Significance
One of the most highly debated questions in the field of transcriptomics is the functionality of antisense transcripts. Are these transcripts merely transcriptional noise and a byproduct of the leakiness of transcriptional repression, or are they functional? Antisense RNAs are being ubiquitously reported, but their functionality remains elusive. Here we report a high-throughput approach to enrich antisense RNAs that are in a double-stranded form with their cognate sense RNAs and thus in a functional complex. This has led to the identification of more than 300 RNase III-dependent potentially functional antisense RNAs in Escherichia coli. These findings reveal a clear picture of the magnitude and degree of functionality of this mostly hidden class of transcripts.
Abstract
Advances in high-throughput transcriptome analyses have revealed hundreds of antisense RNAs (asRNAs) for many bacteria, although few have been characterized, and the number of functional asRNAs remains unknown. We have developed a genome-wide high-throughput method to identify functional asRNAs in vivo. Most mechanisms of gene regulation via asRNAs require an RNA–RNA interaction with its target RNA, and we hypothesized that a functional asRNA would be found in a double strand (dsRNA), duplexed with its cognate RNA in a single cell. We developed a method of isolating dsRNAs from total RNA by immunoprecipitation with a ds-RNA specific antibody. Total RNA and immunoprecipitated dsRNA from Escherichia coli RNase III WT and mutant strains were deep-sequenced. A statistical model was applied to filter for biologically relevant dsRNA regions, which were subsequently categorized by location relative to annotated genes. A total of 316 potentially functional asRNAs were identified in the RNase III mutant strain and are encoded primarily opposite to the 5′ ends of transcripts, but are also found opposite ncRNAs, gene junctions, and the 3′ ends. A total of 21 sense/antisense RNA pairs identified in dsRNAs were confirmed by Northern blot analyses. Most of the RNA steady-state levels were higher or detectable only in the RNase III mutant strain. Taken together, our data indicate that a significant amount of dsRNA is formed in the cell, that RNase III degrades or processes these dsRNAs, and that dsRNA plays a major role in gene regulation in E. coli.
The advent and development of high-throughput sequencing technologies has uncovered the presence of widespread antisense transcription in many bacteria, with the number of annotated genes associated with antisense RNA (asRNA) differing greatly among bacterial species (1, 2). asRNAs are encoded on the DNA strand opposite an annotated gene and overlap a portion of a gene or the entire gene, or span multiple genes with perfect complementarity. asRNAs range in size from tens to thousands of nucleotides. Although numerous chromosomally encoded asRNAs have been identified, few have been confirmed by traditional methods or functionally characterized. Raghavan et al. (3) reported that few asRNAs are conserved between Escherichia coli and Salmonella enterica, and that the predicted promoter sequences of the asRNAs are not conserved between these species, suggesting that most asRNA transcripts are products of spurious transcription and are not biologically functional RNAs.
The majority of functionally characterized asRNAs are found on plasmids, phages, and transposons (4, 5). The mode of regulation by asRNAs can be classified according to molecular mechanism as transcription interference, transcription attenuation, alteration of transcript stability, and translation inhibition (1, 2, 6). With the exception of transcription interference, a physical RNA–RNA interaction between the sense RNAs and asRNAs is necessary for all of these mechanisms, requiring that both RNAs be expressed in the same cell at the same time. The lengths and complete complementarity of the sense RNAs and asRNAs can lead to long double-stranded RNAs (dsRNAs).
Ribonuclease III (RNase III) is a highly conserved endoribonuclease that specifically cleaves dsRNAs and regulates gene expression in E. coli and other bacteria (7–10). Lasa et al. (9) recently demonstrated that RNase III plays a central role in a type of antisense regulation specific for Gram-positive bacteria. Deep sequencing of both short and long RNA fractions in WT and RNase III mutant strains detected a genome-wide RNase III-dependent processing of overlapping transcripts into short, 22-nt RNAs. Three-quarters of sense RNAs from annotated genes appear to be processed via RNase III-dependent asRNA regulation in Staphylococcus aureus. Lasa et al. reported that several other Gram-positive bacteria show a similar pattern of RNase III-dependent short RNAs. However, S. enterica, the sole Gram-negative species tested in the study, did not exhibit the same pattern of short sense and antisense complementary RNAs as the Gram-positive bacteria, suggesting that the mechanism may not exist or may differ in Gram-negative bacteria. In agreement with the foregoing findings, deep sequencing of RNA coimmunoprecipitated with WT or cleavage mutants of RNase III in S. aureus was found to capture low abundant asRNAs that cover 44% of annotated genes (11).
In the present study, we identified functional asRNAs using an in vivo approach in E. coli. We hypothesized that a subset of functional asRNAs would be in dsRNAs, because an RNA–RNA interaction is required for most mechanisms of regulation via known asRNAs. Thus, we developed a method to isolate dsRNAs from total RNA by immunoprecipitation with a monoclonal antibody specific for dsRNA. We expected that dsRNAs would be more abundant in an RNase III mutant strain, and thus we deep-sequenced cDNA libraries of the total RNA (input) and immunoprecipitated dsRNA (IP) from WT and RNase III mutant strains. We have identified and confirmed the expression of numerous asRNAs that are potentially functional and have developed a methodology that is broadly applicable for identifying functional asRNAs in both eukaryotic and prokaryotic organisms.
Results
To identify functional asRNAs that regulate their cognate mRNAs via an RNA–RNA interaction, we designed a protocol to isolate dsRNAs. The monoclonal anti-dsRNA J2 antibody binds dsRNAs at least 40 bp long in a sequence-independent fashion (12–14). The J2 dsRNA monoclonal antibody was previously used to detect viral dsRNA by dsRNA immunoblotting, immunohistochemistry, ELISA, and immunoprecipitation in eukaryotic cells (13–20). In this study, we used the J2 dsRNA antibody in an immunoprecipitation assay to enrich dsRNA from total RNA extracted from E. coli.
