Significance
An experimental system was developed in mice to study the long-term benefits of early exposure to secretory antibodies of the IgA class (SIgA) in breast milk. We found that breast milk-derived SIgA promoted intestinal epithelial barrier function in suckling neonates, preventing systemic infection by potential pathogens. Long-term benefits of early exposure to SIgA included maintenance of a healthy gut microbiota and regulation of gene expression in intestinal epithelial cells. These findings suggest that maternal antibodies provide benefits to the intestinal immune system of the breast-fed infant, which persist into adulthood.
Abstract
Maintenance of intestinal homeostasis requires a healthy relationship between the commensal gut microbiota and the host immune system. Breast milk supplies the first source of antigen-specific immune protection in the gastrointestinal tract of suckling mammals, in the form of secretory IgA (SIgA). SIgA is transported across glandular and mucosal epithelial cells into external secretions by the polymeric Ig receptor (pIgR). Here, a breeding scheme with polymeric Ig receptor-sufficient and -deficient mice was used to study the effects of breast milk-derived SIgA on development of the gut microbiota and host intestinal immunity. Early exposure to maternal SIgA prevented the translocation of aerobic bacteria from the neonatal gut into draining lymph nodes, including the opportunistic pathogen Ochrobactrum anthropi. By the age of weaning, mice that received maternal SIgA in breast milk had a significantly different gut microbiota from mice that did not receive SIgA, and these differences were magnified when the mice reached adulthood. Early exposure to SIgA in breast milk resulted in a pattern of intestinal epithelial cell gene expression in adult mice that differed from that of mice that were not exposed to passive SIgA, including genes associated with intestinal inflammatory diseases in humans. Maternal SIgA was also found to ameliorate colonic damage caused by the epithelial-disrupting agent dextran sulfate sodium. These findings reveal unique mechanisms through which SIgA in breast milk may promote lifelong intestinal homeostasis, and provide additional evidence for the benefits of breastfeeding.
Breast milk provides the first source of antibody-mediated immune protection in the intestinal tract of suckling infants, in the form of secretory IgA (SIgA) (1). IgA produced by plasma cells in the mammary gland is transported across alveolar epithelial cells (ECs) by the polymeric Ig receptor (pIgR). At the apical surface, proteolytic cleavage releases SIgA, in which IgA is covalently attached to secretory component (SC), the extracellular domain of pIgR. SC provides innate immune functions and protects SIgA from degradation by host and microbial proteases (2). In suckling infants, SIgA antibodies shape the composition of the gut microbiota and promote a mutualistic relationship with the host (3, 4). Immunological benefits of breast milk include prevention of infection, protection against inflammation, and promotion of intestinal barrier function (5). After weaning, IgA produced by plasma cells in the intestinal lamina propria is transported across ECs into the gut lumen by pIgR (6). There is a major gap in our knowledge of whether the beneficial effects of breast milk-derived SIgA persist after weaning. Epidemiological studies suggest that early exposure to breast milk reduces the risk of developing inflammatory bowel disease (IBD) in children (7). However, the fact that breast milk contains many bioactive molecules (8, 9) makes it difficult to ascertain the specific effects of SIgA. We developed a paradigm in mice to compare the benefits of early exposure to maternally-supplied (passive) SIgA vs. lifelong production of endogenous (active) SIgA.
Results
Sources of SIgA in the Intestinal Tract.
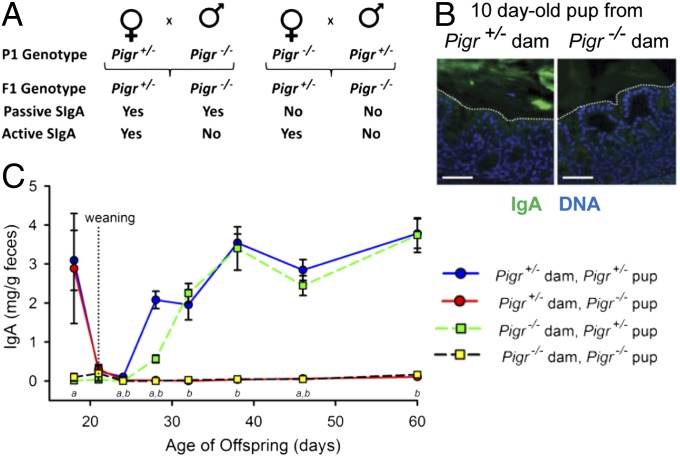
By breeding Pigr−/− dams with Pigr+/− males, and vice versa, we generated offspring that did or did not receive passive SIgA in breast milk (Fig. 1A). Immunohistochemical analysis confirmed that Pigr+/− but not Pigr−/− dams transported SIgA across mammary ECs (SI Appendix, Fig. S1A). Surprisingly, IgA accumulated in the mammary glands and milk of Pigr−/− dams (SI Appendix, Fig. S1 A and B), possibly by paracellular leakage. Although this SC-devoid IgA was found in the stomachs of pups from Pigr−/− dams (SI Appendix, Fig. S1C), it did not survive transit into the colon (Fig. 1B), likely owing to proteolytic degradation in the GI tract (10). By contrast, SIgA was abundant in the colonic lumen of offspring from Pigr+/− dams. In suckling neonates, fecal SIgA was observed only in offspring of Pigr+/− dams and fell to low levels at weaning when passive SIgA from breast milk was no longer available (Fig. 1C). Within 1 wk after weaning, Pigr+/− offspring began to produce SIgA via active intestinal transport. SIgA was not detected in feces of Pigr−/− offspring at any time after weaning, despite the presence of abundant IgA+ plasma cells in the colonic lamina propria of older mice (SI Appendix, Fig. S2), as previously observed in pIgR-knockout mice (11, 12). Thus, intestinal SIgA is derived exclusively from breast milk during the suckling period and by endogenous transport after weaning.
