impaired U12-type splicing on the transcriptome is unknown. Here, we describe a unique zebrafish mutant, caliban (clbn), with arrested development of the digestive organs caused by an ethylnitrosourea-induced recessive lethal point mutation in the rnpc3 [RNA-binding region (RNP1, RRM) containing 3] gene. rnpc3 encodes the zebrafish ortholog of human RNPC3, also known as the U11/U12 di-snRNP 65-kDa protein, a unique component of the U12-type spliceosome. The biochemical impact of the mutation in clbn is the formation of aberrant U11- and U12-containing small nuclear ribonucleoproteins that impair the efficiency of U12-type splicing. Using RNA sequencing and microarrays, we show that multiple genes involved in various steps of mRNA processing, including transcription, splicing, and nuclear export are disrupted in clbn, either through intron retention or differential gene expression. Thus, clbn provides a useful and specific model of aberrant U12-type splicing in vivo. Analysis of its transcriptome reveals efficient mRNA processing as a critical process for the growth and proliferation of cells during vertebrate development.
Splicing, the excision of introns from pre-mRNA, is an essential step in gene expression and a major source of complexity in the transcriptome (1). The process is catalyzed by highly dynamic complexes of small nuclear ribonucleoproteins (snRNPs) called spliceosomes (2). Not widely appreciated is the coexistence of two types of introns in most eukaryotic genomes. The vast majority, the major class or U2-type introns, are marked by GT and AG at their 5′ and 3′ ends, respectively. Minor class or U12-type introns, of which there are ∼700 in the human genome, were initially recognized by the presence of AT and AC in these positions, prompting their original name of AT-AC introns (3). However, we now know that most of these introns contain the same GT-AG termini found in U2-type introns (4, 5) and are instead distinguished from them by two highly conserved motifs: one adjacent to the 5′ splice site (ss) and one corresponding to the branch point sequence (BPS), close to the 3′ ss (6, 7). These introns also lack the 3′ polypyrimidine tract characteristic of U2-type introns. Minor class introns are excised by U12-type spliceosomes, which are analogous in function and similar in composition to U2-type spliceosomes; each comprise five small nuclear RNAs (snRNAs) and hundreds of associated proteins (6). Although the U5 snRNA is shared between the two complexes, the U12-type spliceosome contains four unique snRNAs (U11, U12, U4atac, and U6atac) (7–9) and at least seven unique proteins (10). Since its discovery two decades ago, the existence of this highly conserved, second splicing pathway has been an intriguing aspect of gene expression. Interestingly, U12-type introns are nonrandomly distributed across the genome (11) and their removal is thought to be rate-limiting in the generation of mature mRNAs (12–14).
Aberrant splicing is the basis of many human diseases and up to one-third of disease-causing mutations may lead to splicing defects (15). Although the majority of these defects occur in U2-type introns and splicing factors, perturbations in U12-type splicing have also been described. Most recently, mutations in the U12-type U4atac snRNA were found to underlie the human developmental disorder Taybi-Linder syndrome (TALS)/microcephalic osteodysplastic primordial dwarfism 1 (MOPD1) (16, 17). Other examples of diseases that can arise because of impaired splicing of U12-type introns include spinal muscular atrophy (18, 19) and Peutz-Jeghers syndrome (PJS), an intestinal polyposis syndrome that predisposes to cancer because of mutations in the tumor suppressor gene (TSG) LKB1 (liver kinase B1) (20). The observation that impaired U12-type splicing can affect TSG expression has interesting implications for other TSGs containing U12-type introns, such as PTEN (phosphatase and tensin homolog deleted on chromosome 10), where even a modest loss in PTEN function predisposes mice to cancer (21).
To specifically study the role of U12-type introns in gene expression and development, we exploited a zebrafish mutant, caliban (clbn), which carries a point mutation in rnpc3 [RNA-binding region (RNP1, RRM) containing 3], encoding the zebrafish ortholog of human RNPC3, one of seven proteins unique to the U12-type spliceosome (22). We found aberrant minor class spliceosomal snRNPs in clbn extracts and used microarray and RNA sequencing (RNAseq) to show that impairment of U12-type splicing impacts broadly on gene expression, disrupting multiple genes involved in mRNA processing, transcription, splicing, and nuclear export. As a result, clbn mutants develop a pleiotropic phenotype with early lethality.
Results
The Intestine, Liver, and Pancreas Are Abnormal in clbn.