We assessed the specificity of the J2 monoclonal antibody using artificial in vitro transcribed double-stranded and single-stranded RNA substrates in RNA immunoblots and immunoprecipitations (SI Appendix, Fig. S1 A and B). These data demonstrate that the J2 dsRNA monoclonal antibody specifically recognizes dsRNA in both immunoprecipitation and immunoblot analyses.
Identification and Characterization of dsRNAs.
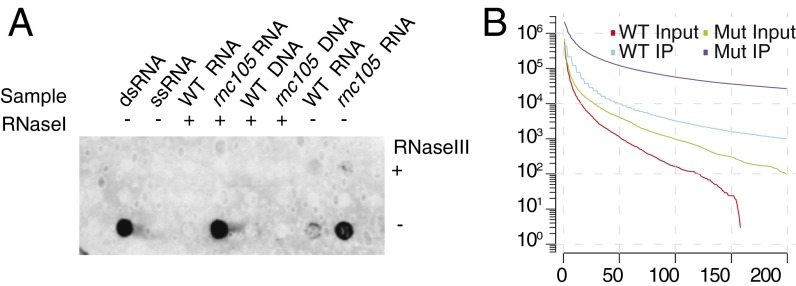
We expected to find that RNase III plays a central role in the degradation and processing of dsRNA substrates in vivo. Thus, to determine the extent of dsRNA present in E. coli and to examine the role of RNase III in regulation of dsRNA levels, we immunodot-blotted RNA extracted from WT and rnc105 mutant strains with the J2 monoclonal antibody. The RNase III enzyme binds dsRNA, but is catalytically inactive in the rnc105 mutant strain. The rnc105 mutant strain has significantly more dsRNA than the WT strain; in addition, the antibody is specific for endogenous dsRNA (Fig. 1A). The signal is absent in the DNA and in the RNase III-treated RNA samples; however, RNase I, an ssRNA-specific endoribonuclease, has no effect on the signal, indicating that the signal comes primarily from dsRNA. These data demonstrate the presence of dsRNA in E. coli and indicate that RNase III plays a central role in its processing.
Fig. 1.
Identification of genome-wide dsRNA. (A) RNA and DNA samples from WT and rnc105 mutant strains were immunodot-blotted with the J2 monoclonal antibody. In addition, artificial ssRNA and dsRNA samples were blotted as controls. The samples in the top row of the dot blot were treated with RNase III, and samples in the bottom row were not. In addition, RNA and DNA samples were either treated with RNase I or untreated, as indicated. (B) The total bases in the genome covered as a function of minimum number of reads in each library, illustrating the amount and enrichment of potential dsRNAs. In other words, at each point the line indicates that y positions in the genome covering both strands in the given library had at least x reads mapped on the less-covered strand.
To identify functional asRNAs in a transcriptome-wide manner, dsRNAs from WT and mutant rnc105 strains of E. coli were immunoprecipitated, depleted of ribosomal RNA (rRNA), converted to cDNA libraries, and deep-sequenced. As an input control for the immunoprecipitation, rRNA-depleted total RNAs from both strains were also converted to cDNA and deep-sequenced. The resulting total and IP libraries were analyzed. In addition, a control experiment was performed to demonstrate that the dsRNAs immunoprecipitated were formed in vivo and not after cell lysis (SI Appendix, Fig. S2).
We obtained 51 million reads of length 41–50 bp from all four libraries and mapped them to the E. coli K12 genome (21), resulting in 8–13 million high-quality mappings for each library. We further analyzed the read coverage of bases with reads mapping to both strands, illustrating the global differences of base coverage at putative dsRNA regions among libraries (Fig. 1B). The double-stranded coverage is greater in the rnc105 mutant strain input library than in the WT library, confirming that RNase III plays a role in the processing of dsRNAs. In addition, the IP libraries from both the WT and rnc105 mutant strains show a marked increase in double-stranded coverage compared with their input controls, indicating that the IP was efficient. The IP libraries had 16,329 potential regions of sufficient length to have been immunoprecipitated by the antibody with reads mapping to both strands at the same position, covering more than 2 MB.
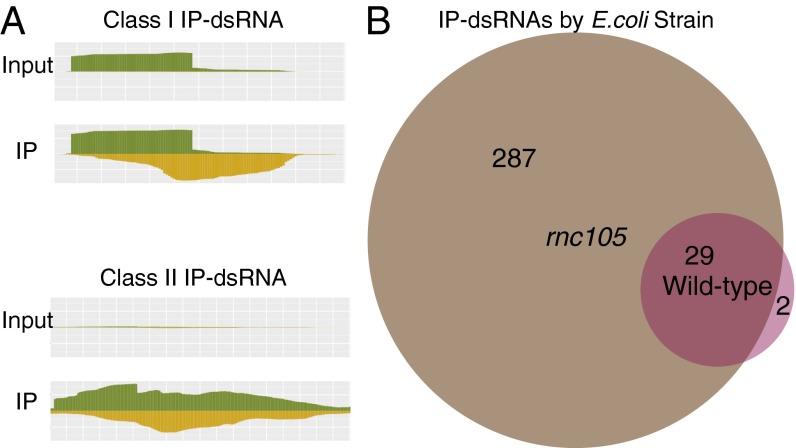
To determine whether these potential double-stranded regions were indeed dsRNAs in the cell, and thus immunoprecipitated, we assigned scores to them based on the pattern of observed and expected coverage after manual inspection of known dsRNAs and putative dsRNA regions of a sample of the genome (Fig. 2A). For many of the known cis-asRNAs, one strand of the dsRNA region was more highly expressed than the other strand. These regions, which we term class I dsRNAs, exhibit the differential expression pattern of known cis-asRNAs in the input libraries. In addition, we identified many dsRNAs with similar expression levels on both strands in the input libraries. We termed these regions class II dsRNAs and designated a separate model to identify them in the data. For both classes, the overall levels in the IP libraries increased, and their coverage on both strands was closer to even.