Fig. 1.
Passive and active delivery of SIgA into the gastrointestinal tract. (A) Breeding scheme for generation of mice receiving passive and/or active SIgA. (B) Sections of colons from representative 10-d-old Pigr−/− pups from Pigr+/− and Pigr−/− dams, stained for IgA (green) and nuclei (blue). Dotted lines indicate the boundary between the epithelium and lumen. (Scale bars, 50 μm.) (C) Levels of fecal IgA from Pigr+/− and Pigr−/− offspring of Pigr+/− and Pigr−/− dams (mean ± SEM, n = 9). Two-way ANOVA: a, significant effect of maternal Pigr genotype; b, significant effect of offspring Pigr genotype (P < 0.05).
Effects of Early Exposure to Passive SIgA on the Gut Microbiota.
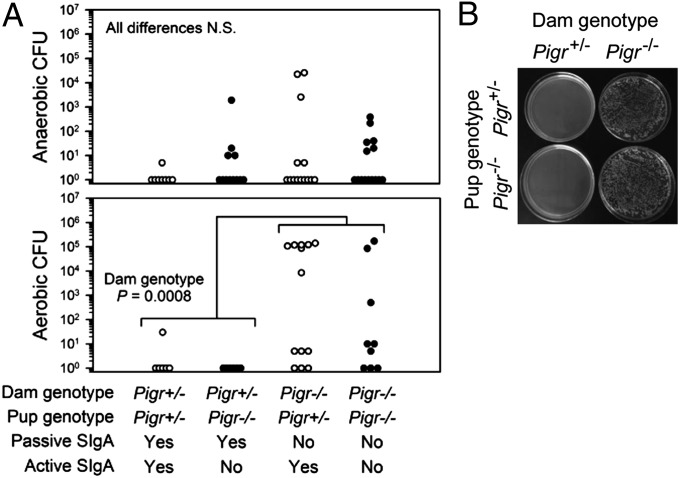
The spatial relationship between the host gut and its resident bacteria was visualized by fluorescence in situ hybridization (SI Appendix, Fig. S3). Colonic bacteria were in close contact with the epithelium of young mice at weaning (21 d), a critical time when cross-talk of host cells with the expanding gut microbiota stimulates development of the epithelial barrier (13). In adult mice, bacteria were spatially segregated from the epithelium, as reported previously (14). To determine whether early exposure to passive SIgA facilitated development of the intestinal barrier, mesenteric lymph nodes (MLNs) were harvested from weanling mice and cultured for the presence of bacteria (Fig. 2). Failure to receive SIgA in breast milk resulted in significantly increased translocation of aerobic bacteria to MLNs (Fig. 2A). Many of the bacteria harvested from MLNs of pups from Pigr−/− dams were slow-growing, obligate aerobes that formed large, mucoid colonies (Fig. 2B). Randomly selected colonies were identified as the alphaproteobacterium Ochrobactrum anthropi by sequencing of the 16S rRNA gene (SI Appendix, Fig. S4A). No differences in fecal levels of O. anthropi were observed between offspring of Pigr+/− and Pigr−/− dams (SI Appendix, Fig. S4B), suggesting that translocation of this bacterium into MLNs in offspring of Pigr−/− dams was due to the absence of SIgA-mediated barrier protection rather than overgrowth of O. anthropi in the gut lumen. We propose that early exposure to passive SIgA in breast milk protects neonates against invasion of potential pathogens across the epithelial barrier into the draining lymphatics.
Fig. 2.
Lack of passive SIgA in breast milk results in translocation of aerobic bacteria to MLNs in nursing mice. (A) MLN homogenates from 21-d-old mice were cultured for 4 d under anaerobic or aerobic conditions for quantification of cfus; each symbol represents an individual mouse. Differences in cfu were analyzed by two-way ANOVA; N.S., not significant. (B) Representative plates showing aerobically grown colonies harvested from MLNs.
