Significance
High-mobility group box (HMGB)1 is a nuclear protein that we have identified as a proinflammatory mediator during infection or sterile tissue injury, which importantly orchestrates the innate immune responses. The mechanisms of HMGB1 release require translocation of HMGB1 from nucleus to cytoplasm and release into the extracellular space. We recently reported that the inflammasome and PKR mediates HMGB1 release from the cytoplasm, but the mechanism of HMGB1 translocation from nucleus to cytoplasm was previously unknown. Here, we describe our discovery that JAK/STAT1 is required for LPS- or interferon-induced HMGB1 nuclear translocation. These findings have significant implications for the field, and for designing therapeutics for potential use in inflammatory diseases.
Keywords: damage-associated molecular pattern, cytokine, pathogen-associated molecular pattern, innate immunity, therapy
Abstract
Extracellular high-mobility group box (HMGB)1 mediates inflammation during sterile and infectious injury and contributes importantly to disease pathogenesis. The first critical step in the release of HMGB1 from activated immune cells is mobilization from the nucleus to the cytoplasm, a process dependent upon hyperacetylation within two HMGB1 nuclear localization sequence (NLS) sites. The inflammasomes mediate the release of cytoplasmic HMGB1 in activated immune cells, but the mechanism of HMGB1 translocation from nucleus to cytoplasm was previously unknown. Here, we show that pharmacological inhibition of JAK/STAT1 inhibits LPS-induced HMGB1 nuclear translocation. Conversely, activation of JAK/STAT1 by type 1 interferon (IFN) stimulation induces HMGB1 translocation from nucleus to cytoplasm. Mass spectrometric analysis unequivocally revealed that pharmacological inhibition of the JAK/STAT1 pathway or genetic deletion of STAT1 abrogated LPS- or type 1 IFN-induced HMGB1 acetylation within the NLS sites. Together, these results identify a critical role of the JAK/STAT1 pathway in mediating HMGB1 cytoplasmic accumulation for subsequent release, suggesting that the JAK/STAT1 pathway is a potential drug target for inhibiting HMGB1 release.
High-mobility group box 1 (HMGB1), a ubiquitous DNA-binding protein, is a promiscuous sensor driving nucleic acid-mediated immune responses and a pathogenic inflammatory mediator in sepsis, arthritis, colitis, and other disease syndromes (1–5). Immune cells actively release HMGB1 after activation by exposure to pathogen-associated molecular patterns or damage-associated molecular patterns, including lipopolysaccharide (LPS) and inflammasome agonists (1, 6, 7). High levels of extracellular HMGB1 accumulate in patients with infectious and sterile inflammatory diseases. Extracellular disulfide HMGB1 stimulates macrophages to release TNF and other inflammatory mediators by binding and signaling through Toll-like receptor (TLR)4. Reduced HMGB1 facilitates immune cell migration by interacting with the receptor for advanced glycation end products (RAGE) and CXCL12 (8–12), a process regulated by posttranslational redox-dependent mechanisms. Administration of neutralizing anti-HMGB1 mAbs or other HMGB1 antagonists significantly reduces the severity of inflammatory disease, promotes bacterial clearance during Pseudomonas aeruginosa or Salmonella typhimurium infection and attenuates memory impairment in sepsis survivors (1, 13–15). Together, these and other findings indicate the importance of a mechanistic understanding of HMGB1 release from activated immune cells and the regulatory signaling pathways controlling these processes.
Most cytokines harbor a leader peptide that facilitates secretion through the endoplasmic reticulum–Golgi exocytotic route. HMGB1, which lacks a leader peptide, is released via unconventional protein secretion pathways (1, 6, 7). In quiescent cells, most HMGB1 is localized in the nucleus. Upon activation of immune cells, efficient HMGB1 release requires acetylation of HMGB1 within the two nuclear localization sequence (NLS) sites and subsequent HMGB1 accumulation in the cytoplasm (1, 6, 16–20). HMGB1 release is mediated by inflammasome activation during pyroptosis, a form of proinflammatory programmed cell death (6, 7, 22–24). Protein kinase (PK)R is a critical regulator of inflammasome-dependent HMGB1 release (6, 25). Pharmacological inhibition of PKR abrogates LPS-induced HMGB1 release by macrophages but does not prevent nuclear translocation of HMGB1 to cytoplasm. This suggests that some other, as yet unknown, inflammasome-independent pathway regulates HMGB1 translocation from nucleus to cytoplasm.