Fig. 2.
(A) Two different models were used to analyze the deep sequencing data and identify class I and II IP-dsRNAs. Class I IP-dsRNAs, characterized by differential coverage in the input library, are scored by their coverage increase in the strand less coverage in the input (yellow), from input to IP libraries. Class II IP-dsRNAs, which have similar coverage on both strands in the input library, were scored by the total coverage increase in both strands (yellow and green). Both classes were also scored higher as the +/− strand coverage ratio approached 1 in the IP library. The scores for the coverage increase and ratio were modeled in a probabilistic framework with a background distribution, resulting in a single overall score, which was used to predict whether the data indicated the presence of an IP-dsRNA. (B) In our models, IP-dsRNAs were found predominantly in the rnc105 strain. IP-dsRNAs identified in the WT libraries were also present in the mutant libraries, with two exceptions.
We developed a scoring method that assigns higher values to regions that exhibit the aforementioned patterns. We then reported the scores of all potential regions based on similarity to the class I or class II patterns, depending on which scored higher. We determined a score threshold based on the scores of known dsRNAs. Taking into account only those regions that scored above our threshold, we applied an algorithm to extend and cluster these high-scoring regions to better define the boundaries of the double-stranded region (Materials and Methods). Our scoring method identified 213 class I and 103 class II putative double-stranded regions covering 55,655 bases. We termed these dsRNA regions immunoprecipitated-dsRNAs (IP-dsRNAs), which refer only to the dsRNA region of the overlapping transcripts. A total of 316 IP-dsRNAs were identified in the rnc105 mutant strain, compared with only 31 in the WT strain (Fig. 2B and SI Appendix and Dataset 1). Moreover, 29 of the 31 WT strain IP-dsRNAs were also identified in the rnc105 mutant strain. Taken together, these data clearly demonstrate the central role of RNase III in the metabolism of dsRNA in E.coli.
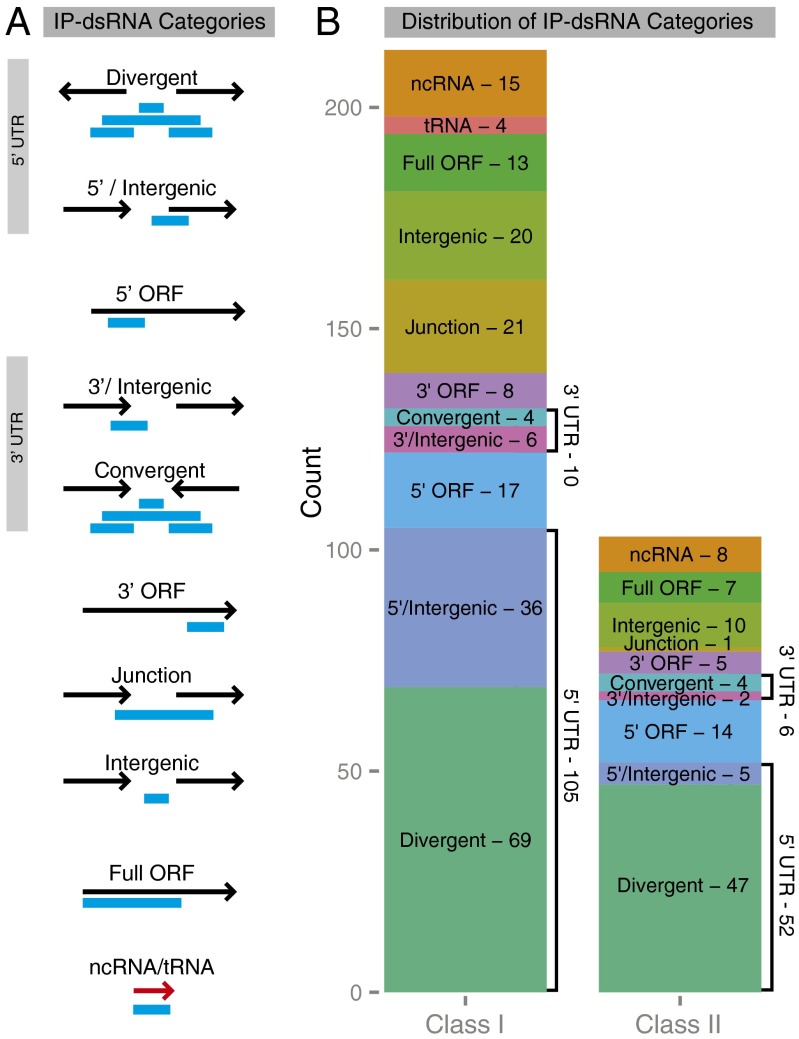
To obtain a global picture of the location of RNase III-dependent IP-dsRNAs relative to genes, we categorized them based on the annotated ORFs, as described in Materials and Methods and outlined in Fig. 3. Our data demonstrate that the majority of IP-dsRNAs are found at the 5′ UTRs of genes, either as overlapping 5′ divergent gene transcripts or as 5′ asRNAs, suggesting that asRNA-dependent gene regulation occurs primarily at the 5′ end of transcripts. A well-studied mechanism of translational regulation involves ncRNAs occluding the ribosome binding site (RBS), thereby blocking translation. Thus, we examined the number of IP-dsRNAs that cover putative RBSs. 147 of the IP-dsRNAs categorized as full ORF, 5′/intergenic, or divergent cover the 10 bases upstream of the start codon of genes, composing 80% or more of the IP-dsRNAs in each of these categories. The 63 IP-dsRNAs identified in both the 5′/intergenic and gene junction categories appear to contain bona fide cis-asRNAs; that is, they do not appear to be a part of any transcript associated with an annotated ORF. In contrast, IP-dsRNAs identified in divergent and convergent genomic loci might not be dedicated cis-asRNAs, because they also may be a part of annotated mRNAs. Regardless, these overlapping transcripts may still regulate one another via antisense mechanisms.