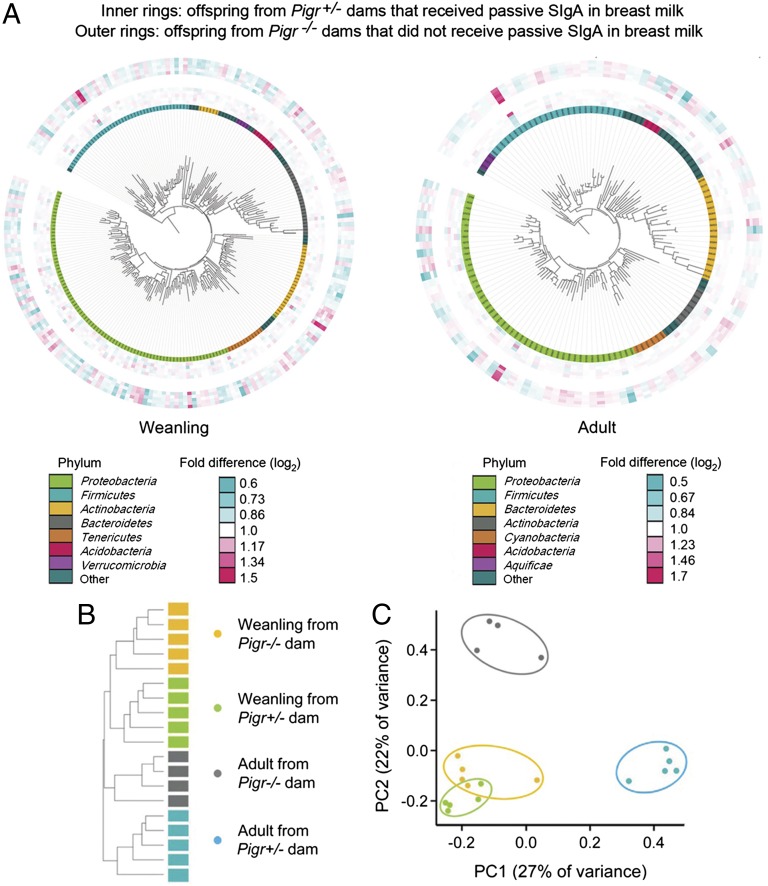
To determine the effects of early exposure to passive SIgA, independent of differences in endogenous (active) SIgA, the composition of the microbiota was assessed in fecal samples from Pigr+/− offspring of Pigr+/− and Pigr−/− dams. Because newborn mice obtain their microbiota from their mothers during parturition (15–17), it was important to control for potential differences in the microbiota of Pigr+/− and Pigr−/− dams. To this end, littermate Pigr+/− and Pigr−/− female offspring of Pigr+/− breeding pairs were cohoused until they were bred with littermate Pigr−/− or Pigr+/− males (which also had been cohoused). Because mice are coprophagic, cohousing results in cross-transfer of the microbiota (17). Because both Pigr+/− and Pigr−/− dams were the offspring of Pigr+/− mothers, dams of both genotypes had been exposed to passive SIgA during suckling. However, we cannot rule out the possibility that differences in the maternal microbiota of Pigr+/− and Pigr−/− dams at the time of parturition and during the suckling period contributed to the phenotypes of their offspring, independent of the presence or absence of passive SIgA in milk. Offspring mice used for experiments were genotyped at weaning and housed in separate cages according to maternal and offspring Pigr genotype, to avoid cross-exposure of the developing microbiota. Fecal samples were collected at 21 d of age (weanling) and from the same mice at 70 d of age (adult). PhyloChip microarray hybridization of fecal DNA was used to identify “operational taxonomic units” (OTUs), defined as a group of bacteria that share >99% sequence similarity in the 16S rRNA gene, roughly equivalent to bacterial species at the phylogenetic level. Whereas overall microbial richness and diversity at high taxonomic ranks were similar among groups (SI Appendix, Fig. S5 A–C), the number of OTUs that changed in abundance from weanling to adult mice was considerably greater in offspring from Pigr−/− dams than in offspring from Pigr+/− dams (SI Appendix, Fig. S5D). Early exposure to passive SIgA in breast milk was found to alter the abundance of 1,047 OTUs (within 177 bacterial families) in weanling mice and 467 OTUs (within 104 families) in adult mice (P < 0.05) (Fig. 3). Gram-negative Proteobacteria of the family Pasteurellaceae and Gram-positive Firmicutes of the family Lachnospiraceae dominated the taxa that were up-regulated in the absence of passive SIgA, and this change persisted into adulthood (SI Appendix, Fig. S6 A–C). In addition, 80 bacterial taxa were identified that were uniquely present in the fecal microbiota of at least 60% of the mice within a given category (maternal Pigr genotype and age at sampling), and absent from all mice in any of the other categories (SI Appendix, Fig. S6 D–F). The majority of unique bacterial taxa that were present only in weanling offspring of Pigr−/− dams were Gram-negative Proteobacteria of the family Comamonadaceae. Hierarchical clustering dendrograms (Fig. 3B) and 2D ordination plots (Fig. 3C), based on Unifrac distance matrices (18), were used to compare the microbial communities of individual mice. Differences in the microbiota of offspring of Pigr+/− and Pigr−/− dams were apparent at weaning, and increased as the mice aged. These findings complement recent findings of differences in the gut microbiota between adult wild-type and Pigr knockout mice, suggesting that both passive and active SIgA shape the composition of the gut microbiota (19).
Fig. 3.
