Significance
Triple-negative breast cancers comprise 10% of invasive breast carcinomas but are responsible for a disproportionate number of deaths and remain poorly understood. Unfortunately, current therapies are only weakly effective, and the median disease-free survival is 4 y among young women. Clinical studies support the relevance of Enhancer of Zeste Homolog 2 (EZH2) overexpression to the progression of triple-negative breast carcinomas. Our study shows that EZH2 acts as an activator of the NOTCH1 promoter and signaling to expand the stem cell pool, leading to accelerated breast cancer initiation and growth. We discovered that this function is independent of EZH2 histone methyltransferase activity and of its Polycomb Repressive Complex 2-binding partners, paving the way for novel therapeutic strategies.
Abstract
Breast cancer is the second-leading cause of cancer-related deaths in women, but the details of how it begins remain elusive. Increasing evidence supports the association of aggressive triple-negative (TN) breast cancer with heightened expression of the Polycomb group protein Enhancer of Zeste Homolog 2 (EZH2) and increased tumor-initiating cells (TICs). However, mechanistic links between EZH2 and TICs are unclear, and direct demonstration of a tumorigenic function of EZH2 in vivo is lacking. Here, we identify an unrecognized EZH2/NOTCH1 axis that controls breast TICs in TN breast carcinomas. EZH2 overexpression increases NOTCH1 expression and signaling, and inhibition of NOTCH1 activity prevents EZH2-mediated stem cell expansion in nontumorigenic breast cells. We uncover a unique role of EZH2 in activating, rather than repressing, NOTCH1 signaling through binding to the NOTCH1 promoter in TN breast cancer cells. EZH2 binding is independent of its catalytic histone H3 lysine 27 methyltransferase activity and of the Polycomb Repressive Complex 2 but corresponds instead to transcriptional activation marks. In vivo, EZH2 knockdown decreases the onset and volume of xenografts derived from TN breast TICs. Conversely, transgenic EZH2 overexpression accelerates mammary tumor initiation and increases NOTCH1 activation in mouse mammary tumor virus-neu mice. Consonant with these findings, in clinical samples, high levels of EZH2 are significantly associated with activated NOTCH1 protein and increased TICs in TN invasive carcinomas. These data reveal a functional and mechanistic link between EZH2 levels, NOTCH1 signaling activation, and TICs, and provide previously unidentified evidence that EZH2 enhances breast cancer initiation.
Invasive breast carcinoma arises in the terminal-duct-lobular unit and progresses through phases of increasing proliferation and altered differentiation to atypical ductal hyperplasia and carcinoma in situ. Anaplasia, which describes cells lacking differentiation, is a hallmark of cancer. Dysregulation of genes governing cell type identity may lead to malignant transformation (1). The transcriptional memory of cells is tightly regulated through epigenetic mechanisms largely by Polycomb and Trithorax group proteins (2). Enhancer of Zeste Homolog 2 (EZH2) is the catalytic subunit of Polycomb Repressive Complex 2 (PRC2), which silences gene transcription through trimethylation of histone H3 on lysine 27 (H3K27me3) (3). EZH2 protein is up-regulated in multiple malignancies (4, 5), where its oncogenic activity is thought to be primarily mediated by silencing tumor suppressor genes (6). Recent evidence implicates EZH2 in transcriptional activation (7–10), but the mechanisms are not well-defined.
EZH2 is up-regulated in clinically aggressive breast carcinomas, where it independently predicts survival (11). EZH2 overexpression is significantly associated with triple-negative (TN) carcinomas, a biologically aggressive group of breast cancer characterized by lack of estrogen and progesterone receptor expression and absence of HER-2/neu overexpression (11). In benign breast tissues, elevated levels of EZH2 protein signal future development of breast cancer up to 12 y before diagnosis, indicating that EZH2 up-regulation precedes morphological atypia or carcinoma (12). Recently, EZH2 has been shown to play a role in self-renewal of breast tumor-initiating cells (TICs) (13). However, direct demonstration that EZH2 promotes breast cancer initiation is lacking, and the responsible mechanisms need further investigation. Our data identify a unique molecular mechanism by which EZH2 promotes breast cancer development and provide support for targeting the gene activating function of EZH2 in TN invasive breast cancer.
Results
EZH2 Knockdown Reduces TICs and Inhibits the NOTCH1 Pathway in Breast Cancer.
To examine the effect of EZH2 on the TIC populations of TN breast cancer, we used primary human breast cancer cells and SUM149 and MDA-MB-231 cell lines. As is the case for clinical samples of TN breast cancer, these cells exhibit high endogenous levels of EZH2 in comparison with benign breast cells (14). EZH2 knockdown (KD) was achieved through stable lentiviral-mediated short hairpin RNA interference (shRNA) previously developed in our laboratory (14). To reconstitute EZH2 expression in KD cells, we used a wild-type EZH2-encoding, myc-tagged adenovirus (11) (Fig. 1A).