We and others have previously established an important role of type 1 and type 2 interferons (IFNs) and downstream JAK/STAT1 signaling activation in mediating HMGB1 release (26–28). Pharmacological inhibition of JAK/STAT, genetic deletion of STAT1, or inhibition of extracellular IFN-β with neutralizing antibodies significantly abrogates LPS-induced HMGB1 release from macrophages (26–28). Importantly, pharmacological inhibition of the JAK/STAT1 pathway, genetic deletion of STAT1, or inhibition of IFN-β expression by genetic deletion of IRF3 significantly promotes survival in both lethal endotoxemia and experimental sepsis (28–30). Accordingly, we reasoned here that JAK/STAT1 may represent a critical signaling mechanism controlling HMGB1 translocation from nucleus to cytoplasm.
Results
Pharmacological Inhibition of the JAK/STAT Pathway Blocks LPS-Induced HMGB1 Nuclear Translocation and Release.
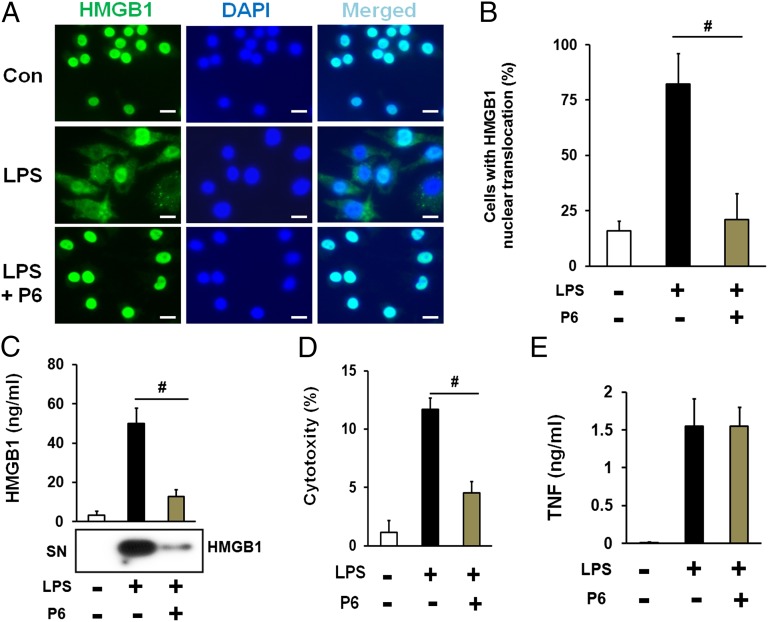
To investigate the role of the JAK/STAT pathway in LPS-induced HMGB1 nuclear translocation, mouse macrophage-like RAW 267.4 cells were stimulated by addition of LPS in the presence of the JAK/STAT inhibitor pyridone 6. HMGB1 cytoplasmic accumulation was assessed by immunostaining and HMGB1 levels in the cell culture medium assessed by Western blot. As expected, LPS induced robust HMGB1 cytoplasmic accumulation. Inhibition of JAK/STAT significantly blocked LPS-induced HMGB1 translocation to cytoplasm (Fig. 1 A and B and SI Appendix, Fig. S1) and HMGB1 release (Fig. 1C), which is correlated with LDH levels in the cell culture medium (Fig. 1D). Inhibition of JAK/STAT failed to decrease LPS-induced secretion of TNF, IL-10, RANTES, IL-12p70, and MCP-1 (Fig. 1E and SI Appendix, Figs. S2 and S3). Next, we sought to determine whether JAK/STAT signaling specifically regulates LPS-induced HMGB1 cytoplasmic accumulation. RAW 267.4 cells were stimulated by rapamycin, which induces HMGB1 cytoplasmic accumulation but does not activate JAK/STAT (31). Notably, pyridone 6 failed to inhibit rapamycin-induced HMGB1 cytoplasmic accumulation (SI Appendix, Fig. S4). Cytoplasmic accumulation of HMGB1 promotes autophagy (31, 32). Pyridone 6 significantly inhibited LPS-induced, but not rapamycin-induced, autophagy in RAW 267.4 cells (SI Appendix, Figs. S5 and S6). Together, these results indicate that JAK/STAT specifically mediates LPS-induced HMGB1 cytoplasmic accumulation.
Fig. 1.
Pharmacological inhibition of the JAK/STAT pathway blocks LPS-induced HMGB1 nuclear translocation and release. (A–D) Mouse macrophage-like RAW 267.4 cells were stimulated with LPS in the absence or the present of pyridone 6 for 16h. The localization of cellular HMGB1 was measured by fluorescent immunostaining analysis. (Scale bars, 20 µm.) Shown in A are representative images of HMGB1 immunostaining, and shown in B are means ± SEM of two independent experiments. The levels of HMGB1 in the culture medium were measured by Western blot analysis (C). Cytotoxicity was determined by LDH assay (D). (E) Mouse macrophages were stimulated with LPS in the absence or the present of pyridone 6 for 6 h. The levels of TNF in the culture medium were assessed by ELISA. Data shown are means ± SEM of two independent experiments. #P < 0.05.