The composition of the intestinal microbiota is shaped by passive SIgA. Fecal samples were collected from Pigr+/− offspring of Pigr+/− and Pigr−/− dams at 21 d of age (weanling) and from the same mice at 70 d of age (adult). (A) Interactive Tree of Life maps showing relative abundance (weighted Unifrac distances) of OTUs as quantified by PhyloChip analysis of the bacterial 16S rRNA gene. OTUs are defined as groups of bacteria that share >99% sequence identity in the 16S rRNA gene. Each of the inner rings represents an individual offspring of Pigr−/− dams, and each of the outer rings represents an individual offspring of Pigr+/− dams. (B) 2D ordination plot generated by principal coordinate analysis of the fecal microbiota of Pigr+/− offspring of Pigr+/− and Pigr−/− dams (sampled at 21 and 70 d of age), based on the presence or absence of 80 individual OTUs (unweighted Unifrac distances) that were uniquely characteristic of each group (i.e., present in the fecal microbiota of 60% of mice in a given group and absent in all mice in every other group). Symbols represent individual mice. (C) Hierarchical clustering of the 80 unique OTUs described in B. Bars represent samples from individual mice. 2D ordination and hierarchical clustering plots based on the abundance of bacterial OTUs (weighted Unifrac distances) are shown in SI Appendix, Fig. S5 E and F.
Effects of Passive and Active SIgA on Gene Expression in Colonic ECs and Responses to Epithelial Damage.
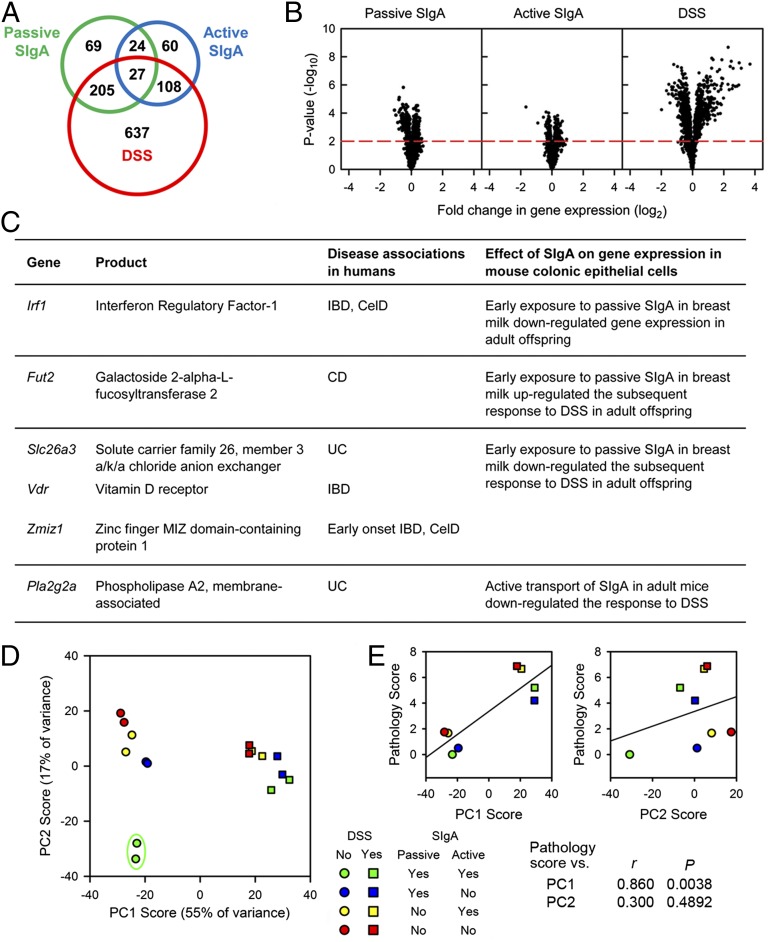
The intestinal epithelium serves as a physical and immunological barrier, producing a broad range of biologically active factors (20). To determine the effects of passive and active SIgA on responses to epithelial damage, adult Pigr+/− and Pigr−/− offspring from Pigr+/− and Pigr−/− dams were given the epithelial-disrupting agent dextran sulfate sodium (DSS) in drinking water for 8 d. Global gene expression was quantified by microarray hybridization of RNA from purified colonic ECs, and genes that were significantly regulated by passive SIgA, active SIgA, DSS, or a combination of factors were identified by three-way ANOVA (P < 0.01) (Fig. 4 and SI Appendix, Fig. S7 and Table S1). Significantly, some of the genes found to be regulated by SIgA in mouse colonic ECs are orthologous to human gene loci that have been associated with inflammatory diseases of the intestine (21–24) (Fig. 4C provides a selected list of highly relevant genes, and SI Appendix, Table S2 gives a complete list). To identify biological pathways that may be regulated by passive and active SIgA and/or DSS, the list of genes identified by microarray analysis was screened for matches in the Reactome pathway database (www.reactome.org) (SI Appendix, Table S3). Graphical representations of pathways that were predicted to be regulated by passive or active SIgA and DSS can be viewed by selecting the corresponding hyperlinks in SI Appendix, Table S3. Selected pathways are illustrated in SI Appendix, Fig. S8. Many of the pathways that seem to be regulated by early exposure to passive SIgA in breast milk involve DNA replication and repair and RNA transcription and splicing, cellular processes that are critical to maintenance of the rapidly dividing epithelial layer. To validate the accuracy of the microarray data, expression levels of 16 selected genes were reanalyzed in RNA from individual mice by Nanostring nCounter hybridization (SI Appendix, Fig. S9). The excellent concordance of the data analyzed by these independent approaches demonstrates that the microarray data based on pooled RNA samples accurately reflect the transcriptional response of mouse ECs. Accordingly, the microarray data for gene expression were used for all subsequent analyses. A multifactorial approach was used to reduce the 1,130 regulated genes identified by microarray analysis to two “supervariables,” or principal components (PCs) (Fig. 4D), which together accounted for 72% of the overall variance in gene expression (details of the analysis are given in SI Appendix, Table S4). Many genes that contributed strongly to PC1 are known to be induced during acute inflammatory responses, including Cxcl2, Nos2, Tnfrsf1b, Pla2g2a, Cd80, Ccl3, and Socs3. By contrast, genes that contributed strongly to PC2 tended to be involved in cell signaling, metabolism, and repair, including Gripap1, Cpt1, Setd1b, Nr1d2, Pex12, Fbxo4, and Cyp27a1. Gene expression in control mice that received both passive and active SIgA, characterized by low scores for both PC1 and PC2, was clearly different from that of mice in all other categories. DSS treatment resulted in increased scores for PC1 in all mice and was significantly correlated with tissue damage (Fig. 4E) (representative histology images are shown in SI Appendix, Fig. S10). However, mice that received passive SIgA had significantly lower pathology scores than did mice that did not receive passive SIgA, falling below the regression line in Fig. 4E (P = 0.0011 by analysis of covariance). In contrast to PC1, scores for PC2 were not correlated with DSS exposure or tissue