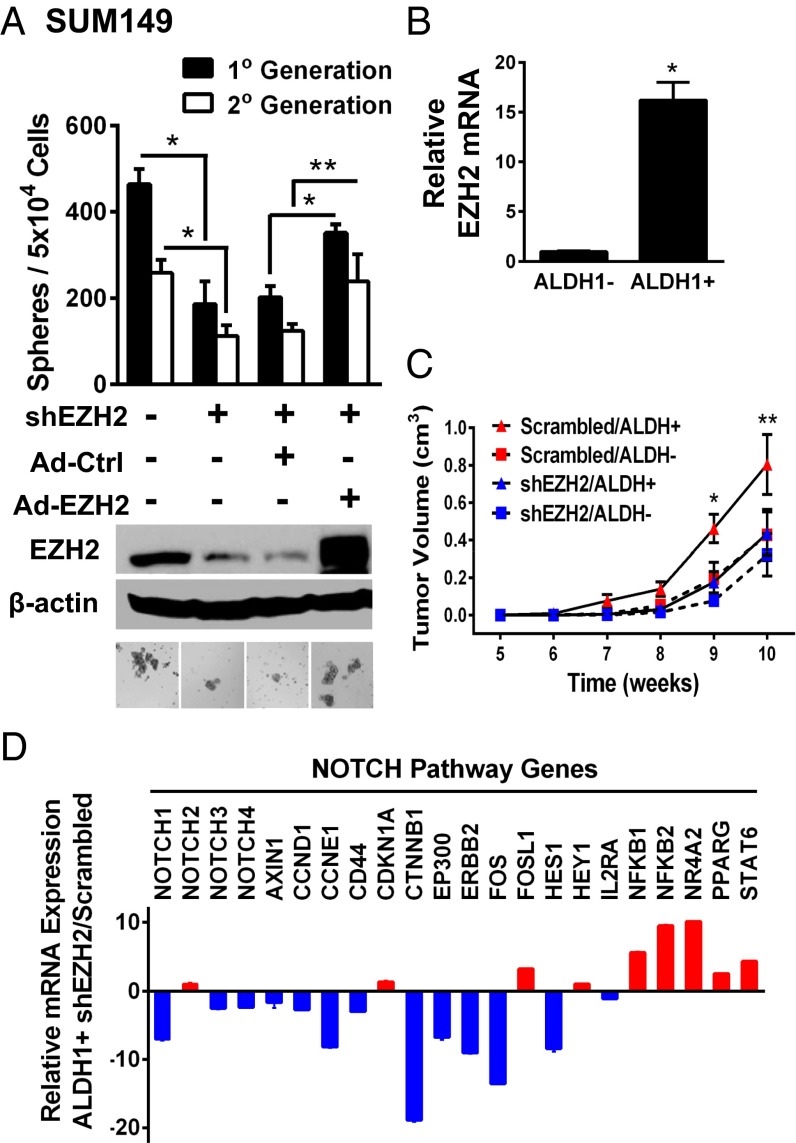
Fig. 1.
EZH2 KD reduces TICs in TN breast cancer and leads to NOTCH1 pathway inhibition. (A) Immunoblots of SUM149 cells with EZH2-targeted shRNA (shEZH2) compared with scrambled shRNA. EZH2 was transiently rescued using a myc-tagged, EZH2-encoding adenovirus. Ad-Ctrl, adenovirus control. Bars show the average sphere numbers ± SD per 5 × 104 plated cells (*P = 0.0001; **P ≤ 0.0004). Representative images of spheres after 7 d in culture. (Magnification: 200×.) (B) EZH2 quantitative RT-PCR in SUM149 ALDH1+ and ALDH1− cells (*P = 0.0001). (C) ALDH1+ and ALDH1− populations of SUM149 EZH2 KD or control cells were isolated by flow cytometry, and 1 × 104 cells were injected into the cleared mammary fat pads of NOD/SCID mice (n = 10 mice per condition). EZH2 KD significantly decreases the volume of tumors formed by SUM149 ALDH1+ cells compared with controls. Average tumor volume ± SEM for weeks 5–10 postinjection for all conditions (mixed regression model, *P = 0.04 and **P = 0.004). (D) Cells described in C were subjected to qRT-PCR for NOTCH family genes.
To understand the role of EZH2 in the regulation of breast TICs, we performed mammosphere assays, based on the property of TICs to survive in nonadherent, serum-free culture conditions (15). EZH2 KD in SUM149 and MDA-MB-231 cells reduced sphere numbers compared with controls (Fig. 1A and Fig. S1A), which was effectively rescued by EZH2 reexpression in SUM149 EZH2 KD cells (Fig. 1A). To identify TICs, we also used the positive activity of aldehyde dehydrogenase 1 (ALDH1) measured by the ALDEFLUOR assay (16) and the cell surface markers CD44+/CD24− (17). In SUM149 cells, EZH2 mRNA expression was higher in the sorted ALDH1+ population vs. the ALDH1− population, indicating that EZH2 is preferentially expressed at higher levels in TICs vs. non-TICs (Fig. 1B). EZH2 KD in SUM149 and MDA-MB-231 cells significantly reduced the percentage of ALDH1+ and CD44+/CD24− cells compared with controls (Fig. S1 A and B). Extending these observations to human breast cancer, EZH2 KD decreased sphere numbers and reduced the ALDH1+ and CD44+/CD24− populations in primary cancer cells derived from two patients with TN invasive carcinomas (Fig. S1 F and G).
The in vivo consequences of decreased TICs attributable to EZH2 KD were investigated by injecting ALDH1+ and ALDH1− populations of SUM149 EZH2 KD and controls into the cleared mammary fat pads of NOD/SCID mice. EZH2 KD in SUM149 cells significantly delayed tumor onset and decreased the tumor volume of ALDH1+ cells compared with controls, whereas it had no significant effect on the ALDH1− populations (Kaplan–Meier, log-rank P = 0.0019; and mixed-regression model, P < 0.05; Fig. 1C, Fig. S1C, and Table S1).
To search for critical genes and pathways mediating the effect of EZH2 on TICs, we used a stem cell signaling focused PCR array comparing the ALDH1+ and ALDH1− populations of SUM149 EZH2 KD and control cells. NOTCH1 was one of the most significantly down-regulated genes by EZH2 KD in the ALDH1+ population compared with the ALDH1− cells (9,687 fold vs. 18 fold, respectively; Student t test P < 0.00001; Fig. S2 A and B). Real-time RT-PCR of NOTCH signaling pathway genes validated these results and showed that EZH2 KD most significantly reduces the mRNA levels of NOTCH1 compared with the other NOTCH receptors and deregulates NOTCH signaling pathway components in the ALDH1+ population (Fig. 1D). This effect was also observed in vivo because xenografts formed by EZH2 KD ALDH1+ cells exhibited decreased NOTCH1 signaling proteins compared with controls (Fig. S2C). Interrogation of publicly available human breast cancer cDNA array datasets using Oncomine confirmed the significant and hitherto unknown association between EZH2 and NOTCH1 mRNA expression in breast carcinomas from six independent datasets (Fig. S3).
EZH2 KD in MDA-MB-231, SUM149, and patient-derived breast cancer cells down-regulated NOTCH1 intracellular domain (NICD1) protein, the activated intracellular form of NOTCH1, and reduced the expression of NOTCH pathway proteins, consistent with the mRNA data (Fig. S1 D, F, and G). Ectopic expression of EZH2 was sufficient to rescue NICD1 protein expression in SUM149 and MDA-MB-231 EZH2 KD cells (Fig. S1E). Collectively, we provide evidence that the TIC-enriched population in TN breast cancer manifests increased NOTCH1 signaling in an EZH2-dependent manner and exhibits gene expression signatures of stemness.