Activation of the JAK/STAT Pathway by Type 1 IFN Induces HMGB1 Cytoplasmic Accumulation and Release.
IFN-β stimulates HMGB1 release, which is dependent, in part, on JAK/STAT (26, 27). To establish the role of the JAK/STAT signaling for HMGB1 cytoplasmic accumulation, RAW 267.4 cells were stimulated with IFN-β in the presence or absence of the JAK/STAT pathway inhibitor pyridone 6. As revealed by fluorescent immunochemical staining, IFN-β stimulation induced cytoplasmic accumulation of HMGB1 (SI Appendix, Figs. S7 and S8); IFN-β and IFN-α stimulation induced HMGB1 release in a time- and dose-dependent manner (Fig. 2 A and B). Pyridone 6 dose-dependently inhibited IFN-β–induced HMGB1 cytoplasmic accumulation, HMGB1 secretion, and LDH release (Fig. 2C and SI Appendix, Figs. S7–S9). This response is cell type-specific, because IFN-β failed to increase HMGB1 release in mouse embryonic fibroblasts (SI Appendix, Fig. S10). Together, this indicates that JAK/STAT is required for IFN-β–induced HMGB1 cytoplasmic accumulation and release in RAW 267.4 cells. We and others recently revealed that secretion of HMGB1 from cytoplasm into the extracellular space requires activation of the inflammasome and PKR (6). Accordingly, we next hypothesized that JAK/STAT regulation of HMGB1 nuclear translocation preceded inflammasome- and PKR-dependent secretion. To test this possibility, we assessed HMGB1 nucleus to cytoplasm translocation in the presence of PKR inhibitors; 2-aminopurine (2-AP) failed to block IFN-β–induced HMGB1 nucleus to cytoplasm translocation (SI Appendix, Fig. S8), even though 2-AP dose-dependently inhibited IFN-β–induced HMGB1 release (Fig. 2D). Thus, JAK/STAT signals mediate mobilization of HMGB1 from the nucleus to the cytoplasm, and PKR-dependent inflammasome activation mediates secretion.
Fig. 2.
Activation of the JAK/STAT pathway by type 1 IFN induces HMGB1 release. (A and B) RAW 267.4 cells were stimulated with IFN-α and IFN-β at various concentrations (A) for the indicated time (B). The levels of HMGB1 in the culture medium were measured by Western blot analysis. (C and D) RAW 267.4 cells were stimulated with IFN-β (100 U/mL) in the absence or the present of pyridone 6 (C) or 2-AP (D) at various concentrations for 16 h. The levels of HMGB1 in the culture medium were measured by Western blot analysis.
JAK/STAT1 Is Required for LPS- or IFN-β–Induced HMGB1 Acetylation Within NLS Sites.
HMGB1 acetylation at two NLS sites is required for the nuclear translocation and cytoplasmic accumulation of HMGB1 (17). LPS stimulation rapidly induces hyperacetylation of HMGB1 (17). To confirm this phenomenon, monocytic THP-1 cells were stimulated with LPS for 3h. High-resolution liquid chromatography–tandem mass spectrometric analysis (LC-MS/MS) revealed that HMGB1 was constitutively acetylated at the lysine residues 111, 113, 171, 172, and 176. Following LPS stimulation, significant acetylation was observed at the lysine residues 2, 6, 7, 11, 27, 28, 29, 42, 43, 179, 181, 183, and 184. Notably, lysine residues 28, 29, 42, and 43 are located within the first NLS of HMGB1, whereas lysine residues 179, 181, 183, and 184 are located within the second NLS of HMGB1 (SI Appendix, Figs. S11–S13). Thus, LPS stimulation induces lysine acetylation within both HMGB1 NLS sites.