Fig. 3.
Genomic location of the dsRNAs. The IP-dsRNAs were categorized based on their location relative to annotated ORFs in that region of the genome (EcoCyc). (A) Schematic illustration of the categories of IP-dsRNAs. IP-dsRNAs identified at the 5′ end of genes in a divergent orientation were classified as divergent, whereas those at the 3′ ends of convergent genes were classified as convergent. IP-dsRNAs near head-to-tail-oriented genes were classified as gene junction if spanning the 3′ end of the upstream gene and the 5′ end of the downstream gene, as intergenic if located between the two genes, or 5′/intergenic or 3′/intergenic if spanning one gene boundary, but not both gene boundaries. If they covered more than one-half of an ORF, they were classified as full ORF. If they were contained in the 3′ or 5′ half of an ORF, they were classified as 3′ or 5′ ORF, respectively. (B) Bar graph summarizing the distribution of the IP-dsRNA categories.
Northern blot analyses were performed on 21 selected IP-dsRNAs to confirm the expression of both strands of RNA and to examine their dependence on RNase III (SI Appendix, Table S1). Generally, the strongest signals detected on the Northern blots and the deep-sequencing data correspond to the overlapping regions of the RNAs. We hypothesize the IP-dsRNAs are extremely stable in the absence of a WT RNase III in the cell.
dsRNAs Located at the 5′/ Intergenic Ends of ORFs.
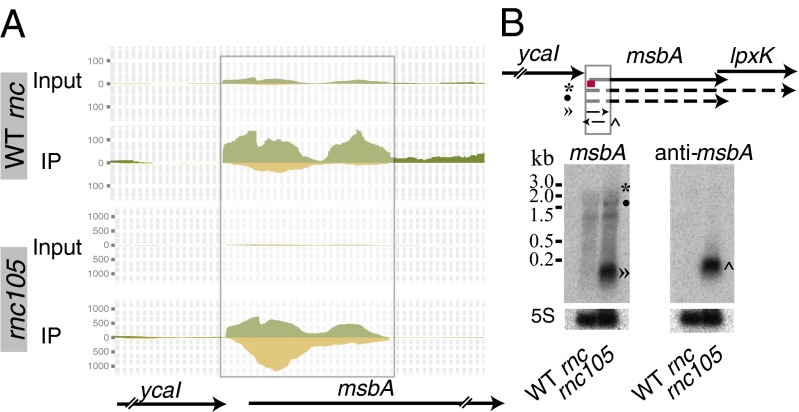
A total of 41 IP-dsRNAs were identified at the 5′/intergenic ends of genes with a head-to-tail orientation. The asRNAs identified in these IP-dsRNAs are bona fide cis-asRNAs, discrete transcripts that do not encode an annotated ORF. Northern blot analyses confirmed the presence of asRNAs opposite two genes (msbA and yrbL) from this category of IP-dsRNAs (SI Appendix, Table S1). The IP-dsRNAs located in the 5′/intergenic ends of genes are in operons or singularly transcribed. The msbA gene is predicted to be in an operon with the upstream and downstream genes ycaI, lpxK, and ycaZ (22). We identified an IP-dsRNA at the 5′ end of the msbA gene extending into the intergenic space. The coverage maps of the four libraries show immunoprecipitation in the rnc105 mutant strain (Fig. 4A). Northern blot analyses confirmed the presence of both sense and antisense transcripts, with the antisense transcript detected only in the mutant strain.
Fig. 4.
Verification of transcripts forming IP-dsRNAs at the 5′ end of transcripts oriented head to tail, termed 5′/intergenic. The IP-dsRNA transcripts were confirmed by Northern blot analyses for msbA. (A) The deep-sequencing results displayed in a coverage map of input and IP libraries in the WT and rnc105 mutant strains. The height at each position indicates the number of reads that mapped to that base. The translucent box indicates the IP-dsRNA. The + strand is always shown on top (in green), and the − strand is on the bottom (in yellow). Note that the y-axis scale differs between the WT and rnc105 mutant strains. (B) Northern blot analyses of total RNA fractionated on a formaldehyde-agarose gel (Right) or a denaturing polyacrylamide gel (Left), blotted to a nylon membrane, and hybridized with oligonucleotides. The red line represents the location of the oligonucleotide probes used for the Northern blot. The predicted transcripts are denoted and marked with the appropriate band in the Northern blot. Three independent experiments were performed, and representative data are shown.
The yrbL gene is a monocistronic transcript with an IP-dsRNA identified at its 5′/intergenic end. We identified a short anti-yrbL RNA at the 5′ end of the gene that is detected only in the strain lacking active RNase III. In addition, the RNA steady-state levels of yrbL are higher in the rnc105 mutant strain compared with the WT strain (SI Appendix, Fig. S3 A and B).
asRNAs Identified Opposite Gene Junctions in Operons.
