Significance
Supramolecular assembly is a powerful strategy to generate structural and functional diversity from a limited set of components. Owing to their chemical and structural versatility, proteins represent particularly attractive building blocks for engineering supramolecular materials with new or improved properties. We previously exploited the strength, directionality, and reversibility of designed metal coordination interactions to arrange a monomeric, redox protein into highly ordered supramolecular architectures. Here we demonstrate that metal-directed self-assembly not only yields very stable architectures but that it also leads to a dramatic stabilization of the individual protein components, whose redox activities can now be utilized for controlled growth of inorganic nanocrystals. These emergent physical and functional properties are attained with minimal modification of the original building blocks.
Keywords: protein self-assembly, supramolecular coordination chemistry, nanomaterials, biomaterials, inorganic nanoparticles
Abstract
The designed assembly of proteins into well-defined supramolecular architectures not only tests our understanding of protein–protein interactions, but it also provides an opportunity to tailor materials with new physical and chemical properties. Previously, we described that RIDC3, a designed variant of the monomeric electron transfer protein cytochrome cb562, could self-assemble through Zn2+ coordination into uniform 1D nanotubes or 2D arrays with crystalline order. Here we show that these 1D and 2D RIDC3 assemblies display very high chemical stabilities owing to their metal-mediated frameworks, maintaining their structural order in ≥90% (vol/vol) of several polar organic solvents including tetrahydrofuran (THF) and isopropanol (iPrOH). In contrast, the unassembled RIDC3 monomers denature in ∼30% THF and 50% iPrOH, indicating that metal-mediated self-assembly also leads to considerable stabilization of the individual building blocks. The 1D and 2D RIDC3 assemblies are highly thermostable as well, remaining intact at up to ∼70 °C and ∼90 °C, respectively. The 1D nanotubes cleanly convert into the 2D arrays on heating above 70 °C, a rare example of a thermal crystalline-to-crystalline conversion in a biomolecular assembly. Finally, we demonstrate that the Zn-directed RIDC3 assemblies can be used to spatiotemporally control the templated growth of small Pt0 nanocrystals. This emergent function is enabled by and absolutely dependent on both the supramolecular assembly of RIDC3 molecules (to form a periodically organized structural template) and their innate redox activities (to direct Pt2+ reduction).
The enormous structural and chemical diversity of proteins makes them highly attractive building blocks for functional materials. Supramolecular protein assemblies constitute the major components of cellular machinery, and many of them [e.g., 0D ferritin (1), 1D silicatein filaments (2, 3), 2D S-layers (4)] have also found a number of applications in nano- and biotechnology (2, 4, 5). Such protein assemblies are typically highly ordered, yet they possess dynamic structures that respond to external stimuli (6). Distinctively, protein assemblies are innately functional: the individual building blocks of these assemblies often perform specific chemical functions. For instance, the monomeric components of silicatein filaments or ferritin cages each have catalytic sites that process the precursors (SiO2 or Fe2+) for the downstream biomineralization processes (1, 3). On self-assembly, these individual components can be significantly stabilized (allowing them to withstand the extracellular environment and perform their functions) (7), and their chemical function takes on an entirely different context within the supramolecular architecture (acting as the template for biomineralization in the cases of silicatein and ferritin). Thus, they display emergent properties, both structurally and functionally.
Such advanced properties of natural supramolecular protein architectures have provided a strong motivation for efforts in designed protein assembly. The ability to choose arbitrary proteins as building blocks and to program their arrangement into desired supramolecular architectures could not only reveal natural design principles for protein self-assembly, but also generate entirely new functions and physical attributes beyond what these proteins evolved into during natural selection. However, this task is complicated by the fact that proteins are complex macromolecules that do not possess many canonical interaction motifs with which to program their organization. A number of strategies have been devised toward this end, including computational design of associative surfaces (8), the construction of chimeric protein assemblies with preprogrammed symmetries (9, 10), and those that use specific ligand–protein interactions (11–14), metal coordination (15–17), disulfide bonds (18–20), or a combination of these strategies (21, 22) to connect protein building blocks. These approaches have produced supramolecular architectures with increasing structural sophistication or improved functions. However, the design of protein assemblies that simultaneously possess well-defined structures, functions, and emergent/collective properties like their evolved counterparts remains a significant challenge, which we address in this study.