damage. Both passive and active SIgA were required for maintenance of the low PC2 score characteristic of control mice, suggesting that absence of either source of SIgA resulted in increased expression of genes associated with epithelial metabolism and repair. These findings are consistent with a role for SIgA in prevention of cellular stress in the harsh environment of the intestinal tract. In this context it is important to note that there were significant differences in the gut microbiota of adult mice resulting from early exposure to passive SIgA in breast milk during the suckling period (Fig. 3). Thus, the mechanisms for amelioration of intestinal inflammation could involve direct effects of SIgA on intestinal ECs, as well as indirect effects of SIgA on shaping the composition of the gut microbiota. In summary, both early exposure to passive SIgA in breast milk and the continuous presence of endogenous, active SIgA affected the pattern of gene expression in the intestinal epithelium of adult animals.
Fig. 4.
Gene expression in colonic ECs and responses to acute inflammation are shaped by both passive and active sources of SIgA. Seventy-day-old Pigr+/− and Pigr−/− offspring of Pigr+/− and Pigr−/− dams were left untreated or given 2% DSS in drinking water for 8 d. Global gene expression was quantified by microarray analysis of RNA from colonic ECs. RNA was collected from six mice in each treatment category, then was pooled into two samples from three mice per sample. The statistical analyses of the microarray data are described in detail in SI Appendix, Fig. S7. (A) Total number of genes significantly altered by each effect or combination (P < 0.01). (B) Relative changes in expression levels for the 1,130 differentially regulated genes described in A. (Left) Specific effects of early exposure to passive SIgA in breast milk were calculated by comparing gene expression in Pigr+/− offspring of Pigr+/− vs. Pigr−/− dams in the absence of DSS. (Center) Specific effects of endogenous, actively transported SIgA were calculated by comparing gene expression in Pigr+/− vs. Pigr−/− offspring of Pigr+/− dams in the absence of DSS. (Right) Specific effects of DSS were calculated by comparing gene expression in Pigr+/− offspring of Pigr+/− dams in the presence or absence of DSS. Dots above the red dashed line represent genes that were significantly altered by the indicated factor (P < 0.01). (C) Selected genes regulated by SIgA in mouse colonic ECs that are orthologous to human gene loci associated with CD, UC, both CD and UC (IBD), early-onset IBD, and CelD. SI Appendix, Table S2 provides a complete list of disease-associated genes that were regulated by passive SIgA, active SIgA, and DSS. (D) Multifactorial PCA was conducted using expression levels of the 1,130 differentially regulated genes described in A. Each sample represents pooled RNA from three mice, for a total of six mice per treatment group. Data for mRNA transcript levels from each sample were reduced to two PCs (PC1 and PC2). Scores were calculated as described in SI Appendix, Table S4. The points within the green oval represent the unique pattern of gene expression in mice that received both passive and active SIgA. (E) Colon pathology scores were determined for individual mice (six mice per group), and the average for each group was plotted against the average PC1 and PC2 scores from D.
Discussion
Maintenance of intestinal homeostasis requires a delicate balance between the 100 trillion resident bacteria in our intestinal tract and the host immune system. During vaginal delivery the newborn mammal leaves the sterile environment of the womb and receives its first microbial exposure from the vaginal and fecal microbiota of the mother (15–17). Unable to produce its own SIgA at this age (Fig. 1C), the newborn relies on passive SIgA in breast milk to shape the composition of its developing gut microbiota and protect against infection with pathogenic microbes (1, 3, 4, 25, 26). Signals received from the microbiota and regulated by SIgA drive the maturation of intestinal ECs and development of barrier function (27). This epithelial barrier is critically important at the time of weaning, when intestinal SIgA concentrations are at their lowest point owing to loss of passive maternal SIgA and immaturity of the weanling’s intestinal immune system (Fig. 1C). We found that epithelial barrier function was compromised in weanling mice that had not received passive SIgA in breast milk, allowing colonization of draining lymph nodes with large numbers of aerobic bacteria, including the opportunistic pathogen O. anthropi (Fig. 2). O. anthropi, which is abundant in environmental sources such as water and soil, has been identified as an opportunistic pathogen in preterm infants and immunocompromised individuals (28–30). By promoting development of the epithelial barrier in neonatal mice, passive SIgA may also prevent sensitization of the immature systemic immune system with microbial antigens, which has been associated with development of allergic and inflammatory diseases (31).