An EZH2/NOTCH1 Axis Regulates Stem Cells.
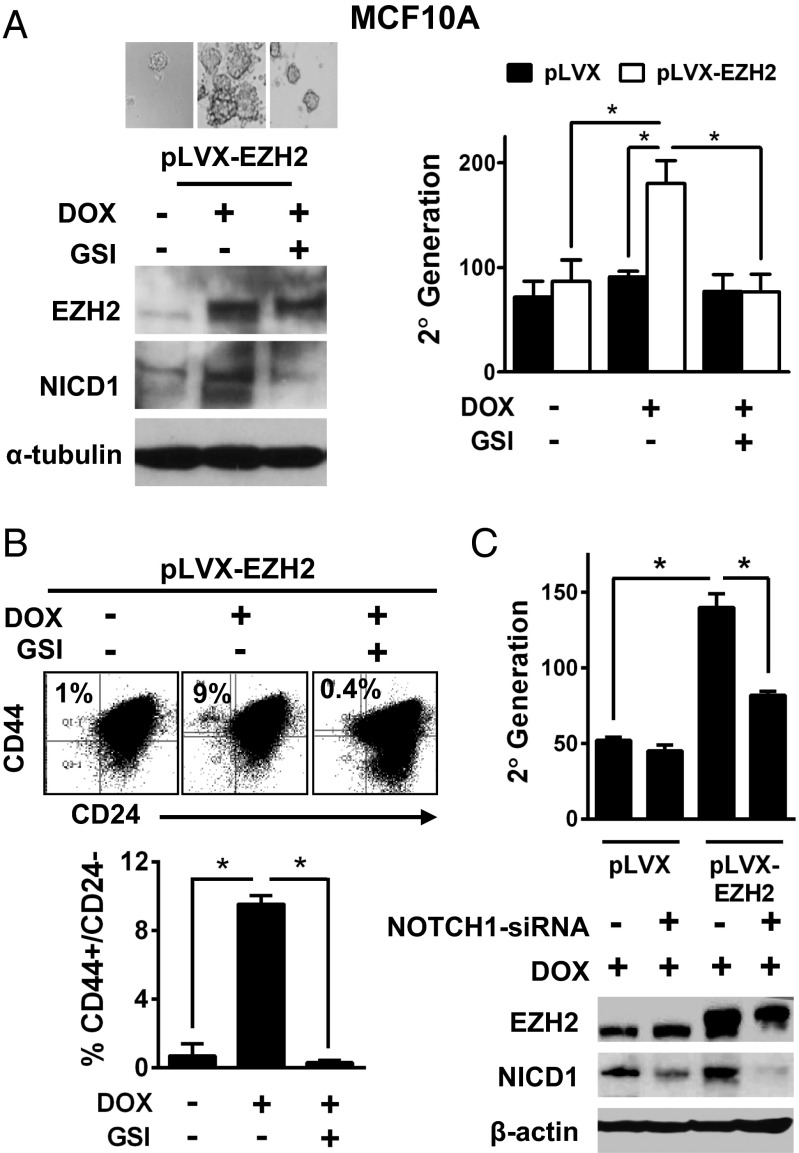
Endogenous EZH2 mRNA levels in primary epithelial cells derived from mammoplasties are higher in the mammosphere forming population compared with adherent cell cultures (Fig. S4A). We used a conditional doxycycline (DOX)-mediated system to overexpress EZH2 in MCF10A cells (18). Whereas DOX treatment of MCF10A-pLVX-EZH2 cells significantly induced EZH2 overexpression in ALDH1+ and ALDH1− cells, EZH2 mRNA levels were significantly higher in ALDH1+ cells (Fig. S4B). Consistently, DOX-induced EZH2 overexpression increased NOTCH1 mRNA levels in ALDH1+ MCF10A cells (Fig. S4C). DOX-mediated EZH2 overexpression in MCF10A-pLVX-EZH2 cells increased NICD1 protein, mammospheres, and the percentage of CD44+/CD24− and ALDH1+ cells compared with untreated cells (Fig. 2 A–C and Fig. S4D). Reduction of NOTCH activation by pretreatment with a γ-secretase inhibitor (GSI) or NOTCH1 siRNA was sufficient to prevent these effects (Fig. 2 A–C and Fig. S4D). Demonstrating the importance of this pathway to human breast cancer, expression of constitutively active NICD1 (19) in patient-derived breast cancer cells effectively rescued the decreased sphere formation due to EZH2 KD (Fig. S1H).
Fig. 2.
NOTCH1 pathway activation is required for EZH2-mediated breast stem cell expansion. (A, Left) Immunoblots of MCF10A pLVX-EZH2 cells DOX-induced and controls probed with anti-EZH2 and anti-NICD1. GSI (17 nM for 3 d) was added 24 h before DOX. Representative images of mammospheres after 7 d in culture. (Magnification: 200×.) (A, Right) mammosphere assay of MCF10A pLVX and pLVX-EZH2 cells DOX-induced and controls, with or without GSI Average sphere number ± SD per 5 × 104 plated cells in the secondary generation (*P < 0.0001). (B) Flow cytometric assays to detect CD44+/CD24− populations in MCF10A pLVX-EZH2 Dox-induced and controls. GSI was added (1.7 nM for 7 d) 24 h before DOX. Percentages are expressed ± SD (*P ≤ 0.005). (C) Immunoblots and mammosphere assays of MCF10A pLVX and pLVX-EZH2 cells DOX-induced and controls, treated with NOTCH1 siRNA or scrambled controls 24 h before DOX. Average sphere number ± SD per 5 × 104 plated cells in the secondary generation (*P < 0.0005).
EZH2 Regulates NOTCH Transcriptional Activity and Expands Stem Cells in a Histone Methyltransferase Activity-Independent Manner.