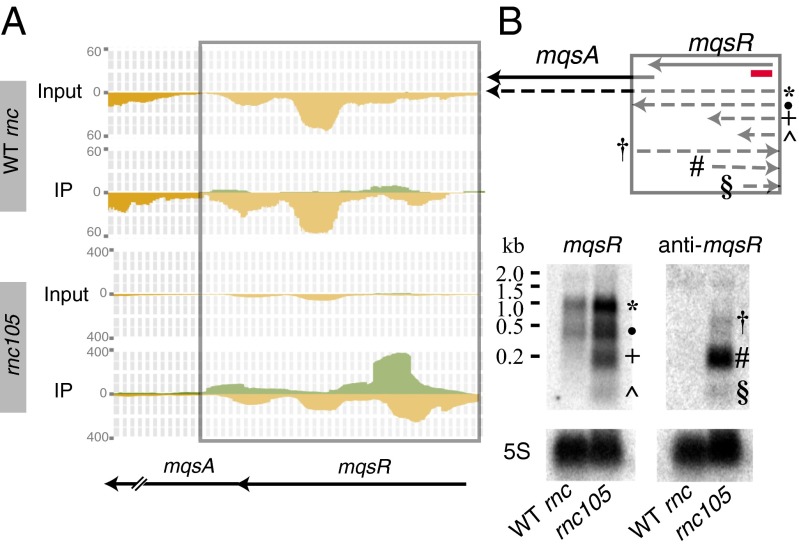
RNase III plays a major role in the maturation of ribosomal RNA and regulates the quantity of mRNAs and ncRNAs (10, 23, 24). Our data suggest that asRNAs transcribed opposite to gene junctions form dsRNA complexes with the mRNA and guide processing and/or degradation of the operon. We identified 22 IP-dsRNAs that span gene junctions within operons (Fig. 3), and confirmed the sense and antisense transcripts by Northern blot analysis for one example (SI Appendix, Table S1). The mqsRA operon encodes a type II toxin/antitoxin system. The deep-sequencing data indicate that the anti-mqsR transcript overlaps the gene junction and covers the entire mqsR gene (Fig. 5A). Northern blot analysis confirmed the deep-sequencing data and demonstrated that both the sense and antisense mqsR steady-state levels are more abundant in the RNase III mutant strain. In addition, the anti-mqsR transcript is undetectable in the WT strain (Fig. 5B).
Fig. 5.
Verification of IP-dsRNA transcripts at gene junctions. The dsRNA transcripts (A) were verified via Northern blot analyses (B) for mqsR as described in the Fig. 4 legend.
asRNAs Transcribed Opposite of Small Regulatory RNAs.
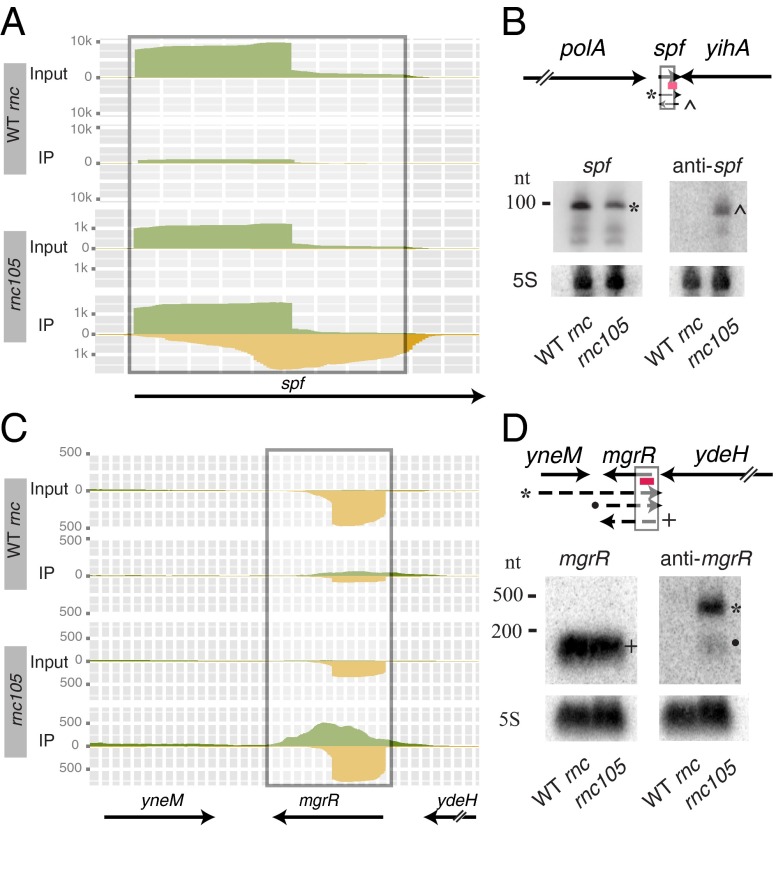
Our deep-sequencing analyses revealed 23 asRNAs transcribed opposite to ncRNAs, mainly small regulatory RNAs (Fig. 3B). We observed that the ncRNAs identified in IP-dsRNA frequently are convergently transcribed with the adjacent gene. We hypothesized that the anti-ncRNAs identified in the IP-dsRNAs were transcribed via read-through of the convergent gene. For example, the spf gene is convergently transcribed with the downstream yihA gene, and the anti-spf RNA could be transcribed via read-through of the yihA transcript (Fig. 6 A and B). However, Northern blot analysis suggested that the anti-spf RNA is not a product of read-through transcription, demonstrated by the lack of an RNA signal the size of the yihA and anti-spf transcript (∼1,000 nucleotides). Instead, a single RNase III-dependent anti-spf transcript at ∼100 nucleotides was observed, corresponding to an RNA that does not include the yihA transcript. In contrast, the asRNA transcribed opposite of the small RNA MgrR appears to be at least partially a product of read-through from the yneM gene (Fig. 6 C and D).
Fig. 6.
Verification of IP-dsRNA transcripts at ncRNAs. The IP-dsRNA transcripts were verified via Northern blot analyses for spf (A and B) and mgrR (C and D) encoding small regulatory RNAs as described in the Fig. 4 legend, but using 8% denaturing polyacrylamide gels.