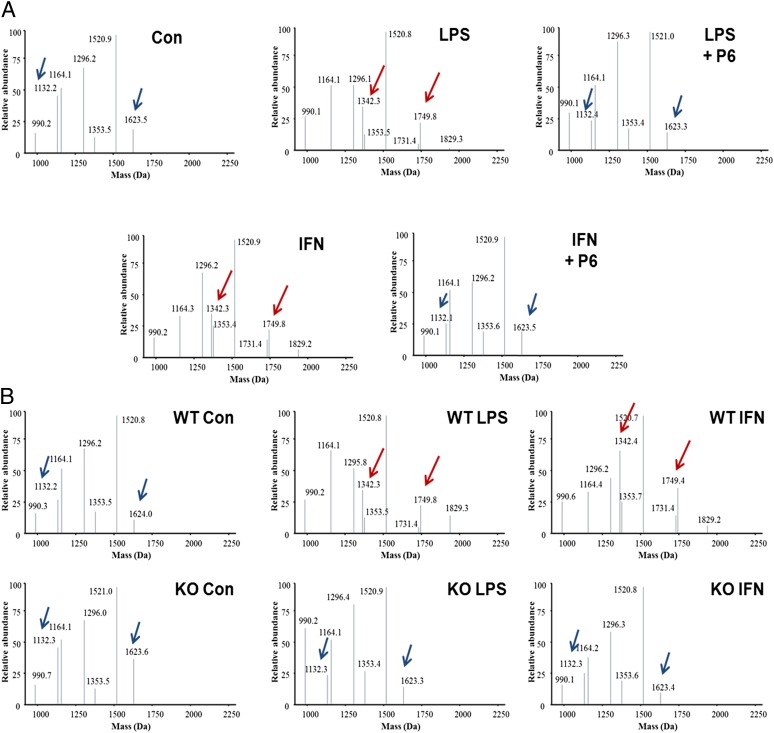
To elucidate the mechanism by which JAK/STAT regulates HMGB1 nucleus to cytoplasm translocation, we next determined whether JAK/STAT is required for LPS- or IFN-β–induced HMGB1 acetylation within NLS sites. We observed the appearance of peptides with molecular masses of 1,749.8 and 1,342.3 Da, corresponding to hyperacetylated NLS1 and NLS2, respectively (Fig. 3A). These peptides were absent from unstimulated macrophages, which contained only peptides of molecular masses 1,623.4 and 1,132.4 Da corresponding to hypoacetylated NLS1 and NLS2, respectively (Fig. 3A). The increased mass shifts of 126 Da for NLS1 and 210 Da for NLS2 in stimulated macrophage samples represent the presence of either 3× acetyl or 5× acetyl modifications on NLS1 and NLS2. Addition of the JAK/STAT inhibitor, pyridone 6, significantly inhibited LPS- and IFN-β–induced HMGB1 acetylation at the NLSs (Fig. 3A). Genetic deletion of STAT1 abrogates LPS- and IFN-induced HMGB1 acetylation within the NLS sites (Fig. 3B). Together, these observations establish that JAK/STAT1 is required for LPS- and IFN-β–induced HMGB1 acetylation within both NLS sites.
Fig. 3.
JAK/STAT1 is required for LPS- or IFN-β–induced HMGB1 acetylation within NLS sites. (A) Mouse peritoneal macrophages were stimulated with indicated stimuli in the absence or the presence of pyridone 6 for 6 h. (B) Mouse peritoneal macrophages isolated from WT or STAT1 KO mice were stimulated with indicated stimuli for 6 h. The acetylation of intracellular HMGB1 protein within the NLS sites was assessed using LC-MS/MS. Shown are representative MS traces.
JAK/STAT1 Is Dispensable for LPS- or IFN-β–Induced HMGB1 Disulfide Bond Formation Between Cys23 and Cys45.
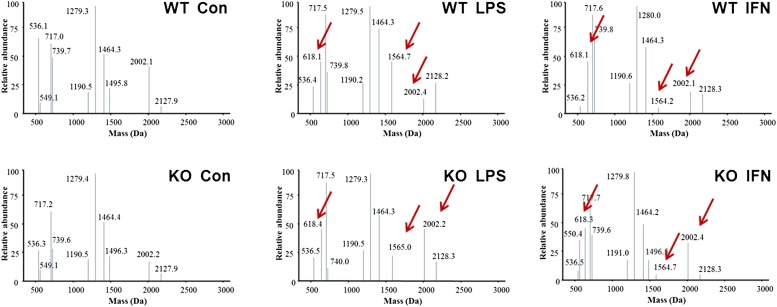
HMGB1 contains three redox-sensitive cysteine residues (Cys): Cys23, Cys45, and Cys106. The redox status of these cysteines determines HMGB1 biological activity (8, 11). In this context, fully reduced HMGB1 forms a complex with CXCL12 and promotes migration of immune cells via CXCR4; partially oxidized HMGB1 with a thiol form of Cys106 and a disulfide bond between Cys23 and Cys45 triggers inflammatory responses via TLR4, whereas cysteine all-oxidized HMGB1 has neither cytokine nor chemotactic activity (8, 11). In resting macrophages, the intracellular HMGB1 pool was dominated by the fully reduced form of HMGB1. However, stimulation of macrophages with LPS markedly induces the formation of HMGB1 bearing disulfide bonds between Cys23 and Cys45 (6). Thus, it remained theoretically possible that JAK/STAT1 signaling is required for LPS-induced disulfide bond formation between Cys23 and Cys45. To test this hypothesis, we stimulated wild-type (WT) or STAT1 knockout (KO) mouse macrophages with LPS or IFN-β in the absence or the presence of JAK/STAT inhibitors. Intracellular HMGB1 was enzymatically digested into small peptides and subjected to LC-MS/MS analysis. Both LPS and IFN-β stimulation induced HMGB1 disulfide bond formation between Cys23 and Cys45, as indicated by the presence of peptide with masses of Cys23 (1,564.7 Da) and Cys45 (618.1 Da) after LPS or IFN-β stimulation (Fig. 4). Cys106 of HMGB1 was present in a thiol form in both stimulated and nonstimulated cells, as indicated by the presence of peptide with masses of Cys106 (2,002.1 Da) (Fig. 4). Analysis of b and y ions generated from these peptides confirmed the amino acid sequence of each peptide, the disulfide bond between Cys23 and Cys45, and the thiol form of Cys106. Notably, LPS or IFN-β–induced HMGB1 disulfide bond formation between Cys23 and Cys45 was not inhibited by pyridone 6, as revealed by LC-MS/MS analysis that the presence of peptide with masses of Cys23 (1,564.7 Da) and Cys45 (618.1 Da) was not affected by the addition of pyridone 6 to macrophages (SI Appendix, Fig. S14). Furthermore, the levels of HMGB1 isoform expressing a disulfide bond were comparable between the STAT1-deficient macrophages and their WT controls (Fig. 4), in contrast to levels of the HMGB1 isoform expressing hyperacetylation within NLS sites (Fig. 3B). Taken together, JAK/STAT1 signals are required for LPS- or IFN-β–induced HMGB1 hyperacetylation within NLS sites and nucleus to cytoplasm translocation of HMGB1 but not disulfide bond formation between Cys23 and Cys45.