Although the benefits of breastfeeding for newborns are well documented, further evidence is required to establish a firm link between breastfeeding and maintenance of intestinal homeostasis during the transition from weaning through childhood and into adulthood. Our present findings are consistent with the hypothesis that alterations in the gut microbiota and intestinal epithelial gene expression associated with failure to receive SIgA in breast milk may increase the susceptibility to intestinal inflammation later in life. Importantly, a review of 79 epidemiological studies suggested that breastfeeding of human infants may reduce the risk of developing pediatric IBD (7). We observed certain parallels between the altered microbiota of mice that did not receive passive SIgA in breast milk and the microbiota of IBD patients. Our finding of increased abundance of bacterial taxa in the families Pasteurellaceae (phylum Proteobacteria) and Lachnospiraceae (phylum Firmicutes) in weanling mice deprived of passive SIgA is not unlike the increase in taxa from these families observed in the gut microbiota of pediatric IBD patients (32). Our finding of a striking increase of unique bacterial taxa in the family Comamonadaceae (phylum Proteobacteria) in weanling mice deprived of passive SIgA may be significant in light of the report documenting increased numbers of Comamonadaceae in the microbiota of IBD patients with chronic pouchitis (33). Expansion of Proteobacteria has consistently been reported in the gut microbiota of adult IBD patients (34), suggesting that dysbiosis that develops early in life may persist throughout life.
We found further parallels with human IBD in the altered patterns of intestinal epithelial gene expression in adult mice that had not received passive SIgA during the neonatal period. The expression of 69 genes was altered in colonic ECs owing to the lack of early exposure to passive SIgA in breast milk, and expression of an additional 256 genes was altered by combined effects of passive SIgA with active SIgA and/or DSS (Fig. 4 A and B). Multifactorial gene analysis suggested that both passive and active SIgA impacted the complex balance of gene expression (Fig. 4D). Importantly, a number of genes regulated by SIgA are homologous to human gene loci associated with increased risk for IBD (Fig. 4C and SI Appendix, Table S2). Recently, an IBD-associated polymorphism was identified in a human gene locus on chromosome 1 that includes the PIGR gene. Down-regulation of PIGR gene expression in mothers and their children could exacerbate IBD by reducing the concentration of passive SIgA in breast milk received early in life, and reducing active transport of SIgA in the intestine throughout life. Taken together, these findings suggest that early exposure to passive SIgA in breast milk, as well as active transport of endogenous SIgA, regulate gene expression in intestinal ECs throughout life.
In conclusion, our findings reveal mechanisms through which SIgA in breast milk may promote development and maintenance of intestinal homeostasis, providing additional support for the benefits of breastfeeding. We further propose that oral administration of purified SIgA could be investigated as a biological therapy for intestinal infections and inflammation, particularly in formula-fed infants or in older children and adults in whom active transport of endogenous SIgA is deficient.
Materials and Methods
Mice.
Mice with a targeted mutation in the Pigr gene (11), back-crossed onto the C57BL/6 strain for eight generations, were obtained from the Mutant Mouse Regional Resource Center at the University of Missouri, Columbia, MO (strain B6.129P2-Pigrtm1Fejo/Mmmh). Littermate mice for experiments were generated by breeding Pigr+/− and Pigr−/− mice as described in Fig. 1A. Mice were housed in microisolator cages in an American Association for the Accreditation of Laboratory Animal Care-accredited facility, and procedures were conducted in compliance with the University of Kentucky Institutional Animal Care and Use Committee. Mice were genotyped for Pigr alleles according to the supplier’s protocol.
Tissue Histology and Immunofluorescence Microscopy.
Mouse tissues were fixed in buffered formalin, embedded in paraffin, and sectioned. Serial sections were stained with hematoxylin and eosin or by immunofluorescence for pIgR/SC and IgA as previously described (35).
Quantification of Fecal IgA.
Freshly collected feces were weighed, homogenized in ELISA buffer [50 mM Tris (pH 7.4), 0.14 M NaCl, 1% BSA, and 0.05% Tween 20] and stored at −20 °C until analysis. IgA was quantified by ELISA as previously described (36).
Quantification of Bacteria in MLNs.
MLNs were homogenized in PBS, plated on Schaedler agar (ThermoFisher Scientific), and cultured under aerobic and anaerobic conditions at 37 °C for 96 h for enumeration of cfus.
Analysis of the Fecal Microbiota by PhyloChip Hybridization.