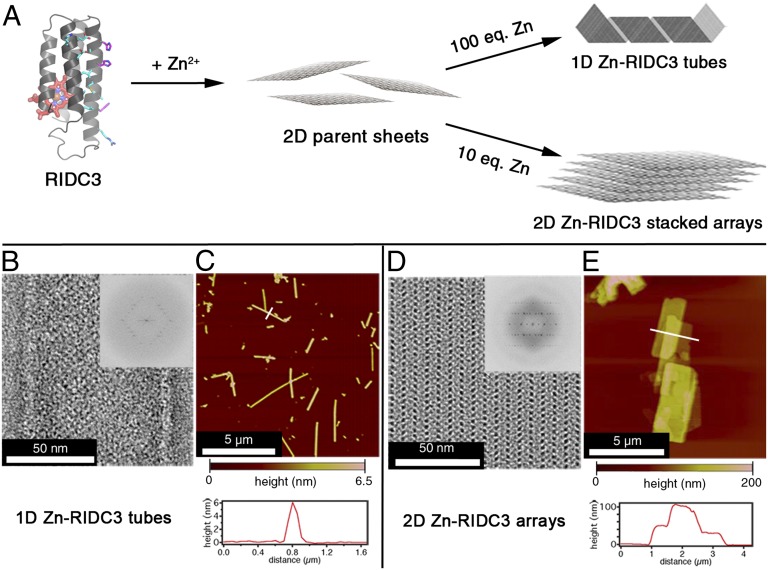
We recently reported that a monomeric heme protein (cytochrome cb562) could be appropriately designed to create a self-assembling variant that we termed RIDC3 (Rosetta Interface Designed Cytochrome 3) (22). With respect to cyt cb562, RIDC3 carries 3 amino acid substitutions on its surface for Zn2+ coordination and 10 additional substitutions to promote its dimerization into a C2-symmetric complex, which possesses coordinatively unsaturated Zn2+ coordination sites to promote the growth of extended arrays (Fig. 1 and Fig. S1) (22). We showed that RIDC3 arranged on Zn2+ coordination into crystalline 1D nanotubes or 2D arrays, whose sizes and morphologies were controllable through pH, RIDC3 concentration, or the molar excess of Zn2+ relative to RIDC3 (Fig. 1A) (22). Under conditions that accelerated Zn-mediated RIDC3 nucleation (high pH, high [RIDC3], and high [Zn]:[RIDC3] ratios), the parent 2D sheets could not grow to large sizes and folded into 50- to 70-nm-wide, hollow nanotubes (Fig. 1 B and C). In contrast, conditions that slowed down Zn-mediated RIDC3 nucleation (low pH, low [RIDC3], and low [Zn]:[RIDC3] ratios) favored the formation of large planar arrays that stacked into multilayered sheets (Fig. 1 D and E) (22).
Fig. 1.
Zn-mediated assembly of RIDC3 into supramolecular arrays and structural characterization of the arrays. (A) Scheme for self-assembly of monomeric RIDC3 into helical nanotubes and multilayered 2D arrays. Amino acids designed to promote dimerization are shown as cyan sticks and those responsible for metal binding as magenta sticks. Addition of excess Zn2+ effects the formation of polymerization-competent nuclei that either grow unidimensionally and fold into 1D helical nanotubes (Upper) or bidimensionally and stack into multilayered 2D periodic arrays (Lower). 1D nanotubes (B and C) and 2D arrays (D and E) were structurally characterized by negative stain TEM (B and D) and AFM (C and E). Fourier transforms (B and D, Insets) show that the arrays are crystalline and have identical unit cell parameters.
Here we take a step beyond these initial structure-building studies and examine in detail the physical and chemical properties of the 1D and 2D Zn-RIDC3 assemblies. We show that these assemblies not only are stable to polar organic solvents and high temperatures because of their metal-mediated frameworks, but also that the individual RIDC3 building blocks themselves are significantly stabilized by virtue of supramolecular self-assembly. Given the innate function of RIDC3 as a one-electron transfer agent, the supramolecular Zn-RIDC3 arrays represent redox-active templates with structural periodicity, which can be used to spatiotemporally control the reductive growth of Pt0 nanoparticles (PtNPs) on their surfaces. Thus, the Zn-RIDC3 assemblies display emergent structural and functional properties not manifested in the individual cyt cb562 building blocks.
Results and Discussion
High-Yield Preparation and Characterization of 1D and 2D RIDC3 Assemblies.