Fig. 4.
JAK/STAT1 signaling is dispensable for LPS- or IFN-induced HMGB1 oxidation. Mouse peritoneal macrophages isolated from WT or STAT1 KO mice were stimulated with indicated stimuli for 6 h. The redox status of intracellular HMGB1 protein was assessed by LC-MS/MS. Shown in the graphs are representative MS traces.
Discussion
Protein shuttling between nucleus and cytoplasm depends upon NLS and nuclear export sequences (NES) (16). Posttranslational modifications of these NLS or NES critically regulate the subcellular localization of intracellular proteins (16). HMGB1 contains two NLS sites, directing its nuclear localization. Inflammation or cell stress induces acetylation of HMGB1 at the NLS sites, preventing nuclear reentry and leading to HMGB1 accumulation in the cytoplasm (17–20). In this study, we identified a key role of JAK/STAT1 in HMGB1 hyperacetylation and cytoplasmic accumulation. Together with recent findings that caspase-1 or caspase-11 mediates HMGB1 release via pyroptosis (6, 7, 21–24), we here propose two critical steps for HMGB1 release from LPS-activated immune cells. In the first step, LPS activates the TLR4/TRIF/IFN-β signaling cascade and the downstream JAK/STAT1 pathway, which is required for LPS-induced HMGB1 hyperacetylation within NLS sites and HMGB1 cytoplasmic accumulation. JAK/STAT1 mediates LPS-induced IRF1 expression in macrophages (33). Recent findings reveal that IRF1 promotes HMGB1 acetylation via physical association with histone acetylases and is required for LPS-induced HMGB1 cytoplasmic accumulation and release (34, 35). LPS also activates calcium/calmodulin-dependent protein kinase (CaMK) IV, which culminates in HMGB1 serine phosphorylation, a posttranslational modification that also promotes HMGB1 cytoplasmic accumulation (19). In the second step, endogenous danger signals or pathogens induce canonical or noncanonical inflammasome activation, which, in turn, activates caspase-1 or caspase-11, respectively. This mediates pyroptosis and cytoplasmic HMGB1 release into to the extracellular space (SI Appendix, Fig. S15). Understanding these mechanisms by which immune cells regulate HMGB1 release may enable targeting therapeutics to attenuate HMGB1-related inflammation by selective inhibition of the JAK/STAT1 pathway or caspase-mediated pathway.
Caspase-11 enhances the production of IL-1β and IL-18 by interacting with caspase-1 (36). However, caspase-11 can also induce pyroptosis and HMGB1 release independently of caspase-1 and other inflammasome components (21, 23). The important interplay between caspase-11 and HMGB1 during systemic inflammation is revealed by the fact that genetic deletion of caspase-11 or administration of neutralizing anti-HMGB1 antibodies significantly promotes survival during lethal endotoxemia, enhances bacterial clearance during P. aeruginosa or S. typhimurium infection, and prevents cognitive decline after experimental polymicrobial sepsis (1, 12–15, 22). In contrast, selective gene deletions for IL-1β and IL-18 fails to promote survival during lethal endotoxemia (22), and loss of NLRP3, a canonical inflammasome component, enhances mortality during bacterial or fungal infections (24, 37). Inhibition of global inflammatory responses by genetic deletion of IKKβ and interleukin 1 receptor type 1 generates severe immune deficiency against bacterial infections (38). Together, these findings emphasize the importance of selectively targeting damage-mediated inflammation while preserving the physiological protective immune responses.