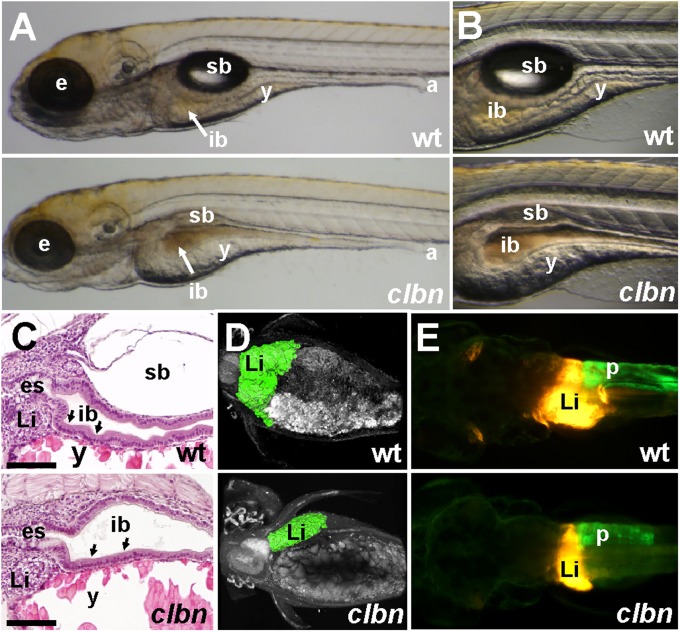
clbns846 was identified in a transgene-assisted ethylnitrosourea mutagenesis screen for zebrafish mutants with defective endodermal organ development (23). Abnormalities in the gross morphology of clbn larvae are first detectable at 108 h postfertilization (hpf) and mutants die between 7 and 10 d postfertilization (dpf). The head, eye, and lens are smaller, the swim bladder fails to inflate, and yolk absorption is delayed. At 120 hpf, the WT intestinal epithelium exhibits a columnar morphology and starts to elaborate folds; in contrast, the intestinal epithelium in clbn remains thin and unfolded (Fig. 1 A–C). At 120 hpf, imaging of the digestive organs on the Tg(gutGFP)s854 (23) and Tg(fabp10:dsRed, ela3l:GFP)gz12 (24) backgrounds shows that the pancreas and liver are markedly smaller in clbn than in WT (Fig. 1 D and E).
Fig. 1.
The clbn phenotype is characterized by abnormalities in the digestive organs (A and B) Brightfield images of left lateral views and (C) histology of WT and clbns846 larvae at 120 hpf show a thick, folded intestinal epithelium (black arrows in C) and inflated swim bladder in WT, whereas clbn displays a thin, unfolded intestinal epithelium, delayed yolk resorption, and smaller eyes. (D) Two-photon microscopy of WT and clbns846 larvae on the Tg(gutGFP)s854 background and (E) epifluorescence microscopy on the Tg(fabp10:RFP,ela3l:EGFP)gz12 background reveal smaller digestive organs in clbns846 compared with WT. The two images in A were taken at the same magnification, as were the two images in each of panels B, D, and E. The clbn phenotype is 100% penetrant and all larvae die between 7 and 10 dpf; however, the severity of the morphological abnormalities may vary between clutches. For example, the digestive organ phenotype is visibly conspicuous in some clutches at around 108 hpf and not in others until 120 hpf. a, anus; e, eye; es, esophagus; ib, intestinal bulb; Li, liver; p, pancreas; sb, swim bladder; y, yolk. (Scale bar in C, 100 μm.)
clbns846 Harbors a Mutation in rnpc3.
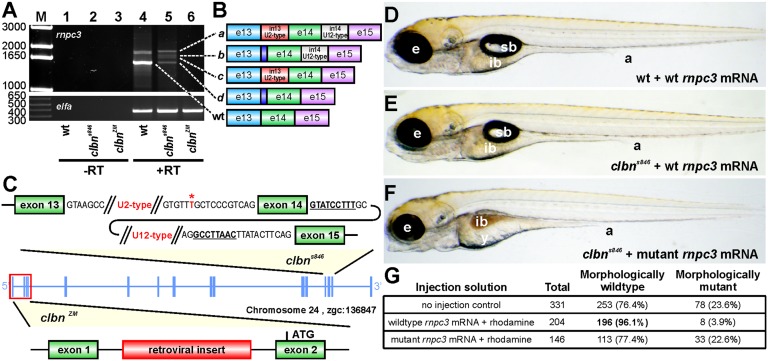
The clbns846 locus was mapped to a region on chromosome 24 containing four genes, col11A1, rnpc3, prmt6, and ntng1 (Fig. S1A). RT-PCR amplification of rnpc3 from clbns846 RNA generated aberrant rnpc3 isoforms (Fig. 2A), which were found by sequencing to correspond to the use of a de novo 3′ ss or retention of one or both of introns 13 and 14 (Fig. 2B). Sequencing of WT and clbns846 genomic DNA revealed a T→A variation, creating a novel 3′ ss (AG) 10 nucleotides upstream of the canonical 3′ ss of intron 13 (Fig. 2C). The shift to the novel 3′ ss in clbns846 is complete, as sequencing of the individual aberrant transcripts gave no evidence of a correctly spliced exon 13–14 junction in any of the products. All of the aberrant transcripts in clbns846 contain premature stop codons after no more than six missense codons, making them likely targets for nonsense mediated decay (Fig. S1B). We confirmed these findings using an in vivo minigene assay in which a WT or mutated minigene was injected into clutches containing WT and clbns846 embryos. The T→A transition in the mutated (clbn) minigene led to a complete shift to the novel splice junction in both WT and clbns846 embryos, as shown by a slightly shifted band in the agarose gel (Fig. S1C, lanes 1 and 3) and confirmed by sequencing.
Fig. 2.