The fact that RIDC3 can be predictably assembled into different supramolecular architectures (with the same underlying molecular pattern) provides a unique opportunity to probe how the physical and chemical properties of proteins may be affected by their supramolecular organization. With this question and downstream materials applications in mind, we first sought to produce monodisperse 1D and 2D RIDC3 assemblies in large quantities. We found that at pH 5.5 (in 5 mM bis-TRIS buffer), the addition of 100 equivalents of Zn2+ to a solution of 50 µM RIDC3 initiated the formation of 1D nanotubes almost immediately, and after 1 wk, these structures were the predominant species present (Figs. 1 B and C and 2). Under these conditions, the 1D nanotubes persisted for at least 1 mo. In contrast, the inclusion of higher concentrations of bis-TRIS (≥50 mM), which weakly coordinates Zn2+, resulted in the rapid formation of the 1D nanotubes and their conversion into 2D arrays after 1 d (Fig. S2). We attribute this phenomenon to the ability of bis-TRIS to accelerate the Zn-dependent equilibration between the 1D nanotubes (kinetic product) and the 2D arrays (thermodynamic product, vide infra). The overall yield for the formation of 1D RIDC3 nanotubes obtained under low bis-TRIS concentrations was nearly quantitative (97.5 ± 0.5%), as determined by measuring the concentration of RIDC3 that remained in solution after isolation of arrays by centrifugation.
Fig. 2.
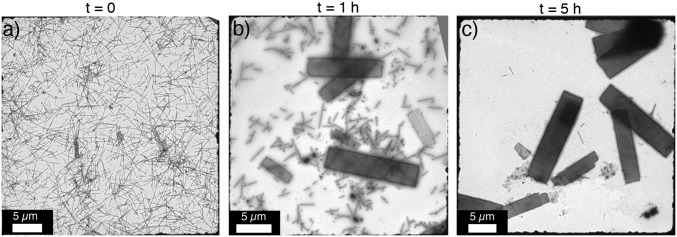
Thermally induced transition of 1D RIDC3 nanotubes into 2D arrays. RIDC3 nanotubes were assembled by the addition of 100 equivalents of Zn2+ in 5 mM bis-TRIS (pH 5.5). Protein arrays were exchanged into MES buffer (pH 5.5) by centrifugation and resuspension, after which the solution was heated to 80 °C. TEM grids were periodically prepared and imaged over the course of 5 h (A–C).
Although the inclusion of high bis-TRIS concentrations provides one route to obtain 2D RIDC3 arrays, we found that the addition of a lower molar excess (10-fold) of Zn2+ over RIDC3 at the same pH (in 20 mM MES buffer) also produced 2D arrays exclusively and in quantitative yield (98.6 ± 0.1%) over 1 wk (Fig. 1 D and E). Both the 1D and 2D RIDC3 assemblies were reproducibly crystalline and displayed the same lattice constants (a ∼ 37 Å, b ∼ 138 Å, α ∼ 90°) as previously reported (22).
Inductively coupled plasma-optical emission spectroscopy (ICP-OES) analyses indicated that 1D and 2D RIDC3 assemblies contained 2.4 ± 0.1 and 3.4 ± 0.2 equivalents of Zn per RIDC3 monomer, respectively. The RIDC3 concentration was deduced from the Fe content of the sample, as each monomer contains a single heme-Fe center. These values are larger than the 1.5 Zn/monomer ratio found within the parent 2D lattice (Fig. S1A), suggesting that excess Zn2+ ions can associate with the surfaces of these assemblies, likely through the many aspartate and glutamate residues (Fig. S1B).
Atomic force microscopy (AFM) measurements revealed that 1D RIDC3 nanotubes, which were flattened on the mica substrate, were 5 ± 1 nm high (Fig. 1C). This value is slightly lower than the expected height (8 nm) of two single layered RIDC3 sheets, which may be due to partial unfolding of the protein nanotubes on deposition on the mica substrate. The 2D RIDC3 arrays were 20–85 nm high, which corresponds to 5–20 stacked sheets (Fig. 1E).
Chemical Stability of 1D and 2D RIDC3 Assemblies.