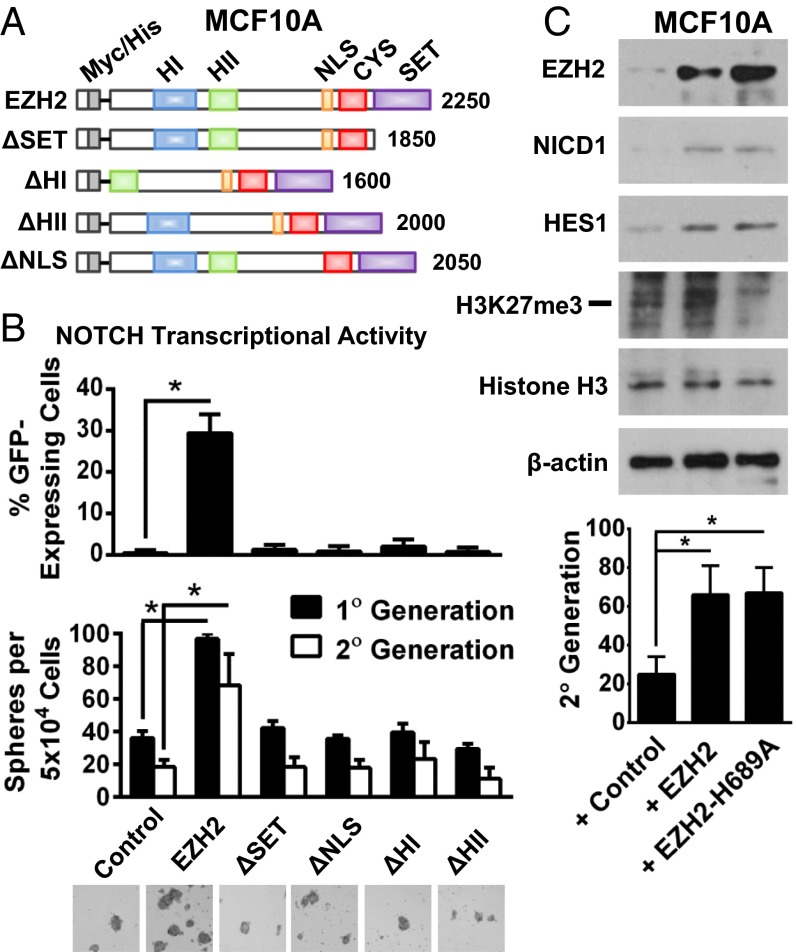
To investigate the effect of EZH2 on NOTCH signaling, we used a lentiviral NOTCH reporter vector that drives the expression of GFP under the minimal essential CMV promoter downstream of NOTCH transcriptional response elements (20). We generated myc-tagged EZH2 deletion mutants involving the amino-terminal homology domains I and II (∆HI and ∆HII), the carboxyl-terminal SET domain (∆SET), and the nuclear localization signal (∆NLS) in adenoviral vectors and expressed them in MCF10A cells (Fig. 3A and Fig. S4E). NICD1 protein was up-regulated by ectopic expression of wild-type EZH2, but not of ∆SET, ∆NLS, ∆HI, or ∆HII (Fig. S4E). Functionally, all deletion mutants blocked the ability of EZH2 to enhance NOTCH transcriptional activity and to increase spheres and the percentage of CD44+/CD24− cells (Fig. 3B and Fig. S4 F and G). To elucidate whether the observed effects of ∆SET are attributable to its enzymatic function, we overexpressed the EZH2-H689A mutant, which has reduced histone methyltransferase (HMT) activity, in MCF10A cells (21). Our data show that EZH2-H689A increased NOTCH1 signaling and sphere numbers to levels similar to wild-type EZH2, suggesting that the HMT activity may not be required for these functions (Fig. 3C).
Fig. 3.
EZH2 regulates NOTCH transcriptional activity and expands stem cells in an HMT activity-independent manner. (A) Schematic diagram of myc-tagged, EZH2 deletion mutants: ΔSET, ΔHI (homology domain I), ΔHII (homology domain II), and ΔNLS (nuclear localization signal). (B, Top) GFP-NOTCH promoter reporter assay of MCF10A cells overexpressing full-length EZH2, EZH2 deletion mutants, or controls. Percentages of GFP-expressing cells ± SD (*P = 0.0004). (B, Middle and Bottom) Mammosphere assays and representative images after 7 d. (Magnification: 200×.) Average number of mammospheres ± SD (*P < 0.0001). (C, Upper) Immunoblot of MCF10A cells transduced with EZH2 and EZH2-H689A mutant probed with anti-NICD1, anti-HES1, anti-H3K27me3, and anti-histone H3. (C, Lower) Mammosphere assay. Average sphere numbers per 5 × 104 plated cells in the second generation ± SD (*P ≤ 0.0001).
EZH2 Binds to the Proximal NOTCH1 Promoter to Activate Transcription.