Paired fecal samples were collected from littermate Pigr+/− offspring of Pigr+/− and Pigr−/− dams at the ages of 21 d (weanlings) and 70 d (adults). Fecal DNA was extracted with the QIAamp DNA stool kit (Qiagen) and analyzed by Second Genome, Inc. (www.secondgenome.com) for microbial richness and diversity by the PhyloChip microarray method (37). Bacterial 16S rRNA genes were amplified by PCR using the degenerate forward primer 5′-AGRGTTTGATCMTGGCTCAG-3′ and the nondegenerate reverse primer 5′-GGTTACCTTGTTACGACTT-3′, fragmented, biotin labeled, and hybridized to the PhyloChip Array, version G3. Stained arrays were scanned with a GeneArray scanner (Affymetrix, Inc.) and analyzed by Affymetrix software (GeneChip Microarray Analysis Suite). Profiles were compared in a pair-wise fashion to determine a binary UniFrac distance metric, which uses the phylogenetic distance between OTUs to determine the dissimilarity between communities (18). For weighted UniFrac, both the abundance and dissimilarities among OTUs were considered. Phylogenetic trees based on bacterial abundance (Fig. 3A) were generated using the “Interactive Tree of Life” software tool (38). A representative 16S rRNA gene from each of 231 differentially expressed OTUs for weanling mice and 132 differentially expressed OTUs for adult mice was aligned to infer the phylogenetic trees. The rings around the tree constitute a heatmap in which the inner rings represent five offspring of Pigr−/− dams and the outer rings represent five offspring of Pigr+/− dams. Heatmap intensities indicate the fold difference in sample signal for the given OTU compared with the mean abundance signal for offspring from Pigr−/− dams (inner rings) for the same OTU. Hierarchical clustering (average linkage) of the fecal microbiota (Fig. 3B) and principal coordinate analysis (Fig. 3C) were based on the unweighted Unifrac distance between samples, given the presence or absence of 80 OTUs present in at least 60% of samples from one category and absent in all of the other categories (age or maternal Pigr genotype).
Induction of Experimental Colitis.
Acute colitis was induced in 8- to 10-wk-old adult mice by oral administration of 2% (wt/vol) DSS (molecular mass 36–50 kDa; MP Biomedicals) in the drinking water for 8 d. Mice were distributed into treatment groups (six mice per group) in a 2 × 2 × 2 factorial design, with the main factors comprising maternal genotype (Pigr+/− or Pigr−/−), offspring genotype (Pigr+/− or Pigr−/−), and DSS (oral DSS or plain drinking water). Littermate mice of both genders were distributed evenly into the treatment groups, and all mice were identified by litter of origin to facilitate subsequent statistical analysis of potential maternal effects (other than Pigr genotype, which was controlled). After treatment, colons were harvested for histological analysis and isolation of ECs. Colon pathology was assessed in a blinded fashion as previously described (35), and a score on a scale of 0–16 (the sum of scores from four sites along the colon) was assigned for each mouse.
Isolation of Colonic ECs and Analysis of Gene Expression.
ECs were isolated from freshly dissected colons by dissociation with DTT and EDTA and separation on a Percoll gradient, as previously described (39). Using this protocol, we typically achieve 99% purity of ECs, with contaminating CD45+ hematopoietic cells at levels of 0.4–1.3% (35). Detailed methods for purification of RNA and analysis of gene expression by hybridization to GeneChip Mouse Gene 1.0 ST arrays (Affymetrix) are described in the SI Appendix, Supplementary Methods.
Identification of Genes Regulated by SIgA in Mouse Colonic ECs That Are Orthologous to Human Disease-Associated Gene Loci.
Orthologous mouse genes were selected by comparing the list of regulated genes from the microarray analysis (SI Appendix, Table S1) with databases of human gene loci identified by genome-wide association studies of human single-nucleotide polymorphisms to be associated with Crohn’s disease (CD) (24), ulcerative colitis (UC) (22, 24), both CD and UC (IBD), early-onset IBD (21), and celiac disease (CelD) (23). A complete list of mouse genes identified in our study that are orthologous to human disease-associated gene loci is shown in SI Appendix, Table S2, and a selected list of highly relevant genes is shown in Fig. 4C.
Statistical Analyses.
Statistical differences in IgA concentrations, cfu, and gene expression were determined by multiple analysis of variance and Fisher’s protected least significant difference test. For calculation of fold differences among OTUs in the phylogenetic trees (Fig. 3A), a Welch t test was performed across the two genotypes using abundance metrics. Principal coordinate analysis was used to position points on 2D ordination plots according to dissimilarity values (Fig. 3C). PC analysis (PCA) was used for multifactorial clustering of gene expression data, as described in SI Appendix, Table S4.
Supplementary Material
Acknowledgments
We thank Dr. Beth Garvy, University of Kentucky; Dr. Subbarao Bondada, University of Kentucky; and Dr. Brad Pearce, Emory University, for their helpful comments. This work was supported by National Institutes of Health (NIH) Grant AI069027 (and an associated American Recovery and Reinvestment Act supplement); a Senior Research Award from the Crohn’s and Colitis Foundation of America (CCFA), a grant from the Kentucky Bioinformatics Research Infrastructure Network (to C.S.K.); a Senior Research Award from the CCFA (to D.A.C.); and NIH Grants NCATS UL1TR000117, NCRR 5P20RR016481-12, and NIGMS 8 P20 GM103436-12 (to A.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.I.M. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54371).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315792111/-/DCSupplemental.