The clbn phenotype is caused by a mutation in rnpc3. (A) RT-PCR of full-length rnpc3 coding sequence amplifies a predominant 1.6-kb product in WT larvae (lane 4) and four aberrantly spliced transcripts in clbns846 (lane 5). A retroviral insertion in clbnZM abolishes generation of full-length rnpc3 mRNA (lane 6). Lanes 1–3 show –RT controls. (Lower) Elongation factor alpha (elfa) control. Dashed lines link the cDNA products in lanes 4 and 5 to the corresponding transcripts, shown schematically in B. The dark blue region denotes retention of 10 intronic nucleotides through use of a de novo 3′ ss. (C) Two noncomplementing clbn alleles. An asterisk over T marks the position of the T→A variation, 12 nucleotides upstream of exon 14 of rnpc3, which causes the clbns846 phenotype. The conserved 5′ ss and BPS in intron 14 (U12-type) of rnpc3 are underlined and in bold. clbnZM carries a retroviral insertion in the first intron of rnpc3. (D–F) Brightfield microscopy images of left lateral views of 5-dpf larvae derived from a clbns846 heterozygous incross that were injected at the one- to two-cell stage with 380 pg of either WT rnpc3 mRNA or a mutant mRNA retaining introns 13 and 14, corresponding to the most abundant form of rnpc3 mRNA in clbns846 (see B). (D) No significant overexpression phenotype in WT larvae injected with WT rnpc3 mRNA. (E) Rescue of the mutant phenotype in genotyped clbns846 homozygous mutant larvae injected with WT rnpc3 mRNA. (F) No rescue of the clbn mutant phenotype injected with mutant rnpc3 mRNA. (G) Noninjected control larvae from the same clutch show a near-perfect Mendelian ratio of WT to clbn larvae, whereas 96% of larvae injected with WT rnpc3 RNA appear WT at 5 dpf (P < 0.0001, Fisher’s exact test). There was no difference between uninjected and mutant rnpc3-injected groups (P = 0.9065). a, anus; e, eye; ib, intestinal bulb; sb, swim bladder; y, yolk. The brightfield images in D, E, and F were taken at the same magnification.
We provided firm evidence that rnpc3 is the mutated gene in clbn using rescue assays and a complementation test. Injection of one-cell stage clbn embryos with in vitro synthesized RNA encoding WT zebrafish Rnpc3 rescued the clbn phenotype (Fig. 2 D–G). To perform a complementation test, we purchased a commercially available zebrafish line harboring a retroviral insertion in intron 1 of rnpc3 (clbnZM) (Fig. 2C). We found that maternal deposits of correctly spliced, full-length rnpc3 mRNA in clbnZM mutant embryos are largely depleted by 24 hpf (Fig. S1D). clbns846 and clbnZM mutant larvae are morphologically indistinguishable at 120 hpf (Fig. S1 F and G). Upon crossing clbns846 and clbnZM heterozygotes, one-quarter of the offspring exhibit a phenotype indistinguishable from that of clbns846 and clbnZM mutants (Fig. S1H). Taken together, these data show that rnpc3 is the mutated gene in clbn and suggest that both alleles are functionally null. We used whole mount in situ hybridization to show widespread rnpc3 mRNA expression in WT embryos until 24 hpf (Fig. S2 A and B). Thereafter, the expression of rnpc3 becomes restricted to proliferating tissues, including the eye, lens, and digestive organs (Fig. S2 C–F). These tissues, which are abnormal in clbn, show reduced expression of rnpc3 in mutant larvae at 120 hpf (Fig. S2H).
clbn Contains Incompletely Spliced Transcripts.
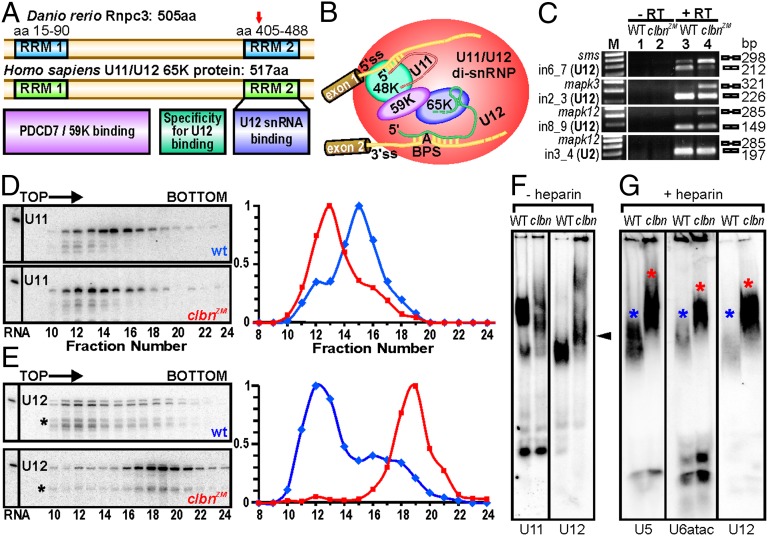
The zebrafish rnpc3 gene encodes a 505-aa nuclear protein with two RNA-recognition motifs (RRM) (25) and the same domain structure as its human ortholog (Fig. 3A). Whereas the excision of U2-type introns is initiated by the binding of individual U1 and U2 monosnRNPs, U12-type introns in human cells are recognized and bound by a preformed U11/U12 di-snRNP (26). This di-snRNP comprises U11 and U12 snRNAs, RNPC3/65K and two other proteins unique to the U12-type spliceosome, the U11/U12 59-kDa and 48-kDa proteins. Together, these components form a “molecular bridge” between the 5′ ss and BPS of U12-type introns (27, 28) (Fig. 3B). We used RT-PCR to assess the consequences of rnpc3 deficiency on the processing of mRNAs harboring U2- and U12-type introns in clbn and found that the U12-type introns in sms, mapk3, and mapk12 were retained in clbn larvae (Fig. 3C), whereas the excision of the U2-type intron in mapk12 was unaffected.
Fig. 3.