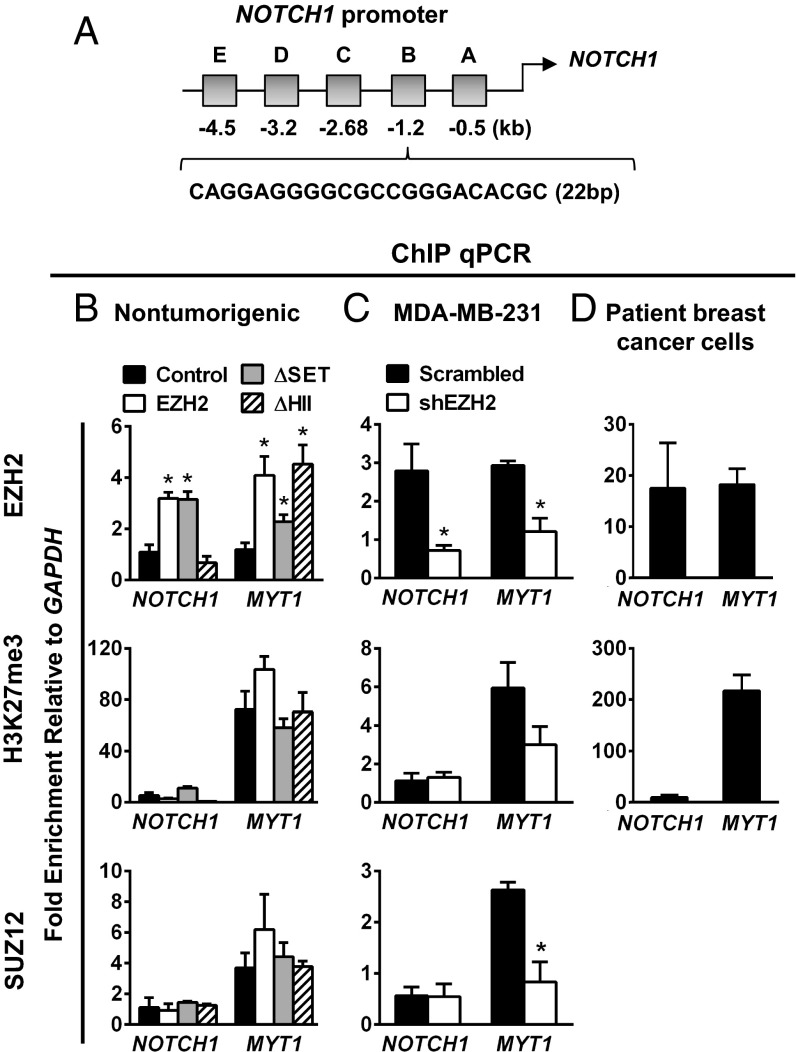
EZH2 has been reported to function as a transcriptional repressor, but there is recent evidence supporting an activating role by yet unclear mechanisms (7–10). In TN breast cancer, EZH2 has been shown to form a complex with RelA and RelB to activate transcription (9). We hypothesized that EZH2-induced NOTCH1 up-regulation may be linked directly to an ability to bind to the NOTCH1 promoter. We performed chromatin immunoprecipitation (ChIP) assays on primary nontumorigenic breast epithelial cells transduced with adenoviral vectors containing wild-type EZH2 or the ΔSET and ΔHII mutants. These mutants were selected because the SET domain is required for HMT activity and the HII domain has been reported to promote gene activation (5, 7). We used primers targeting the NOTCH1 promoter region from −532 to −4510 base pairs upstream of the transcription start site (Fig. 4A). Primers flanking the GAPDH promoter and the MYT1 promoter, a known direct transcriptional repression target of EZH2 through H3K27me3 (22), were used as negative and positive binding controls, respectively. Upon overexpression of wild-type EZH2, we observed a significant increase in EZH2 binding at −1.2 kb, which was blocked by ΔHII (Fig. 4B). EZH2 overexpression was not associated with enrichment for the PRC2 complex repression mark H3K27me3 or with SUZ12 binding but coincided with increased RNA Polymerase II binding, suggesting an activated area in the NOTCH1 promoter (Fig. 4B and Fig. S5 A and C). Consistently, EZH2 overexpression was associated with binding of SET1 and its activation mark, H3K4me2 (23, 24) (Fig. S5).
Fig. 4.
EZH2 binds to the NOTCH1 promoter in benign and in breast cancer cells. (A) Diagram of the NOTCH1 promoter regions analyzed for EZH2 binding in ChIP assays. EZH2 binds to region “B” at −1.2 kb; subsequent experiments were performed in this area. (B) ChIP assays using qRT-PCR in nontumorigenic primary breast cells overexpressing full-length EZH2, ΔSET, ΔHII, or control adenovirus. Primers flanking the GAPDH promoter region are negative binding controls. Primers flanking MYT1, a known direct EZH2 transcriptional repression target through H3K27me3, are positive binding controls. (C) ChIP assays (as in B) in MDA-MB-231 cells show that endogenous EZH2 protein binds the NOTCH1 promoter. EZH2 KD reduces EZH2 binding. (D) ChIP assays (as in B) in patient-derived breast cancer cells reveal that endogenous EZH2 protein binds the NOTCH1 promoter. *P < 0.05 (B and C). All ChIP assays are representative of three independent experiments.
Demonstrating the existence of this mechanism in human TN breast cancer, ChIP assays showed enrichment for endogenous EZH2 protein at the NOTCH1 promoter (Fig. 4 C and D). EZH2 recruitment was associated with enrichment for RelA/RelB, SET1, and H3K4me2, but not for SUZ12 and H3K27me3. Stable EZH2 KD in MDA-MB-231 cells decreased the binding of EZH2, RelA/RelB, SET1, and the H3K4me2 mark at the NOTCH1 promoter, validating the specificity of the interaction (Fig. 4C and Fig. S5 A and B). Re-ChIP experiments demonstrate the presence of EZH2/RelA/RelB complex at the NOTCH1 promoter (Fig. S5B). Taken together, these data identify a unique function of EZH2 epigenetic activation at the NOTCH1 promoter, which may have therapeutic implications.
Transgenic EZH2 Overexpression Up-Regulates NOTCH1, Increases Stem Cells, and Accelerates Tumor Initiation.