The single-layered RIDC3 sheets, which are the parent substructures for both the 1D nanotubes and the 2D arrays (Fig. 1A), are formed by a network of strong Zn coordination interactions between RIDC3 monomers (Fig. S1A). Therefore, we envisioned that both the 1D and 2D RIDC3 assemblies may be particularly stable under materials or biotechnological processing conditions such as in organic solvents and at high temperatures, which normally disrupt protein structures held together by noncovalent interactions. The chemical stability of RIDC3 assemblies was determined by their resistance to morphological changes on incubation in four polar organic solvents that covered a wide range of hydrophobicities [i.e., octanol/water partition coefficients, Pow): tetrahydrofuran (THF, log Pow = 0.46), isopropanol (iPrOH, 0.05) methanol (MeOH, –0.74) or N,N-dimethylformamide (DMF, –1.01)] (23). 1D RIDC3 nanotubes, which are a single-sheet thick and thus represent the minimal stabilizing unit, remained well dispersed and retained their native morphologies and crystalline order, as measured by transmission electron microscopy (TEM), for at least 12 h in up to 40% (vol/vol) THF, 40% iPrOH, 70% DMF, and 60% MeOH (Fig. S3A). Changes to the supramolecular structure of the protein arrays on further increases in organic solvent were dependent on solvent polarity. In less polar solvents (THF and iPrOH), the tubes formed dense aggregates above organic solvent concentrations of ∼50%, but individual tubes remained crystalline (Fig. S3B). In more polar solvents (DMF and MeOH), there was a sharp transition from well-dispersed tubes with intact lattices to partially disassembled, noncrystalline tubes (Fig. S3C). The tendency of nanotubes to aggregate in less polar solvents is likely a result of unfavorable interactions of polar surface amino acids with solvent molecules, which leads to the bundling of tubes. In contrast, the more polar solvents (DMF and MeOH) are likely able to promote the dispersion of RIDC3 tubes through H-bonding up to high concentrations, at which point they also act as protein denaturants and cause disassembly.
Owing to additional interlayer stacking interactions, 2D RIDC3 arrays showed even greater structural stability than the 1D nanotubes, with no changes in either their overall morphology or underlying crystalline lattice after incubation for 12 h in up to 90% (vol/vol) of all four solvents tested (Fig. S4). In fact, even after 2 mo at room temperature in these solvents or after repeated isolation of the arrays by centrifugation and resuspension, we found no evidence for the dissolution of the 2D arrays, for the loss of their crystalline order, or for their aggregation.
Supramolecular Stabilization of RIDC3 Monomers in Organic Solvents.
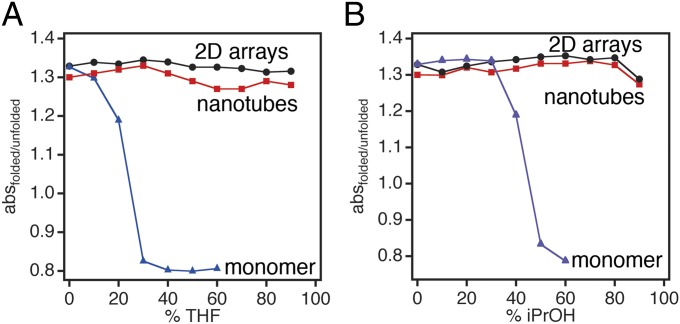
We next probed whether individual RIDC3 monomers are stabilized on arrangement into 1D and 2D supramolecular arrays, which would represent an emergent physical property. Toward this end, we monitored the Soret band of the integrated heme cofactor, which is highly sensitive to changes in solvation and Fe coordination and therefore frequently used to assess the structural integrity/foldedness of hemeproteins (24). It is important to note here that the heme cofactor of RIDC3 is covalently attached to the protein backbone via c-type linkages to engineered Cys residues and does not dissociate on unfolding (24). Because RIDC3 monomers are already thermostable (Tm ∼80 °C), the differential stabilization effect was most apparent in organic solvents, in particular those with high hydrophobicities (i.e., THF and iPrOH). As isolated in solution, RIDC3 monomers unfolded at <30% THF and ∼50% iPrOH (Fig. 3 A and B and Fig. S5A) as indicated by a blue-shift of the Soret maximum from 415 to 408 nm. RIDC3 was more stable in solvents with higher polarities, remaining partially folded in up to 80% DMF and 90% MeOH (Fig. S5B), as has been observed for other monomeric proteins such as α-chymotrypsin (25). In contrast, once assembled in 1D nanotubes and 2D arrays, RIDC3 did not display any unfolding transition at up to 90% THF or iPrOH (Fig. 3 A and B and Fig. S5A). At high concentrations of DMF and MeOH (80–90%), RIDC3 nanotubes, and to a lesser extent 2D arrays, began to show signs of unfolding (Fig. S5B). The unfolding of nanotubes correlates well with the point at which they began to disassemble, as visualized by TEM (Fig. S3C).