Notably, pharmacological inhibition of the JAK/STAT1 pathway or genetic deletion of STAT1 did not decrease the production of other proinflammatory cytokines and chemokines. Thus, it may be possible to target the JAK/STAT1 pathway to selectively inhibit HMGB1 release without significantly compromising innate immune responses. It is also important to note that the modulation of the JAK/STAT1 pathway by either pharmacological or genetic manipulation altered the HMGB1 redox-dependent isoform pattern when cells were exposed to IFN-β but not to LPS stimulation. These data indicate that the mechanisms that regulate HMGB1 release and nuclear translocation are intimately linked to the biological functions of the secreted HMGB1 (11). Recent studies reveal that the TLR4/TRIF/IFN-β signaling cascade and the downstream JAK/STAT1 pathway are important for caspase-11 expression and activation (23, 36), which further emphasizes the critical role of the JAK/STAT1 pathway in inflammatory responses. In line with this, pharmacological inhibition of the JAK/STAT1 pathway, genetic deletion of STAT1, or inhibition of IFN-β expression by knock out of IRF3, significantly inhibits HMGB1 release and enhances survival in both lethal endotoxemia and experimental sepsis (28–30). Together, these findings indicate that targeting the JAK/STAT1 pathway may be beneficial in treating inflammation.
Materials and Methods
Reagents.
LPS (Escherichia coli; 0111:B4) and PKR inhibitor 2-AP were purchased from Sigma-Aldrich. JAK inhibitor pyridone 6 was obtained from Calbiochem and Cell Signaling Technology.
Recombinant mouse IFN-α and IFN-β were purchased from R&D Systems. Mouse anti-HMGB1 mAb IgG2b 2G7 (noncommercial antibody) was originally from Critical Therapeutic (available upon request). Anti-LC3B antibodies were obtained from Cell Signaling Technology (catalog no. 2775).
Cell Isolation and Culture.
Murine macrophage-like RAW 264.7 cells (American Type Culture Collection) were cultured in RPMI medium 1640 supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were used at 90% confluence and treatment was carried out in reduced-serum Opti-MEM I medium. Peritoneal mouse macrophages were isolated as described previously. Cells were cultured in RPMI medium 1640 supplemented with 10% (vol/vol) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Macrophages were gently washed with, and cultured in, reduced-serum OPTI-MEM I medium before stimulation with LPS or type 1 IFNs. At 16 h after stimulation, levels of various cytokines in the culture medium were determined by ELISA.
ELISA.
TNF and IL-10 levels in the culture medium were determined using quantitative ELISA kits (MTA00; R&D Systems), with reference to standard curves of purified recombinant TNF or IL-10 at various dilutions.
HMGB1 Western Blotting Analysis.
The relative levels of HMGB1 in the culture medium were determined by Western blotting analysis as described previously (6, 26, 39). The relative band intensity was quantified by using the NIH image 1.59 software to determine HMGB1 levels, with reference to standard curves generated with purified HMGB1 as described previously (6, 24, 40).
Immunohistochemical Staining.
Immunohistochemical staining was performed as described previously (26, 39). Briefly, mouse macrophage-like RAW 264.7 cells were plated in 24-well chamber slides in DMEM supplemented with 10% (vol/vol) FBS. After 16 h of stimulation, cells were washed twice with ice-cold PBS. Cells were fixed and permeabilized using 0.2% of Triton X-100, blocked with 10% (vol/vol) normal goat serum, and incubated with primary antibodies followed by Alexa Fluor 488 goat anti-mouse IgG (Invitrogen) before mounting. Images were captured by a confocal laser-scanning microscope or a Zeiss Axiovert microscope. HMGB1 cytoplasmic accumulation was assessed by counting the percentage of cells with visible cytoplasmic HMGB1 staining from at least four randomly selected microscopic fields. (The average cell number in each microscopic field is around 70.)
Autophagy Assays.