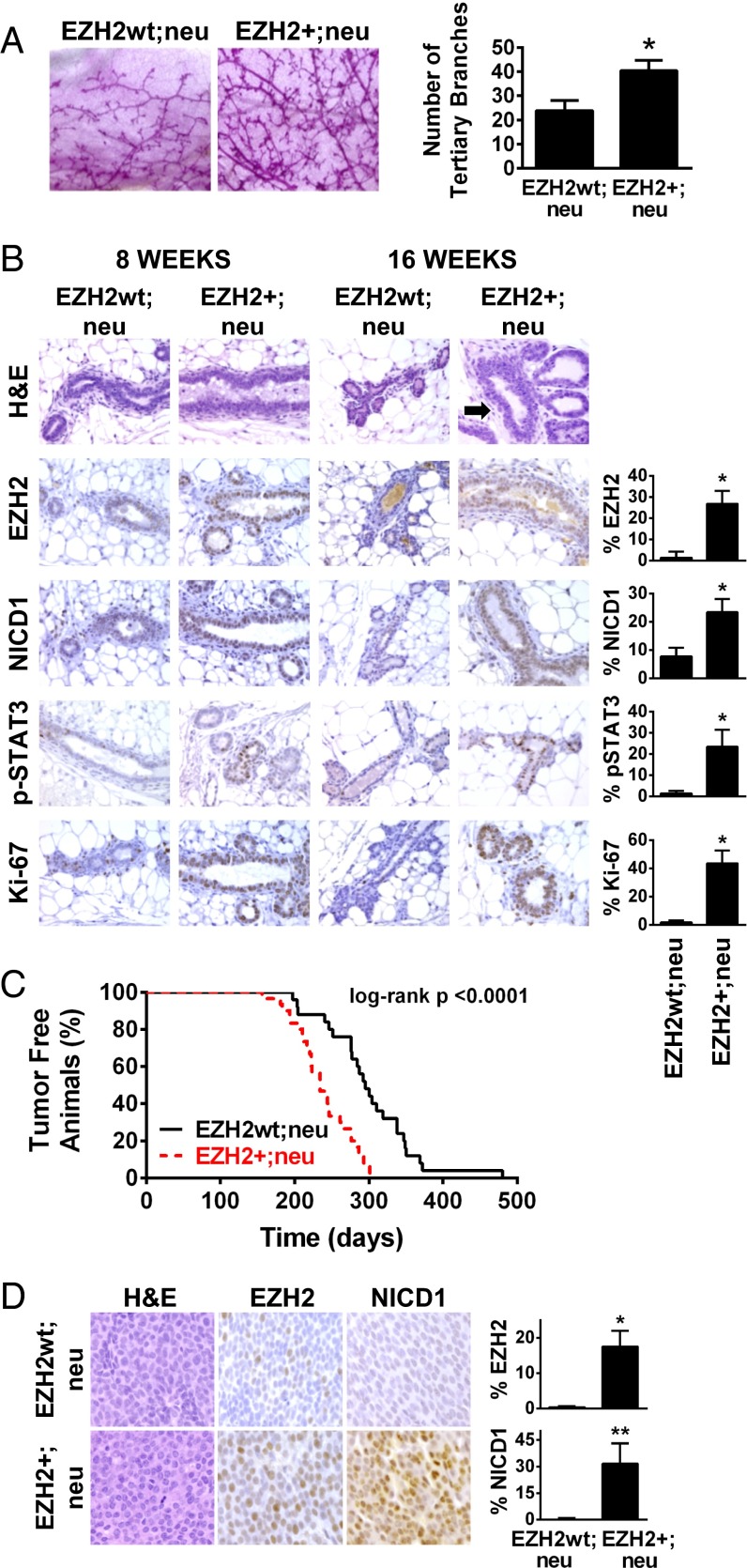
Because of tumor latency, the mouse mammary tumor virus (MMTV)-neu mouse model is well-suited to test the effect of EZH2 overexpression on accelerating tumor initiation (25). Mammary-specific EZH2 transgenic mice developed in our laboratory (26) were crossed with MMTV-neu mice (25). Female 8-wk-old virgin EZH2+;neu mice exhibited ductal hyperbranching compared with EZH2wt;neu mice (Fig. 5A). Virgin EZH2+;neu mice developed atypical intraductal hyperplasia similar to human disease, had up-regulation of NOTCH1 signaling proteins, and increased cell proliferation, compared with EZH2wt;neu mice (Fig. 5B). EZH2+;neu mice formed invasive mammary carcinomas significantly earlier than EZH2wt;neu mice (Kaplan–Meier, log-rank P < 0.0001; Fig. 5C). Although no notable histopathological differences were apparent, EZH2+;neu tumors exhibited increased EZH2 and NICD1 (Fig. 5D). Collectively, these data provide direct in vivo evidence that precancerous EZH2 up-regulation promotes atypical epithelial hyperplasia with heightened NOTCH1 pathway activation and that EZH2 overexpression is sufficient to accelerate tumor initiation in MMTV-neu mice.
Fig. 5.
Transgenic mammary-specific EZH2 overexpression up-regulates NOTCH1 and accelerates tumor initiation in MMTV-neu mice. (A) Whole mounts of mammary glands from 16-wk-old virgin female EZH2+;neu and EZH2wt;neu mice (n = 10 mice per group). (Magnification: 200×.) Average number of tertiary branches ± SD (*P = 0.01). (B) Representative H&E-stained histological sections of mammary glands of EZH2+;neu and EZH2wt;neu mice at 8 and 16 wk of age. Arrow shows atypical intraductal hyperplasia in EZH2+;neu mice. Immunohistochemical detection of EZH2, NICD1, phosphorylated-STAT3 (p-STAT3), and Ki-67 proteins. (Magnification: 400×.) Bar graphs show the percentages of relative expression ± SD at 16 wk quantified using Framework for Image Dataset Analysis (FRIDA) software (*P < 0.0004). (C) Kaplan–Meier curve shows that EZH2+;neu mice (n = 30) formed mammary carcinomas significantly earlier than EZH2wt;neu mice (n = 25) (median time to tumor initiation: 243 d vs. 295 d, respectively; log-rank P < 0.0001). (D) Representative pictures of mammary tumors from EZH2+;neu and EZH2wt;neu mice stained for H&E and immunostained for EZH2 and NICD1. (Magnification: 400×.) Percentage of relative expression ± SD was quantified using FRIDA software (*P < 0.003, **P = 0.009).
Flow cytometric analyses in the lineage negative (Lin‒) population using ESA and CD49f, shown to delineate cellular subsets in MMTV-neu mice and in the human breast (27, 28), demonstrated that EZH2+;neu glands had increased stem cells (Lin− ESAmed CD49f high) and progenitors (Lin− ESAhigh CD49f med) compared with EZH2wt,neu glands (Fig. S6A). The transition from preneoplasia to tumor formation in MMTV-neu mice is characterized by an increase in ESA-, CD49f-, and CD61-expressing cells detected by flow cytometry (28). Using dual immunohistochemistry, we detected areas of increased numbers of tumor cells expressing these markers in EZH2+;neu tumors compared with EZH2wt;neu tumors (Fig. S6D).
Transplantation experiments (29) showed that stem cells, but not progenitor cells, isolated from preneoplastic EZH2+;neu and EZH2wt;neu mammary glands exhibited in vivo gland-reconstituting activity. Of note, stem cells from EZH2+;neu glands formed hyperplastic outgrowths with increased number and size of terminal end buds, higher NICD1 and Ki-67 expression, and elevated percentages of stem cells compared with EZH2wt;neu outgrowths (Fig. S6 B and C).
EZH2/NICD1 Axis in TN Breast Cancer Tissues.