Northern blot analyses revealed two sizes of anti-MgrR transcripts, both dependent on RNase III. The 300-nt band visualized on the Northern blot is consistent with a transcript consisting of yneM and the anti-MgrR RNA. A smaller, ∼90-nt band corresponds to an antisense transcript overlapping only with MgrR (Fig. 6 C and D). We confirmed the sense and antisense strands of seven ncRNA IP-dsRNAs by Northern blot analysis (SI Appendix, Table S1 and Figs. S4 and S5), and anti-MgrR was the sole apparent product of transcriptional read-through from the adjacent gene.
5′-End Divergently Transcribed mRNAs Form dsRNAs.
The E. coli genome is compact, and numerous transcripts partially overlap neighboring transcripts in their 5′ and 3′ ends. Our data suggest that a subset of these 5′ divergent overlapping mRNAs form dsRNA in a potential mechanism to coordinate gene regulation. We identified 116 IP-dsRNAs at 5′ divergent genomic loci and verified the transcripts of three examples (SI Appendix, Table S1). Of note, the deep-sequencing and Northern blot analysis data for this category of IP-dsRNAs suggest unannotated 5′ ends of numerous transcripts in double-stranded regions. The lptF/pepA and ydbH/ldhA divergent gene pairs represent the 5′ divergent category of IP-dsRNAs (SI Appendix, Fig. S6). The coverage maps demonstrate the high scoring IP-dsRNA and illustrate the enrichment observed in the IP library. The Northern blot analysis results suggest that the IP-dsRNA transcripts from these regions include the downstream ORFs of the divergent genes. The lptF/pepA and ydbH/ldhA steady-state transcript levels are higher in the rnc105 mutant strain, suggesting that dsRNAs are regulated by RNase III. In addition, we observed that the divergent transcripts overlapped either in the intergenic space or within one of the genes, as illustrated by the lptF/pepA and ydbH/ldhA transcripts, respectively.
Discussion
The reported estimates of asRNA transcripts vary greatly among bacteria, from 1% to 75% of annotated genes, and recent reports suggest that some, if not most, antisense transcripts are nonfunctional products of pervasive transcription (1–3, 9, 11). In contrast, we have identified a subset of asRNAs that are physiologically relevant and base pair with their complementary target RNAs, forming dsRNAs. We have demonstrated that the dsRNAs are processed by RNase III and thus are more abundant in an RNase III mutant strain. The RNase III dependence of the transcripts, as well as the in vivo immunoprecipitation of dsRNAs, indicate that the IP-dsRNAs that we have identified are dsRNAs in vivo. Furthermore, our data indicate that in the absence of an active RNase III, the dsRNA regions of transcripts are stable and more abundant than the single-stranded regions of the same transcripts. We hypothesize that other RNases degrade and process the single-stranded regions of the transcripts, whereas the dsRNA regions remain in the mutant strain. In contrast, in the WT strain, the dsRNA regions are mostly undetectable or similar in abundance to the single-stranded regions of transcripts.
To identify a functional subset of asRNAs, we developed an immunoprecipitation method to isolate dsRNA in both WT and RNase III mutant strains and used stringent statistical modeling to identify IP-dsRNAs. Initial inspection of the data revealed two coverage patterns among IP-dsRNAs; thus, we used two models to assign scores to the potential dsRNA regions, resulting in two classes of IP-dsRNAs (Fig. 2). Class I IP-dsRNAs are formed by two transcripts that are differentially expressed in the input libraries. The sense strand is highly abundant, whereas the antisense strand is absent or hardly detectible. We expect that under certain environmental conditions, the asRNA may be up-regulated and repress expression of the sense RNA. On the other hand, class II IP-dsRNAs are formed by transcripts expressed equally on both strands in the input libraries, implying that they are coregulated.
Applying these two models to our data, we have identified many previously reported types of functional asRNAs, including asRNAs transcribed opposite noncoding sRNAs and asRNAs resulting from overlapping long 5′ UTRs of divergently transcribed genes. In addition, we have identified a large category of novel chromosomal asRNAs transcribed opposite the 5′/intergenic ends of nondivergently transcribed genes and gene junctions within operons. These cis-asRNAs appear to be ncRNAs that do not code for any annotated ORFs. As proof of principle, we identified 9 of the 18 known and characterized dsRNAs in E. coli. Six of the known dsRNAs that we did not identify in our analyses had very low coverage in both the input and the IP libraries. The other three were not identified as IP-dsRNAs because their plus and minus strand coverages were too different. Although our data suggest that a dsRNA could have been pulled down in our experiment in these regions, given that reads were found in both strands, these regions did not stand sufficiently apart from the background to be identified as IP-dsRNAs.
Most of the known RNA-based mechanisms of gene regulation, including transacting sRNAs, influence regulation at the 5′ end of genes, likely because it is energetically favorable. In agreement with this, 50% of the IP-dsRNA regions are located in the 5′ UTR of genes, whereas only 0.5% of the IP-dsRNAs are located in the 3′ UTR, suggesting that the main mechanism of asRNA gene regulation via dsRNA intermediates occurs at the 5′ end of transcripts.
The genomic organization of genetic elements has long been thought to play a role in the coordinate regulation of genes, whether coexpressed or differentially expressed. Coordinate regulation of overlapping transcripts from divergently transcribed genes has been described in S. aureus and Listeria monocytogenes (11, 25). Lasa et al. (9) reported involvement of RNase III in the formation of short RNA fragments (∼22 nt) mapping to overlapping transcripts. They observed similar expression patterns in other Gram-positive bacteria but not in S. enterica, indicating a Gram-positive–specific mechanism, possibly owing to different collections of RNases, helicases, and other RNA-binding proteins. We have identified RNase III-dependent IP-dsRNAs localizing to overlapping 5′ UTRs of numerous divergent genes, suggesting a basis for their coregulation. Because our approach does not allow for reliable identification of dsRNA fragments shorter than 40 bp, we cannot exclude the possibility of RNase III-processed short RNA fragments in E. coli. The molecular mechanism involving asRNAs, formation of dsRNAs, and RNase III remains to be elucidated.