Autophagy was evaluated in cells by fluorescence Zeiss Axiovert microscope or Western blot. In fluorescence microscopy experiments, macrophage-like RAW 264.7 cells stably transfected with GFP-LC3 (provided by N. Tony Eissa, Department of Medicine, Baylor College of Medicine, Houston) were stimulated with LPS or rapamycin in the absence or presence of pyridone 6 for 16 h, and cells were examined for the presence of GFP-LC3 punctate structures under a Zeiss Axiovert microscope as described previously (39). Quantitation of autophagy was performed based on the percentage of cells that contain GFP-LC3–positive autophagic punctate dots. In the Western blot experiments, the levels of 18-kDa cytosolic LC3-I and 16-kDa lipidated autophagosome-bound LC3-II were determined by Western blot as described previously.
HMGB1 Preparation for LC-MS/MS Analysis.
All chemicals and solvents were of the highest available grade (Sigma). Culture supernatants were precleared with 50 μL of protein G–Sepharose beads for 1 h at 4 °C. Supernatant HMGB1 was immunoprecipitated with 5 mg of a polyclonal rabbit anti-HMGB1 antibody (Abcam; ab18256) for 16 h at 4 °C. Free thiol groups within HMGB1 were alkylated for 90 min with 10 mM iodoacetamide at 4 °C. Cysteine residues in disulfide bonds were then reduced with 30 mM DTT at 4 °C for 1 h, followed by alkylation of newly exposed thiol groups with 90 mM NEM at 4 °C for 10 min. Samples were subjected to trypsin (Promega) or GluC (New England Biolabs) digestion according to the manufacturer’s instructions for redox or acetyl modification analysis, respectively, and desalted using ZipTip C18 pipette tips (Millipore). Characterization of cysteine bonds and absolute quantification of acetylated HMGB1 was determined as described (6), whereas redox analysis was performed as described previously (11, 40, 41) using an AB Sciex TripleTOF 5600 (Sciex). Analysis of b and y ions generated during MS/MS from these peptides confirmed the amino acid sequence of each peptide and the presence of posttranslational modifications.
Statistical Analysis.
Data are expressed as means ± SD of at least three independent experiments (n = 3–5). One-way analysis of variance was used for comparison among all different groups. A P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank N. Tony Eissa for providing RAW 264.7 cells stably transfected with GFP-LC3. This work was supported, in part, by National Institutes of Health Grants R01 GM62508 (to K.J.T.) and R01 GMO98446 (to H.Y.). B.L. is supported by the foundation of The Elmezzi Graduate School of Molecular Medicine.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 2866.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316925111/-/DCSupplemental.
References
- 1.Yanai H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462(7269):99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 2.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venereau E, Schiraldi M, Uguccioni M, Bianchi ME. HMGB1 and leukocyte migration during trauma and sterile inflammation. Mol Immunol. 2013;55(1):76–82. doi: 10.1016/j.molimm.2012.10.037. [DOI] [PubMed] [Google Scholar]
- 4.Maroso M, et al. Toll-like receptor 4 and high-mobility group box-1 are involved in ictogenesis and can be targeted to reduce seizures. Nat Med. 2010;16(4):413–419. doi: 10.1038/nm.2127. [DOI] [PubMed] [Google Scholar]
- 5.Urbonaviciute V, et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: Implications for the pathogenesis of SLE. J Exp Med. 2008;205(13):3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488(7413):670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu B, Wang H, Andersson U, Tracey KJ. Regulation of HMGB1 release by inflammasomes. Protein Cell. 2013;4(3):163–167. doi: 10.1007/s13238-012-2118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci USA. 2010;107(26):11942–11947. doi: 10.1073/pnas.1003893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittal D, et al. TLR4-mediated skin carcinogenesis is dependent on immune and radioresistant cells. EMBO J. 2010;29(13):2242–2252. doi: 10.1038/emboj.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manfredi AA, et al. Maturing dendritic cells depend on RAGE for in vivo homing to lymph nodes. J Immunol. 2008;180(4):2270–2275. doi: 10.4049/jimmunol.180.4.2270. [DOI] [PubMed] [Google Scholar]
- 11.Venereau E, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 2012;209(9):1519–1528. doi: 10.1084/jem.20120189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouhiainen A, Kuja-Panula J, Tumova S, Rauvala H. RAGE-mediated cell signaling. Methods Mol Biol. 2013;963:239–263. doi: 10.1007/978-1-62703-230-8_15. [DOI] [PubMed] [Google Scholar]