Zebrafish Rnpc3 functions in the U12-type spliceosome. (A) Domain structure of zebrafish Rnpc3 and its human ortholog RNPC3, or U11/U12 di-snRNP 65-kDa protein (www.uniprot.org, accession no. Q96LT9). Red arrow in the C-terminal RRM denotes the approximate position of the premature stop codons in all clbns846 isoforms. (B) Schematic of the human U11/U12 di-snRNP, with RNPC3 (65 K) bridging U12 snRNA and the 59 K protein (28). BPS, branch point sequence; “A” represents the branch point adenosine. (C) RT-PCR analysis of three different transcripts (sms, mapk3, mapk12) shows retention of U12-type introns (lane 4, Top three panels) in clbns846 (5 dpf), but not of a U2-type intron (Bottom). Lanes 1 and 2 show –RT controls. (D and E) Glycerol gradient/Northern analysis revealing differential sedimentation of U11 (D) and U12 (E) snRNA-containing snRNPs in WT and clbnZM extracts. Direction of sedimentation is left to right. Lane 1 contains total RNA. Full-length U11 or U12 signals were quantified using ImageQuant and expressed as a percentage of the fraction with the highest intensity. U12 snRNA showed persistent specific degradation (asterisks) without affecting its migration profile. (F) Northern analysis of WT and clbns846 extracts resolved on 4% (80:1) native polyacrylamide gels and probed for U11 and U12 snRNAs. The predominant U11 snRNPs are disrupted in clbn compared with WT and the U12-containing particles are heavier and migrate more slowly (black arrowhead). (G) Northern analysis of WT and clbns846 extracts resolved on native gels and probed for U5, U6atac, and U12 snRNAs shows retarded bands in clbn (red asterisks) compared with WT (blue asterisks). The same larval lysate was used in F and G. Data are representative of a total of 10 (D and E) and 5 (F and G) biological replicates.
clbn Contains Aberrant Minor Spliceosomal snRNPs.
To determine a biochemical basis for the observed specific defect in U12-type splicing, we sought to compare the size and conformation of zebrafish U11- and U12-containing snRNPs in WT and clbn larvae using glycerol gradient sedimentation followed by Northern blot analysis for U11 and U12 snRNAs. Extracts of WT larvae gave rise to U11 peaks in fractions 15 and 16 (Fig. 3D, Upper), whereas the U11 peak in clbn extracts was found two fractions closer to the top of the gradient, corresponding to a lighter U11 species (Fig. 3D, Lower, and Fig. S3 A and B). In contrast, glycerol gradient analyses of U12 snRNA sedimentation in clbn showed a dramatic shift of the predominant U12 signal toward a heavier particle than in WT (Fig. 3E). We further investigated these findings using native gel electrophoresis followed by Northern blotting for U11 and U12 snRNAs (Fig. 3F). Again, loss of Rnpc3 was found to disrupt the predominant U11 snRNPs and cause the accumulation of a slower-migrating U12-containing particle. This unexpected finding suggests a function for Rnpc3 beyond its known role in the U11/U12 di-snRNP, potentially at later stages of the splicing cycle. U12-containing complexes that might be stalled at any point after spliceosome activation would be expected to contain U6atac or U5 snRNAs (2). To test this theory we used native gel electrophoresis followed by Northern blotting for U12, U5, and U6atac snRNAs (Fig. 3G). To reduce potential nonspecific interactions and facilitate characterization of the stable complex accumulating in clbn embryos, heparin was added to the extracts before separation. Once again, we observed a strong signal corresponding to a heavier U12 snRNP complex in clbn larvae from both alleles (Fig. S3C), which appeared to comigrate with signals for both U5 and U6atac.
Transcriptome Analysis Reveals a Specific U12-Type Splicing Defect in clbn.
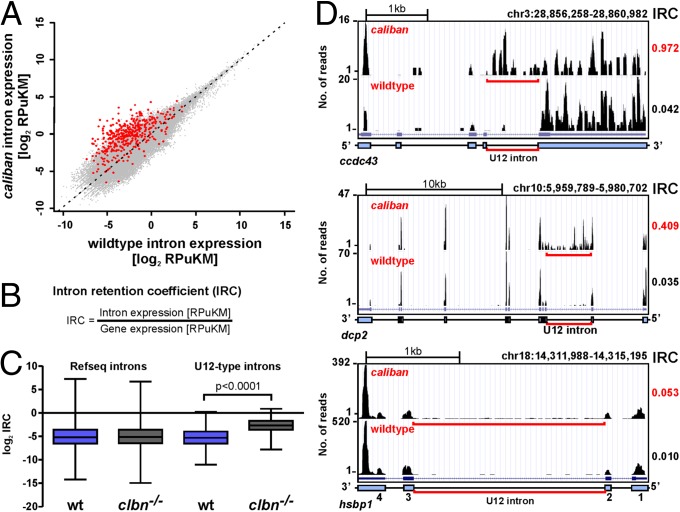
To determine the global impact of the clbn mutation on gene expression and U2- and U12-type splicing, we applied RNAseq to 108-hpf WT and clbns846 mutants (Table S1). RNAseq tags were aligned to the annotated zebrafish (Zv9) genome assembly (Fig. S4A), which contains 606 manually curated U12-type introns (SI Methods and Dataset S1). To normalize for the more common occurrence of nonunique sequences—such as repetitive elements—in introns compared with exons, we generated a zebrafish uniqueome (29) and calculated all expression levels as reads per unique kilobase per million reads (RPuKM). Plotting expression levels of all introns that satisfy minimum expression requirements (RPuKM > 0.05) in both WT and clbn reveals a clear bias toward retention of U12-type introns in clbn (Fig. 4A). We found that a significantly higher fraction of U12-type introns (397 of 606 = 65.5%) show some degree of intron retention [intron retention coefficients (IRC) > 0.05; defined in Fig. 4B] in WT larvae, compared with 49.6% of U2-type introns. This finding is consistent with U12-type introns being processed more slowly than their U2-type counterparts (12, 13). Remarkably, an additional 148 of 606 (24.6%) of U12-type introns are detectable only in clbn (total = 90.1%), highlighting the specificity of the splicing defect. The defect in U12-type splicing in clbn is statistically significant (Fig. 4C); however, IRCs vary markedly between transcripts (Fig. 4D) and do not appear to be influenced by expression levels (Fig. S4B), U12 intron length (Fig. S4C), and AT-AC vs. GT-AG subtype (Fig. S4D).