Recent transcriptome-wide studies of L. monocytogenes identified a dual functional group of long antisense transcripts (lasRNAs), termed excludons, which negatively regulate one ORF via an antisense mechanism while simultaneously contributing to the transcription of adjacent, divergently transcribed ORFs. Some of the divergent and full ORF class I differentially expressed asRNAs may be excludons. Experimental validation of specific examples is needed to determine whether the excludon paradigm of asRNA-mediated gene regulation occurs in E. coli.
asRNAs transcribed opposite to noncoding sRNAs (anti-ncRNAs) also have been reported previously (11), but few examples have been functionally characterized or validated by traditional methods, such as Northern blot analysis or quantitative RT-PCR. We have identified 23 anti-ncRNAs and validated the sequencing data by Northern blot analysis for seven examples. All of the anti-ncRNAs that we tested were regulated by RNase III and were detected by Northern blot analyses only in the rnc105 mutant strain. In a recent study, anti-ncRNAs were identified through an RNase III coimmunoprecipitation (11); however, the authors did not detect many of the anti-ncRNAs by Northern blot analysis, and they suggested these ncRNAs may be a result of pervasive transcription. In contrast to that finding, our data suggest that some anti-ncRNAs are biologically relevant and base pair with the ncRNAs. We hypothesize that the levels of anti-ncRNAs may increase and regulate the levels of the ncRNAs under certain stress or recovery conditions.
We also have identified eight dsRNA regions overlapping or neighboring phage and transposase genes on the chromosome. Most cis-asRNAs were first identified and characterized in plasmids, phages, and transposons and are responsible for the repression of these elements; thus, the identification of asRNA transcripts opposite transposase and phage genes was not surprising. However, most of the observed dsRNA regions associated with transposase genes are found downstream of the gene in the intergenic space. We confirmed two such examples by Northern blot analyses (SI Appendix, Fig. S7). tRNA, tmRNA, and several sRNAs, located adjacent to attachment and integration sites of phages or transposons, have been proposed to be acquired through horizontal gene transfer (26). The small IP-dsRNA regions observed downstream of the phage and transposase genes may be novel horizontally acquired sRNAs. In addition, the sRNAs identified in IP-dsRNAs may have been associated with transposases when they entered the genome.
Surprisingly, we found asRNAs encoded opposite of five type II toxin-antitoxin (TA) systems in E. coli (SI Appendix and Dataset S1). The type II TA systems consist of a toxin and an antitoxin protein expressed from two tandem genes. The toxin and antitoxin form a stable protein complex that results in inhibition of the toxin. In contrast, type I TA systems regulate synthesis of the toxin by inhibiting its efficient translation via an asRNA. The type II TA systems have not been shown to include such an asRNA regulation mechanism, however. Northern blot analyses confirmed the presence of the asRNA for both mqsR and yoeB toxins (Fig. 5 and SI Appendix, Fig. S8). Our identification of asRNAs encoded opposite the type II TA systems suggests an additional level of regulation. The mqsR/mqsA and yefM/yoeB TA steady-state transcripts are more abundant in the rnc105 mutant strain, suggesting that RNase III plays a role in their regulation. However, many TA systems are activated by cell stress. The absence of an active RNase III may stress the cell, and thus the increased number of TA transcripts observed may be attributed to increased transcription, rather than to derepression via RNase III. Further mechanistic analyses are needed to understand the role of asRNAs in regulation of type II TA systems.
Finally, the amount of dsRNA identified in our study suggests that many sense/antisense pairs of RNAs in the cell base pair and form dsRNAs. The binding kinetics of two folded RNAs is worth considering. The ability of the two RNAs to form a dsRNA depends on several factors, including the individual secondary structures of both RNAs (accessibility of certain nucleotides), the relative amounts of the two RNAs in the cell, the presence of ribosomes on the mRNA occluding nucleotides, and the presence of proteins that may interact with both RNAs. The RNA chaperone Hfq is required for many trans-encoded sRNAs that regulate target mRNA through partially complementary base pairing. Recently, Ross et al. (27) demonstrated that Hfq also regulates the binding of a cis-asRNA RNA-OUT with RNA-IN of the Tn10/IS10 transposition system in E. coli. The authors suggested that Hfq may be involved in regulating other asRNA-dependent gene regulation systems. If Hfq does not play a role in facilitating antisense/sense RNA pairing, then another RNA chaperone likely does so.
The majority of functionally characterized regulatory RNAs are differentially expressed and regulated by environmental signals. We expect that applying our protocol to bacteria grown under different environmental conditions will identify more functional asRNAs.
The IP-dsRNA sequences identified in this study could regulate gene expression by several known antisense mechanisms, including transcript stabilization or destabilization, ribosome binding site accessibility, and transcription attenuation. We hypothesize that RNase III plays a direct role in antisense gene regulation by processing or degrading the dsRNA region altering the stability, structure, or RBS availability of the mRNA. In addition, we postulate that the dsRNA region alone may alter the mRNA via the same mechanisms, but that RNase III degrades the complex only after the regulation has occurred. Regardless of the role of RNase III, our data suggest that asRNAs via dsRNA constitute a broad mechanism of gene regulation.
Materials and Methods
Bacterial Strains and RNA Isolation.