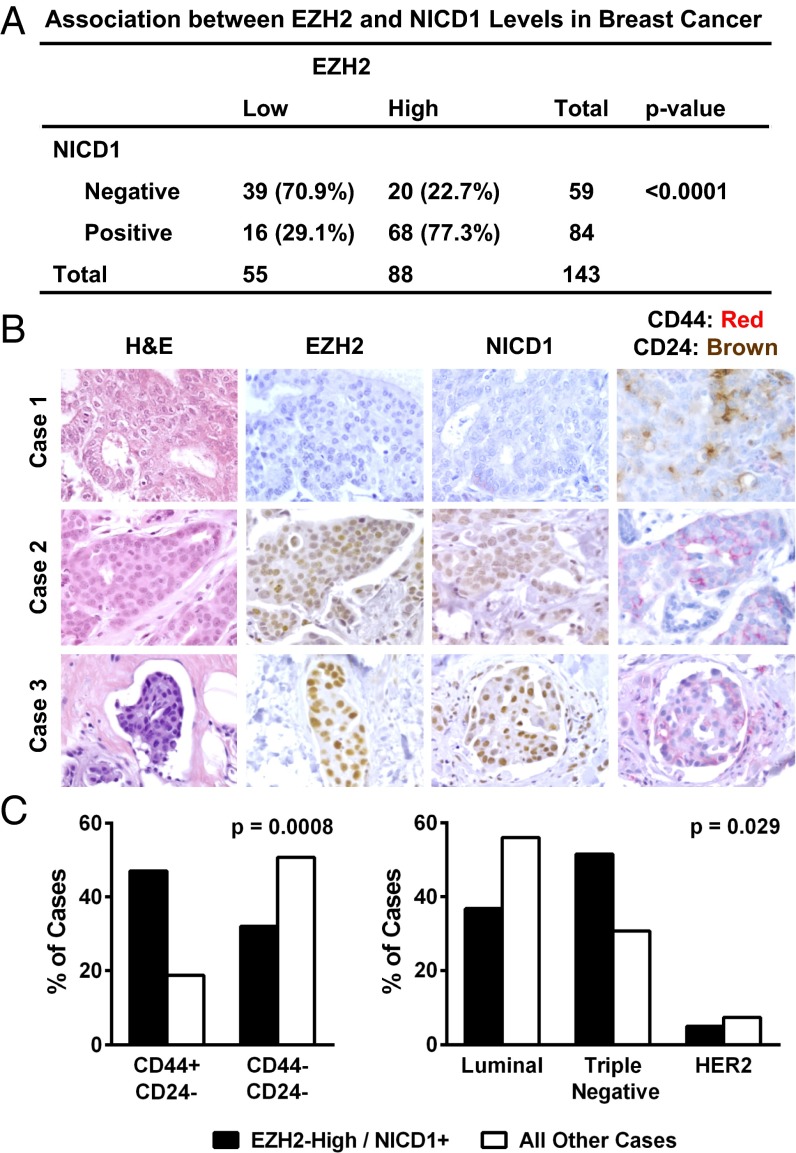
To examine whether EZH2-mediated regulation of NOTCH1 and TICs exists in tumor tissues, tissue microarrays from 143 primary invasive breast carcinoma patients (67 luminal, 9 HER-2/neu over-expressing, 58 TN, and 9 unknown subtype) were interrogated for EZH2 in tandem with NICD1 and the presence of CD44+/CD24− populations by immunohistochemistry. Segregating the patient cohort into EZH2low vs. EZH2high and NICD1+ vs. NICD1− groups, a statistically significant association was identified wherein 77% of the EZH2high scoring patients were also NICD1+ (χ2 test, P < 0.0001; Fig. 6 A and B). Concordant EZH2high and NICD1+ expression was significantly associated with higher histological grade, a measure of poor tumor differentiation, and with the presence of CD44+/CD24− cancer cells (χ2 test, P = 0.006 and P = 0.0008, respectively; Fig. 6 B and C and Table S2). Further characterization of the dataset indicates that of the 58 tumors categorized as TN breast cancers, 83% (48 patients) fall into the EZH2high subgroup. Tumors with concordant EZH2high and NICD1+ are more likely to be TN, whereas those tumors without EZH2high and NICD1+ expression are more likely to be luminal (χ2 test, P = 0.029; Fig. 6C and Table S2).
Fig. 6.
EZH2 overexpression is associated with high NICD1 and increased TICs in human breast cancer tissues. (A) Distribution of EZH2 and NICD1 expression in the patient cohort (n = 143) (χ2 test, P < 0.0001). (B) Human breast cancer tissue samples immunostained for EZH2 and NICD1 and coimmunostained for CD44 (red) and CD24 (brown). Case 1 is a luminal-type invasive carcinoma with low EZH2 expression, negative NICD1, and CD44−/CD24+. Case 2 is a TN invasive carcinoma with high EZH2 expression and positive NICD1 and contains CD44+/CD24− cancer cells. Case 3 is a representative picture of a tumor embolus in a breast lymphatic with high EZH2 and high NICD1 and contains CD44+/CD24− cancer cells. (Magnification: 400×.) (C) Percentage of invasive carcinomas with concordant EZH2high and NICD1+ expression according to the presence of CD44+/CD24− cancer cells (Left) and according to breast cancer subtypes (Right). We found a significant association between the EZH2high/NICD1+ phenotype and the presence of CD44+/CD24− cells and the TN breast cancer subtype (χ2 test: P = 0.0008 and P = 0.029, respectively).
Discussion
EZH2 is an independent marker of recurrence and metastasis in women with breast cancer, where EZH2 overexpression occurs mainly in TN compared with luminal tumors (11). Despite major advances in diagnosis and treatment, there is a considerable gap in our understanding of the mechanisms that induce breast cancer development and progression. In this study, we identify a previously undescribed role for EZH2 in regulating NOTCH1-dependent breast TIC expansion and show that EZH2 has a direct role in breast cancer progression.
We found that EZH2 expression regulates the abundance of TICs in vitro and in vivo. Ectopic EZH2 expression increased the stem cell pool in nontumorigenic breast cells, whereas EZH2 down-regulation reduced the breast TIC population in vitro and in xenograft studies. Our data strengthen those from an earlier study showing that EZH2 can promote breast TIC expansion (13) and further demonstrate the consequences of EZH2 levels on breast cancer initiation. EZH2 down-regulation in TN breast cancer cells retarded breast cancer initiation. Providing previously unidentified in vivo evidence for a role of EZH2 in breast cancer initiation in transgenic models, overexpression of EZH2 in MMTV-neu mice decreased the latency to breast cancer onset. The role of EZH2 in promoting breast stem cell expansion and cancer initiation sheds light on our previous study showing that EZH2 is increased in histologically normal breast tissues from women up to 12 y before they develop breast cancer (12). From a clinical perspective, blocking EZH2 may prevent or ameliorate breast cancer initiation in women with high EZH2 protein expression levels in their breast epithelium.