Fig. 4.
Intron retention in clbn is specific to U12-type introns and highly variable. (A) Scatterplot of normalized intron expression derived by RNAseq for reference (Refseq) introns (gray dots) and U12-type introns (red dots). U12-type introns lie almost entirely above the midline representing equal expression, indicating enhanced intron retention in clbn. Intron expression is given in RPuKM. (B) To quantitate U12-type intron retention in individual transcripts, we defined an intron retention coefficient, which calculates intron retention relative to transcript level. (C) Comparison of the median log2 of the IRCs for Refseq introns and U12-type introns between WT and clbn demonstrates that the clbn splicing defect is specific to U12-type introns. See also Fig. S4. Statistical significance was determined by Student t test. (D) University of California at Santa Cruz genome browser views of three genes with high, intermediate, and low IRCs. ccdc43 shows a high IRC of 0.97, dcp2 an intermediate IRC of 0.41, and hsbp1 shows a low IRC of 0.05.
Activation of Cryptic Splicing Occurs Rarely in clbn.
A Peutz-Jeghers syndrome family has been described in which a mutation in the 5′ ss of the U12-type intron in LKB1 (20) leads to activation of cryptic splice sites and LKB1 haploinsufficiency. Activation of cryptic splice sites also occurs in human cells upon knockdown of the U11/U12 48-kDa protein with RNAi (30). To determine whether cryptic splice sites are activated in clbn, we sequenced the spliced transcripts obtained by RT-PCR for sms, mapk3, and mapk12 mRNAs (Fig. 2C) and found only the annotated exon–exon junctions. We then used RT-PCR to amplify three more U12-type introns with low IRCs, namely fkbp3, snrpe, and ppp2r2c. We found very low levels of U12-type intron retention in snrpe and ppp2r2c in clbn and no evidence of cryptic splicing for all three genes (Fig. S4E). Furthermore, we also found no evidence of activation of cryptic splice sites in clbn when we aligned RNAseq reads against a custom-built library of exon junctions within 100 bp of the annotated splice junctions of all manually curated zebrafish U12-type introns (Fig. S4F and Dataset S2). Collectively these data show that impaired U12-type splicing in clbn is rarely circumvented by activation of cryptic splicing.
Rnpc3 Deficiency Impacts Broadly on Gene Expression.
We combined datasets from microarray analysis and RNAseq to identify high-confidence differentially expressed genes (DEGs) between WT and clbns846 larvae (Datasets S3–S5). Fold-changes for 203 DEGs (113 down, 90 up) were significantly correlated for the two methods (Fig. S5A). As expected, rnpc3 mRNA levels were reduced, consistent with predicted nonsense mediated decay of rnpc3 transcripts carrying the clbns846 mutation. Of the down-regulated genes, many exhibit tissue-specific expression in tissues that develop abnormally in clbn, including the liver, pancreas, intestine, and lens (Dataset S3). Consistent with a prior study in Drosophila (31), a substantial overrepresentation of U12-type intron-containing genes was observed among the DEGs, with a higher proportion of U12-type intron-containing genes in the down-regulated DEGs (35%; 40 of 113) than in the up-regulated set (11%; 10 of 90). Even so, about 90% of U12-type intron-containing genes were not differentially expressed (Fig. S5B) and most of the DEGs do not contain U12-type introns; hence, Rnpc3 deficiency impacts broadly on gene expression and is not limited to U12-type intron-containing genes.
Expression of mRNA Quality Control and Cell Cycle Genes Is Altered in clbn.