E. coli strains SDF204 (W3110rnc+ TD1-17::Tn10) and SDF205 (W3110rnc105 TD1-17::Tn10) were grown in LB medium to log phase (OD600 ∼0.5). For Northern blot analyses, total RNA was isolated using a hot phenol protocol described by Jahn et al. (28), unless stated otherwise. Total RNA isolated for immunodot blots and immunoprecipitations followed the hot phenol protocol described above with several modifications to minimize dsRNA artifacts, as described in detail in SI Appendix, Materials and Methods.
Immunoprecipitation Assays.
Total RNA or in vitro transcribed RNAs were incubated with J2 monoclonal anti-dsRNA antibodies at different ratios at 4 °C overnight in 1× PBS and 0.1% Tween 20 with 2 units of RNasin (Promega). Dynabeads Protein A (Invitrogen) were used to immunoprecipitate the antibody–RNA complexes. The Dynabeads were prepared as suggested by the manufacturer. The antibody–RNA solutions were then added to the beads, gently mixed, and incubated for 10 min at room temperature while rotating. The tubes were then moved to a magnetic stand, after which the supernatant was removed. The beads were washed four times and then resuspended with 1× PBS and 0.1% Tween-20. The beads with the antibody–RNA complexes bound were then subjected to phenol/chloroform/isoamyl alcohol (25:24:1) extraction using a Phase Lock Gel tube. The aqueous phase was ethanol-precipitated and analyzed either by SDS/PAGE and SYBR Safe staining or with an Agilent 2100 Bioanalyzer on a picoRNA chip.
cDNA Library Preparation.
Directional (strand-specific) RNA-seq cDNA libraries were constructed following a ligation-based protocol described in detail in SI Appendix, Materials and Methods.
Deep-Sequencing Analyses.
Reads were mapped to the E.coli K12 genome using Bowtie 2 (29) in local mode with default parameters. Scores for the class I and class II models determined by scoring models, which are described in detail, along with the remainder of the analysis, in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Scott Samuels, Vitaly Sedlyarov, and members of the Schroeder laboratory for thoughtful and critical readings of the manuscript; Andreas Sommer, Arnt von Haeseler, Micheal Wolfinger, Hakim Tafer, and members of the Schroeder laboratory for useful discussions; and Johanna Stranner for excellent technical assistance. This work was supported by the Austrian Science Fund (Grants FWF I538-B12, F4301, and F4308) and the University of Vienna.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences have been deposited at the National Center for Biotechnology Information Sequence Read Archive (Study accession no. SRP028119: experiment accession nos. SRX326854, SRX326853, SRX326852, and SRX326842).
See Commentary on page 2868.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315974111/-/DCSupplemental.
References
- 1.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75(2):286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomason MK, Storz G. Bacterial antisense RNAs: How many are there, and what are they doing? Annu Rev Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghavan R, Sloan DB, Ochman H. Antisense transcription is pervasive but rarely conserved in enteric bacteria. mBio. 2012;3(4):e00156-12. doi: 10.1128/mBio.00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10(2):102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 6.Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: A new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol. 2013;11(2):75–82. doi: 10.1038/nrmicro2934. [DOI] [PubMed] [Google Scholar]
- 7.Stead MB, et al. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2011;39(8):3188–3203. doi: 10.1093/nar/gkq1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand S, Gilet L, Bessières P, Nicolas P, Condon C. Three essential ribonucleases-RNase Y, J1, and III-control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 2012;8(3):e1002520. doi: 10.1371/journal.pgen.1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lasa I, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA. 2011;108(50):20172–20177. doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arraiano CM, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34(5):883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 11.Lioliou E, et al. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012;8(6):e1002782. doi: 10.1371/journal.pgen.1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonin M, et al. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA. 2000;6(4):563–570. doi: 10.1017/s1355838200992318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schönborn J, et al. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19(11):2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukács N. Detection of virus infection in plants and differentiation between coexisting viruses by monoclonal antibodies to double-stranded RNA. J Virol Methods. 1994;47(3):255–272. doi: 10.1016/0166-0934(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 15.Mueller S, et al. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci USA. 2010;107(45):19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Targett-Adams P, Boulant S, McLauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol. 2008;82(5):2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Vaheri A, Weber F, Plyusnin A. Old World hantaviruses do not produce detectable amounts of dsRNA in infected cells and the 5′ termini of their genomic RNAs are monophosphorylated. J Gen Virol. 2011;92(Pt 5):1199–1204. doi: 10.1099/vir.0.029405-0. [DOI] [PubMed] [Google Scholar]
- 18.Overby AK, Popov VL, Niedrig M, Weber F. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J Virol. 2010;84(17):8470–8483. doi: 10.1128/JVI.00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triantafilou K, et al. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125(Pt 20):4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 20.Zangger H, et al. Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl Trop Dis. 2013;7(1):e2006. doi: 10.1371/journal.pntd.0002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: Specific transcription in dense regions of overlapping promoter-like signals. J Mol Biol. 2003;333(2):261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 23.Babitzke P, Granger L, Olszewski J, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175(1):229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava RK, Miczak A, Apirion D. Maturation of precursor 10Sa RNA in Escherichia coli is a two-step process: The first reaction is catalyzed by RNase III in presence of Mn2+ Biochimie. 1990;72(11):791–802. doi: 10.1016/0300-9084(90)90188-m. [DOI] [PubMed] [Google Scholar]
- 25.Wurtzel O, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottesman S, Storz G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3(12):a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross JA, Ellis MJ, Hossain S, Haniford DB. Hfq restructures RNA-IN and RNA-OUT and facilitates antisense pairing in the Tn10/IS10 system. RNA. 2013;19(5):670–684. doi: 10.1261/rna.037747.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahn CE, Charkowski AO, Willis DK. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods. 2008;75(2):318–324. doi: 10.1016/j.mimet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.