Despite interest in the association between EZH2 functions, breast TICs, and TN breast cancer (13), the molecular mechanisms underlying the tumorigenic function of EZH2 in this cancer subtype and the relationship to NOTCH1 signaling have not yet been considered. Furthermore, whereas a role for NOTCH1 in breast tumorigenesis has been established in vivo (30), the factors regulating increased NOTCH1 expression and signaling in breast cancer cells are largely unknown (31, 32). We show that EZH2 is a regulator of NOTCH1 expression and pathway activation in TN breast cancer and that NOTCH signaling activation is required for EZH2-dependent stem cell expansion. The association and mechanistic link between EZH2 and NOTCH1 was validated in vitro, in vivo, and in human breast cancer samples. The relevance of our findings is further supported by a research study, which, by using the NOTCH-GFP reporter assay used here, demonstrated that NOTCH activity identifies the cancer stem cell population in lung carcinomas (20).
Substantial studies show that the canonical function of EZH2 is exerted via transcriptional repression through its HMT activity on H3K27 (4, 11, 13, 22). Most studies have focused on PRC2-mediated repression of tumor suppressor genes as the main oncogenic mechanism of EZH2 (6). More recently, EZH2 was shown to activate transcription via non–PRC2-mediated mechanisms, including interaction with RelA/RelB to activate NF-κB targets (9). Our study defines a unique role and mechanism for EZH2 in TN breast cancer, whereby EZH2 binds to the NOTCH1 promoter and induces epigenetic activation. We demonstrate that the amino-terminal HII domain of EZH2 mediates binding to the NOTCH1 promoter and that the HMT activity of EZH2 is not required for NOTCH1 transcriptional activation. Our results are consistent with a recent study documenting the importance of the amino-terminal domain of EZH2 in enhancing gene transactivation through an HMT-independent mechanism in breast cancer (7). Together, our data strengthen the emerging notion that overexpressed EZH2 may function through noncanonical mechanisms leading to activation of target genes in an HMT-independent manner.
In clinical invasive breast cancer samples, high EZH2 and NICD1 are significantly coexpressed in TN compared with luminal tumors, supporting the contention that the EZH2/NICD1 axis is operative in vivo and in humans. Furthermore, EZH2high/NICD1+ tumors are more frequently poorly differentiated and exhibit high numbers of TICs compared with tumors without this phenotype. In conclusion, our findings establish a previously unrecognized link between EZH2, NOTCH1 signaling activation, and TICs in TN invasive carcinomas. These data advance the current understanding of the mechanisms of EZH2 in breast cancer and lend support to the emerging transcriptional activating role of EZH2. By providing previously unidentified direct evidence that EZH2 overexpression accelerates breast cancer initiation in vivo our work paves the way to targeting EZH2 to halt breast cancer progression.
Materials and Methods
Detailed protocols regarding cell culture, vectors, pharmacologic treatments, Western blot analyses, antibodies, microarrays, mammosphere assays, flow cytometry, ChIP analyses, immunohistochemistry, and animal studies are described in SI Materials and Methods. The breast cancer patient cohort has been described previously (see SI Materials and Methods). Original Western blots are shown in Fig. S7.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants CA107469, CA125577, and CA154224 (to C.G.K.), Department of Defense Breast Cancer Research Program Predoctoral Traineeship Award BC093828 (to H.M.M.), and in part through the University of Michigan's Cancer Center Support Grant P30 CA046592.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.L.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308953111/-/DCSupplemental.
References
- 1.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 2.Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol. 2001;2(6):409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 4.Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106(2):243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min J, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16(3):286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi B, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27(14):5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee ST, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Asangani IA, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol Cell. 2013;49(1):80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100(20):11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding L, Erdmann C, Chinnaiyan AM, Merajver SD, Kleer CG. Identification of EZH2 as a molecular marker for a precancerous state in morphologically normal breast tissues. Cancer Res. 2006;66(8):4095–4099. doi: 10.1158/0008-5472.CAN-05-4300. [DOI] [PubMed] [Google Scholar]
- 13.Chang CJ, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19(1):86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez ME, et al. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28(6):843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez ME, et al. Histone methyltransferase EZH2 induces Akt-dependent genomic instability and BRCA1 inhibition in breast cancer. Cancer Res. 2011;71(6):2360–2370. doi: 10.1158/0008-5472.CAN-10-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11(3):299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 20.Hassan KA, et al. Notch pathway activity identifies cells with cancer stem cell-like properties and correlates with worse survival in lung adenocarcinoma. Clin Cancer Res. 2013;19(8):1972–1980. doi: 10.1158/1078-0432.CCR-12-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27(58):7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang G, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci USA. 2004;101(19):7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.South PF, Harmeyer KM, Serratore ND, Briggs SD. H3K4 methyltransferase Set1 is involved in maintenance of ergosterol homeostasis and resistance to Brefeldin A. Proc Natl Acad Sci USA. 2013;110(11):E1016–E1025. doi: 10.1073/pnas.1215768110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89(22):10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, et al. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am J Pathol. 2009;175(3):1246–1254. doi: 10.2353/ajpath.2009.090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim E, et al. kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 28.Lo PK, et al. CD49f and CD61 identify Her2/neu-induced mammary tumor-initiating cells that are potentially derived from luminal progenitors and maintained by the integrin-TGFβ signaling. Oncogene. 2012;31(21):2614–2626. doi: 10.1038/onc.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu C, et al. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168(3):973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haughian JM, et al. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proc Natl Acad Sci USA. 2012;109(8):2742–2747. doi: 10.1073/pnas.1106509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clementz AG, Rogowski A, Pandya K, Miele L, Osipo C. NOTCH-1 and NOTCH-4 are novel gene targets of PEA3 in breast cancer: Novel therapeutic implications. Breast Cancer Res. 2011;13(3):R63. doi: 10.1186/bcr2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.