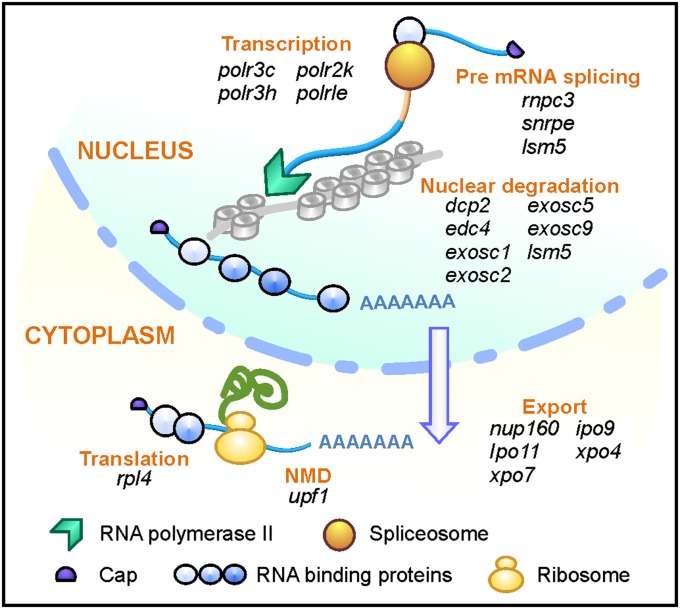
U12-type introns are enriched in “information processing” genes encoding functions in DNA replication, repair, transcription, mRNA processing, and translation (11). In clbn, many such genes, particularly those encoding components of mRNA quality control pathways, such as exosome factors, are either differentially expressed (dcp2, edc4, exosc1, exosc5, exosc9) or exhibit varying degrees of intron retention (edc4, exosc1, exosc2, exosc5) (Fig. 5, Fig. S5C, and Table S2). Although the primary defect caused by the lack of Rnpc3 is in the processing of U12-type introns, the resulting widespread disruption to the expression of genes regulating RNA processing is likely to have a secondary impact on the expression of U2-type intron-containing genes, explaining why they feature so strongly in the list of DEGs. clbn mutants at 108 hpf also exhibit elevated expression of cell-cycle repressor genes, including prohibitin, cyclin G1, e2f5, and ∆113p53 (Fig. S5 D–F). ∆113p53 is a Tp53 target gene encoding an N-terminally truncated isoform of Tp53 thought to antagonize its proapoptotic activity (32).
Fig. 5.
Expression of mRNA processing genes is disrupted in clbn. Schematic diagram showing multiple genes involved in various steps of mRNA processing, including transcription, splicing, nuclear export, and nuclear and cytoplasmic mRNA quality control, are disrupted in clbn, either through intron retention or differential gene expression. See also Fig. S5C.
Discussion
U12-type splicing was a surprise discovery two decades ago, and its biological significance has remained something of an enigma. Here, we describe a unique vertebrate model system with a mutation in a core component that is specific to the U12-type spliceosome, and exploit it to gain new insights into the role of U12-type splicing in the regulation of gene expression during development and disease.
Rnpc3 Deficiency Leads to a Pleiotropic Phenotype in Developing Zebrafish.
The loss of Rnpc3, either because of an intronic point mutation or a retroviral insertion into the rnpc3 locus, leads to a severe and pleiotropic phenotype in developing zebrafish larvae, culminating in death of all larvae by 10 dpf. The most severely affected tissues are those that are highly proliferative between 48 and 96 hpf, including the digestive organs, eye, and lens. Our observation that Rnpc3 deficiency impacts broadly on gene expression, extending well beyond the small subset of genes containing U12-type introns, makes it difficult to pinpoint precisely the molecular basis for the clbn phenotype. As yet, we do not know whether the abnormalities arise directly from perturbations in the expression of a small number of U12-type intron-containing genes, such as the dysregulated cell-cycle repressor genes we identified, or are the result of complex secondary effects emanating from a larger network of genes whose expression is affected by impaired U12-type splicing.
Comparison of the clbn Phenotype with MOPD.
The severe human developmental disorder known as TALS/MOPD1 (16, 17, 33, 34) is caused by impaired U12-type splicing. Here, the affected infants inherit two recessive point mutations in U4atac snRNA and suffer extreme intrauterine growth retardation, severe microcephaly, central nervous system abnormalities, dysmorphic facial features, skin and skeletal abnormalities, and early death (34). Although an impact on the growth of proliferating tissues is a common link in the zebrafish and human conditions, most MOPD1 features are not conspicuous in clbn. One possible explanation for this lack of concordance is that many of the severe defects observed in MOPD1 patients, such as microcephaly, most likely arise early in development. In zebrafish, adverse events early in embryogenesis are frequently circumvented by the availability of maternally derived WT mRNAs. In the particular case of clbn, we have shown that rnpc3 mRNA is maternally deposited, and it is reasonable to expect that this will also be the case for at least a proportion of the genes dependent on efficient minor class splicing, as well as their downstream effectors. If this hypothesis is correct, maternal-zygotic mutants of our clbn alleles would be expected to exhibit a more extensive phenotype that more closely mimics MOPD1.
Rnpc3 Deficiency Disrupts U11/U12 di-snRNP Assembly and Splicing.
We examined the distribution of minor class snRNPs in rnpc3-deficient zebrafish larvae to provide a molecular understanding of the specific U12-type splicing defects observed. Although the dynamic nature of snRNP interactions throughout the splicing cycle is relatively well characterized in mammalian cells, very little is known about this process in zebrafish. Previous studies have shown that the sedimentation profiles of zebrafish snRNPs in glycerol gradients differ in some aspects to those obtained from human cells (35, 36). Similarly, our glycerol gradient and native gel electrophoresis experiments reveal signals for U11 and U12 snRNPs that do not clearly correspond to a distribution into U11 and U12 mono-snRNPs and a U11/U12 di-snRNP, as described in human cells. Notwithstanding that, our data clearly demonstrate a specific role for Rnpc3 in maintaining the integrity and proper distribution of U11 and U12 snRNPs. This result contrasts with our analysis of another zebrafish splicing mutant (cph), where a mutation in the shared core splicing factor, prpf8, disrupts U2-type U4/U6/U5 tri-snRNP formation with negligible impact on U11/U12 snRNPs (35). In this study, the strong accumulation in clbn of a heavy, potentially stalled, complex containing U12, U5, and U6atac snRNAs, which does not seem to be a stable intermediate in WT, suggests a hitherto unsuspected role for Rnpc3 in spliceosome disassembly or recycling. Further studies are required to confirm this.