- 13.Patel VS, et al. High Mobility Group Box-1 mediates hyperoxia-induced impairment of Pseudomonas aeruginosa clearance and inflammatory lung injury in mice. Am J Respir Cell Mol Biol. 2013;48(3):280–287. doi: 10.1165/rcmb.2012-0279OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Entezari M, et al. Inhibition of high-mobility group box 1 protein (HMGB1) enhances bacterial clearance and protects against Pseudomonas Aeruginosa pneumonia in cystic fibrosis. Mol Med. 2012;18:477–485. doi: 10.2119/molmed.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavan SS, et al. HMGB1 mediates cognitive impairment in sepsis survivors. Mol Med. 2012;18:930–937. doi: 10.2119/molmed.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao N, Budnik BA, Gunawardena J, O’Shea EK. Tunable signal processing through modular control of transcription factor translocation. Science. 2013;339(6118):460–464. doi: 10.1126/science.1227299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonaldi T, et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22(20):5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evankovich J, et al. High mobility group box 1 release from hepatocytes during ischemia and reperfusion injury is mediated by decreased histone deacetylase activity. J Biol Chem. 2010;285(51):39888–39897. doi: 10.1074/jbc.M110.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, et al. Calcium/calmodulin-dependent protein kinase (CaMK) IV mediates nucleocytoplasmic shuttling and release of HMGB1 during lipopolysaccharide stimulation of macrophages. J Immunol. 2008;181(7):5015–5023. doi: 10.4049/jimmunol.181.7.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youn JH, Shin JS. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J Immunol. 2006;177(11):7889–7897. doi: 10.4049/jimmunol.177.11.7889. [DOI] [PubMed] [Google Scholar]
- 21.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479(7371):117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 22.Lamkanfi M, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol. 2010;185(7):4385–4392. doi: 10.4049/jimmunol.1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rathinam VA, et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150(3):606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willingham SB, et al. NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J Immunol. 2009;183(3):2008–2015. doi: 10.4049/jimmunol.0900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hett EC, et al. Chemical genetics reveals a kinase-independent role for protein kinase R in pyroptosis. Nat Chem Biol. 2013;9(6):398–405. doi: 10.1038/nchembio.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rendon-Mitchell B, et al. IFN-gamma induces high mobility group box 1 protein release partly through a TNF-dependent mechanism. J Immunol. 2003;170(7):3890–3897. doi: 10.4049/jimmunol.170.7.3890. [DOI] [PubMed] [Google Scholar]
- 27.Jiang W, Pisetsky DS. The role of IFN-alpha and nitric oxide in the release of HMGB1 by RAW 264.7 cells stimulated with polyinosinic-polycytidylic acid or lipopolysaccharide. J Immunol. 2006;177(5):3337–3343. doi: 10.4049/jimmunol.177.5.3337. [DOI] [PubMed] [Google Scholar]
- 28.Kim JH, et al. Bacterial endotoxin induces the release of high mobility group box 1 via the IFN-beta signaling pathway. J Immunol. 2009;182(4):2458–2466. doi: 10.4049/jimmunol.0801364. [DOI] [PubMed] [Google Scholar]
- 29.Herzig D, et al. STAT1-deficient mice are resistant to cecal ligation and puncture-induced septic shock. Shock. 2012;38(4):395–402. doi: 10.1097/SHK.0b013e318265a2ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaguchi S, et al. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem Biophys Res Commun. 2003;306(4):860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- 31.Tang D, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190(5):881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang D, Kang R, Livesey KM, Zeh HJ, 3rd, Lotze MT. High mobility group box 1 (HMGB1) activates an autophagic response to oxidative stress. Antioxid Redox Signal. 2011;15(8):2185–2195. doi: 10.1089/ars.2010.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohmori Y, Hamilton TA. Requirement for STAT1 in LPS-induced gene expression in macrophages. J Leukoc Biol. 2001;69(4):598–604. [PubMed] [Google Scholar]
- 34.Pan PH, Cardinal J, Li ML, Hu CP, Tsung A. Interferon regulatory factor-1 mediates the release of high mobility group box-1 in endotoxemia in mice. Chin Med J (Engl) 2013;126(5):918–924. [PubMed] [Google Scholar]
- 35.Dhupar R, et al. Interferon regulatory factor 1 mediates acetylation and release of high mobility group box 1 from hepatocytes during murine liver ischemia-reperfusion injury. Shock. 2011;35(3):293–301. doi: 10.1097/SHK.0b013e3181f6aab0. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, et al. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92(4):501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 37.Gross O, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459(7245):433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 38.Hsu LC, et al. IL-1β-driven neutrophilia preserves antibacterial defense in the absence of the kinase IKKβ. Nat Immunol. 2011;12(2):144–150. doi: 10.1038/ni.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, et al. EGCG stimulates autophagy and reduces cytoplasmic HMGB1 levels in endotoxin-stimulated macrophages. Biochem Pharmacol. 2011;81(9):1152–1163. doi: 10.1016/j.bcp.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoine DJ, et al. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J Hepatol. 2012;56(5):1070–1079. doi: 10.1016/j.jhep.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Yang H, et al. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1) Mol Med. 2012;18:250–259. doi: 10.2119/molmed.2011.00389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.