References
- 1.Brandtzaeg P. The mucosal immune system and its integration with the mammary glands. J Pediatr. 2010;156(2) Suppl:S8–S15. doi: 10.1016/j.jpeds.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4(6):603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev. 2012;245(1):132–146. doi: 10.1111/j.1600-065X.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- 4.Janoff EN, Gustafson C, Frank DN. The world within: Living with our microbial guests and guides. Transl Res. 2012;160(4):239–245. doi: 10.1016/j.trsl.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker A. Breast milk as the gold standard for protective nutrients. J Pediatr. 2010;156(2) Suppl:S3–S7. doi: 10.1016/j.jpeds.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 6.Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: New advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4(6):598–602. doi: 10.1038/mi.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barclay AR, et al. Systematic review: The role of breastfeeding in the development of pediatric inflammatory bowel disease. J Pediatr. 2009;155(3):421–426. doi: 10.1016/j.jpeds.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 8.Lönnerdal B. Bioactive proteins in human milk: Mechanisms of action. J Pediatr. 2010;156(2) Suppl:S26–S30. doi: 10.1016/j.jpeds.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Hettinga K, et al. The host defense proteome of human and bovine milk. PLoS One. 2011;6(4):e19433. doi: 10.1371/journal.pone.0019433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chintalacharuvu KR, Morrison SL. Production of secretory immunoglobulin A by a single mammalian cell. Proc Natl Acad Sci USA. 1997;94(12):6364–6368. doi: 10.1073/pnas.94.12.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansen FE, et al. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J Exp Med. 1999;190(7):915–922. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimada S, et al. Generation of polymeric immunoglobulin receptor-deficient mouse with marked reduction of secretory IgA. J Immunol. 1999;163(10):5367–5373. [PubMed] [Google Scholar]
- 13.Moens E, Veldhoen M. Epithelial barrier biology: Good fences make good neighbours. Immunology. 2012;135(1):1–8. doi: 10.1111/j.1365-2567.2011.03506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen CH, et al. Patterns of early gut colonization shape future immune responses of the host. PLoS ONE. 2012;7(3):e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reikvam DH, et al. Epithelial-microbial crosstalk in polymeric Ig receptor deficient mice. Eur J Immunol. 2012;42(11):2959–2970. doi: 10.1002/eji.201242543. [DOI] [PubMed] [Google Scholar]
- 20.Carvalho FA, Aitken JD, Vijay-Kumar M, Gewirtz AT. Toll-like receptor-gut microbiota interactions: Perturb at your own risk! Annu Rev Physiol. 2012;74:177–198. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 21.Imielinski M, et al. Western Regional Alliance for Pediatric IBD International IBD Genetics Consortium NIDDK IBD Genetics Consortium Belgian-French IBD Consortium Wellcome Trust Case Control Consortium Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41(12):1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trynka G, et al. Spanish Consortium on the Genetics of Coeliac Disease (CEGEC) PreventCD Study Group Wellcome Trust Case Control Consortium (WTCCC) Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43(12):1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jostins L, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman L, Moss RL. Necrotizing enterocolitis: an update. Semin Fetal Neonatal Med. 2011;16(3):145–150. doi: 10.1016/j.siny.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Camacho-Gonzalez A, Spearman PW, Stoll BJ. Neonatal infectious diseases: evaluation of neonatal sepsis. Pediatr Clin North Am. 2013;60(2):367–389. doi: 10.1016/j.pcl.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandtzaeg P. Gate-keeper function of the intestinal epithelium. Benef Microbes. 2013;4(1):67–82. doi: 10.3920/BM2012.0024. [DOI] [PubMed] [Google Scholar]
- 28.Stiakaki E, et al. Ochrobactrum anthropi bacteremia in pediatric oncology patients. Pediatr Infect Dis J. 2002;21(1):72–74. doi: 10.1097/00006454-200201000-00018. [DOI] [PubMed] [Google Scholar]
- 29.Duran R, Vatansever U, Acunaş B, Başaran UN. Ochrobactrum anthropi bacteremia in a preterm infant with meconium peritonitis. Int J Infect Dis. 2009;13(2):e61–e63. doi: 10.1016/j.ijid.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 30.Naik C, Kulkarni H, Darabi A, Bhanot N. Ochrobactrum anthropi: A rare cause of pneumonia. J Infect Chemother. 2013;19(1):162–165. doi: 10.1007/s10156-012-0436-1. [DOI] [PubMed] [Google Scholar]
- 31.Brandtzaeg P. The gut as communicator between environment and host: Immunological consequences. Eur J Pharmacol. 2011;668(Suppl 1):S16–S32. doi: 10.1016/j.ejphar.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Saulnier DM, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannock GW, et al. Comprehensive analysis of the bacterial content of stool from patients with chronic pouchitis, normal pouches, or familial adenomatous polyposis pouches. Inflamm Bowel Dis. 2012;18(5):925–934. doi: 10.1002/ibd.21936. [DOI] [PubMed] [Google Scholar]
- 34.Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9(4):219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- 35.Frantz AL, et al. Multifactorial patterns of gene expression in colonic epithelial cells predict disease phenotypes in experimental colitis. Inflamm Bowel Dis. 2012;18(11):2138–2148. doi: 10.1002/ibd.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frantz AL, et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5(5):501–512. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loy A, Pester M, Steger D. Phylogenetic microarrays for cultivation-independent identification and metabolic characterization of microorganisms in complex samples. Methods Mol Biol. 2011;688:187–206. doi: 10.1007/978-1-60761-947-5_13. [DOI] [PubMed] [Google Scholar]
- 38.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 39.Weigmann B, et al. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc. 2007;2(10):2307–2311. doi: 10.1038/nprot.2007.315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.