Regulation of U12-Type Splicing.
The results of our transcriptome analysis show that approximately two-thirds of U12-type intron-containing mRNAs exhibit some degree of U12-type intron retention even in developing WT zebrafish, much higher than the fraction of U2-type introns retained. At first sight, U12-type splicing appears to be a relatively inefficient process that may limit the full expression of U12-type intron-containing genes. However, recent findings suggest that regulation of U12-type splicing may be more complicated than previously thought. For example, p38MAPK-mediated up-regulation of the otherwise rate-limiting levels of U6atac in HeLa cells appears to constitute a sophisticated regulatory mechanism to facilitate the rapid mobilization of critical U12-type intron-containing transcripts in response to growth stimuli (14). Because many of these transcripts encode genes with functions in mRNA production, such an amplification of U12-type splicing activity is likely to generate a robust transcriptome-wide response.
U12-Type Splicing and Disease.
Recent breakthroughs that firmly link defects in U12-type splicing to human disease (16, 17, 19, 20) provide a strong impetus to explore its potential involvement in other pathological conditions. Interestingly, we have noted that U12-type introns are found in a number of cancer genes, including oncogenes, such as BRAF, C-RAF, and several members of the MAPK family. Extrapolating from our results in clbn, where the survival and integrity of rapidly proliferating cells is dependent on U12-type splicing, it is tantalizing to speculate that the efficiency of minor class splicing could influence the growth of tumors, particularly those that are dependent on the RAS-MAPK pathway (37, 38). In this regard, it is intriguing that p38MAPK has recently emerged as a key regulator of U12-type splicing activity (16), suggesting a potential regulatory mechanism to achieve efficient levels of U12-type splicing in tumors that are reliant on the MAPK pathway. Future investigations will determine whether efficient U12-type splicing is an important contributor to the posttranscriptional RNA processing mechanisms governing gene expression in cancer.
Methods
Ethics Statement.
Zebrafish experiments were approved by the Ludwig Institute for Cancer Research/Department of Surgery, University of Melbourne Animal Ethics Committee, and the Walter and Eliza Hall Institute Animal Ethics Committee.
Gene Expression Analysis by Quantitative RT-PCR.
Total RNA from zebrafish larvae was prepared using TRIzol (Life Technologies). cDNA was prepared from 0.5 to 1 μg of DNase-treated total RNA with the SuperScript III First Strand Synthesis System (Life Technologies). Quantitative RT-PCR assays were performed using the SensiMix SYBR Kit (Bioline) or by TaqMan Gene Expression Assay (Life Technologies). Expression data were normalized to elongation factor-α.
Analysis of snRNP Size and Conformation.
Extracts from 6-dpf larvae were prepared in buffer G (20 mM Hepes pH 7.9 KOH, 150 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 0.02% Nonidet P-40) containing 8% (vol/vol) glycerol, layered onto 10–30% (vol/vol) glycerol gradients and centrifuged for 19 h at 28,000 × g in an SW40 rotor at 4 °C in an Optima L-90K ultracentrifuge (Beckman Coulter). Next, 30 × 1 mL fractions were collected from the top. RNA was extracted in phenol-chloroform and separated by electrophoresis on 10% polyacrylamide/8 M urea-TBE gels. For native gels, lysates were prepared in buffer G containing 20% glycerol and incubated with or without 5 mg/mL heparin for 10 min on ice before loading on 4% native polyacrylamide gels. Nucleic acids were transferred to Hybond-N nylon membranes (GE Healthcare) and hybridized with 32P-radiolabeled cRNA (U11, U6atac, and U5) or cDNA (U12) probes to zebrafish snRNAs.
Microarray Analysis.
Total RNA was extracted from triplicate pools of 20 genotyped clbn and homozygous WT larvae. Amplification, labeling, and hybridization to the Agilent Zebrafish V2 gene expression platform were performed at the Australian Zebrafish Phenomics Facility. Data were analyzed using R in combination with the Linear Models for Microarray Data (LIMMA) package (http://bioinf.wehi.edu.au/limma). After background correction and normalization, differentially expressed genes were ranked using the B-statistic (39).
RNA Sequencing.
Pools of poly(A)-enriched RNA from clbn and homozygous WT larvae were used to generate stranded cDNA libraries for SOLiD3 sequencing. Fifty-base pair single-end reads were mapped to the zebrafish Zv9 genome assembly using the X-MATE recursive mapping pipeline (40), allowing for five color space mismatches per 50-bp read (CS-50-5). Intron retention and gene expression were analyzed using Galaxy (41).
Data Deposition and Statistical Methods.
Raw microarray and sequencing data are available in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession no. GSE53935. Statistical analyses were performed using Prism5/Graphpad. For oligonucleotides and further experimental details, see SI Methods.
Supplementary Material
Acknowledgments
We thank Gabriel Kolle and Ivonne Petermann for technical expertise with RNAseq; Minni Anko, Oliver Sieber, Anuratha Sakthianandeswaren, Chris Love, and Dmitri Mouradov for valuable scientific discussions; Tyler Alioto for providing a scan of the zebrafish Zv8 genome assembly for U12-type introns; Cameron Nowell for microscopy; Val Feakes for histology; Janna Taylor for graphics; and Dora McPhee, Kelly Turner, Mark Greer, Tyson Blanch, and Lysandra Richards for expert fish husbandry. This work was supported by the National Health and Medical Research Council of Australia [Project Grants 433614 and 1024878 (to J.K.H.) and 637395 (to G.J.L.), Program Grant 487922 (to J.K.H.), and Enabling Grant 455871], Australian Research Council Grant DK060322 (to G.J.L.), National Institutes of Health Grant DK060322 (to D.Y.R.S.), a Boehringer Ingelheim Fonds PhD fellowship and a University of Melbourne International Postgraduate Research Scholarship (to S.M.), the Australian Cancer Research Foundation, and a Victorian State Government Operational Infrastructure Support grant.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE53935).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305536111/-/DCSupplemental.
References
- 1.Licatalosi DD, Darnell RB. RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet. 2010;11(1):75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Jackson IJ. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991;19(14):3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietrich RC, Incorvaia R, Padgett RA. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1(1):151–160. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 5.Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35(Database issue):D110–D115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurica MS, Moore MJ. Pre-mRNA splicing: Awash in a sea of proteins. Mol Cell. 2003;12(1):5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 7.Hall SL, Padgett RA. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271(5256):1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 8.Tarn WY, Steitz JA. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273(5283):1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 9.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84(5):801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 10.Turunen JJ, Niemelä EH, Verma B, Frilander MJ. The significant other: Splicing by the minor spliceosome. Wiley Interdiscip Rev RNA. 2013;4(1):61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2(6):773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 12.Patel AA, McCarthy M, Steitz JA. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 2002;21(14):3804–3815. doi: 10.1093/emboj/cdf297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16(11):1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Younis I, et al. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. Elife. 2013;2:e00780. doi: 10.7554/eLife.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18(8):472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edery P, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332(6026):240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- 17.He H, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332(6026):238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulisfane N, et al. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet. 2011;20(4):641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- 19.Lotti F, et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151(2):440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hastings ML, et al. An LKB1 AT-AC intron mutation causes Peutz-Jeghers syndrome via splicing at noncanonical cryptic splice sites. Nat Struct Mol Biol. 2005;12(1):54–59. doi: 10.1038/nsmb873. [DOI] [PubMed] [Google Scholar]
- 21.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42(5):454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Will CL, et al. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10(6):929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442(7103):688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 24.Farooq M, et al. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008;317(1):336–353. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Zhao E, et al. Cloning and identification of a novel human RNPC3 gene that encodes a protein with two RRM domains and is expressed in the cell nucleus. Biochem Genet. 2003;41(9–10):315–323. doi: 10.1023/b:bigi.0000006032.04031.d0. [DOI] [PubMed] [Google Scholar]
- 26.Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev. 1999;13(7):851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benecke H, Lührmann R, Will CL. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J. 2005;24(17):3057–3069. doi: 10.1038/sj.emboj.7600765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tidow H, Andreeva A, Rutherford TJ, Fersht AR. Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5′ splice site of U12-type introns. Structure. 2009;17(2):294–302. doi: 10.1016/j.str.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Koehler R, Issac H, Cloonan N, Grimmond SM. The uniqueome: A mappability resource for short-tag sequencing. Bioinformatics. 2011;27(2):272–274. doi: 10.1093/bioinformatics/btq640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turunen JJ, Will CL, Grote M, Lührmann R, Frilander MJ. The U11-48K protein contacts the 5′ splice site of U12-type introns and the U11-59K protein. Mol Cell Biol. 2008;28(10):3548–3560. doi: 10.1128/MCB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pessa HK, et al. Gene expression profiling of U12-type spliceosome mutant Drosophila reveals widespread changes in metabolic pathways. PLoS ONE. 2010;5(10):e13215. doi: 10.1371/journal.pone.0013215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, et al. p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev. 2009;23(3):278–290. doi: 10.1101/gad.1761609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdel-Salam GM, et al. A homozygous mutation in RNU4ATAC as a cause of microcephalic osteodysplastic primordial dwarfism type I (MOPD I) with associated pigmentary disorder. Am J Med Genet A. 2011;155A(11):2885–2896. doi: 10.1002/ajmg.a.34299. [DOI] [PubMed] [Google Scholar]
- 34.Nagy R, et al. Microcephalic osteodysplastic primordial dwarfism type I with biallelic mutations in the RNU4ATAC gene. Clin Genet. 2012;82(2):140–146. doi: 10.1111/j.1399-0004.2011.01756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keightley MC, et al. In vivo mutation of pre-mRNA processing factor 8 (Prpf8) affects transcript splicing, cell survival and myeloid differentiation. FEBS Lett. 2013;587(14):2150–2157. doi: 10.1016/j.febslet.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trede NS, et al. Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc Natl Acad Sci USA. 2007;104(16):6608–6613. doi: 10.1073/pnas.0701919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26(22):3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 38.McCormick F. Cancer therapy based on oncogene addiction. J Surg Oncol. 2011;103(6):464–467. doi: 10.1002/jso.21749. [DOI] [PubMed] [Google Scholar]
- 39.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 40.Wood DL, Xu Q, Pearson JV, Cloonan N, Grimmond SM. X-MATE: A flexible system for mapping short read data. Bioinformatics. 2011;27(4):580–581. doi: 10.1093/bioinformatics/btq698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007;Chapter 10:5. doi: 10.1002/0471250953.bi1005s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.