Fig. 3.
Stability of monomeric and self-assembled RIDC3 in THF (A) and iPrOH (B) monitored by changes in the heme absorption spectrum. The foldedness of RIDC3 was assessed by calculating the absorbance ratio at 415 nm (Soret maximum for folded RIDC3) over 408 nm (Soret maximum for unfolded RIDC3) for monomeric RIDC3 and at 418 nm over 408 nm for Zn-directed assemblies.
A number of strategies have been devised to improve the stabilities of proteins and enzymes to make them suitable for biotechnological processes, particularly those requiring organic solvents (26). These strategies include computational design (27–29), rational (30) or random mutagenesis (31), chemical modification with polymers (32, 33), immobilization on solid supports (34), or covalent cross-linking of crystals (35, 36). Stabilization of proteins through metal-mediated supramolecular arrays, such as demonstrated here, perhaps combines the best of these approaches: it is minimally invasive, the monomers can be recovered and recycled in their native forms by metal chelation, and the 1D and 2D crystalline arrays boast very high surface area/volume ratios, a near-unity protein weight fraction, and high dispersity/stability in polar organic solvents.
Thermal Stability and Interconversion of 1D and 2D RIDC3 Assemblies.
1D and 2D RIDC3 assemblies were also found to be highly thermostable, maintaining their supramolecular architecture and crystallinity on incubation at 70 °C for 12 h (Fig. S6A). Above 80 °C, some disorder in the supramolecular lattice of the 2D arrays was apparent, although the lattice constants did not change (Fig. S6B). This disorder is ascribed to the partial unfolding/dissociation of proteins on the surface layers, leaving the inner layers unaffected. Accordingly, we observed a slight increase in the fraction of dissolved RIDC3 (from 2.5% to 8% after 12 h).
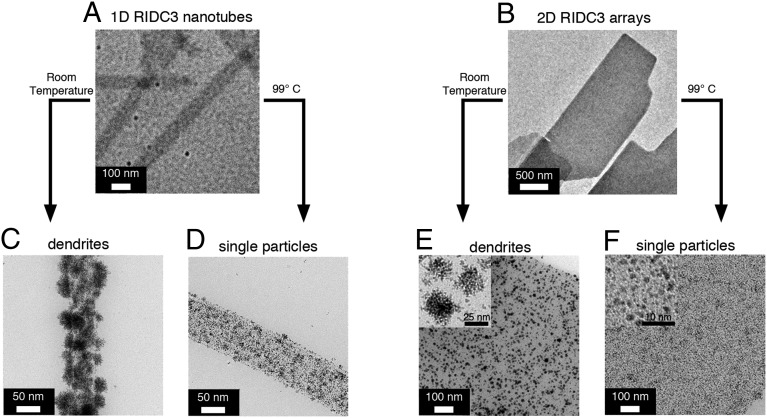
Interestingly, incubation of 1D nanototubes at 80 °C resulted in their gradual conversion into 2D arrays over 5 h (Fig. 2), with no concomitant increase in the free monomer concentration in solution. This observation is consistent with the higher stability of the 2D arrays. Temperature-induced morphological transformations have also been reported for tobacco mosaic virus (TMV) (Table S1), an atypically stable tubular virus (37). However, the clean thermal conversion of the 1D RIDC3 nanotubes into the 2D RIDC3 sheets represents, to our knowledge, a previously unidentifed example of a designed biomolecular assembly that shows a crystalline-to-crystalline structural transition. A plausible mechanism involves the thermally induced unraveling of the 1D tubes into sheets that subsequently aggregate into the thermodynamically more stable, stacked 2D arrays. In an alternative scenario, the 1D tubes may fully dissociate into monomers, which then assemble on Zn2+ coordination into the 2D arrays.
Redox-Controlled Growth of Platinum Nanoparticles on RIDC3 Assemblies.
Having established the robustness of Zn-directed RIDC3 superstructures and the stabilization of the monomers therein, we set out to examine whether these advantages could be coupled with the built-in redox activity of RIDC3 monomers to control the templated growth of inorganic materials. We chose Pt0 nanoparticles (PtNPs) as the proof-of-principle target because the synthesis of these often-used catalysts has been demonstrated under various solution conditions and in the presence of different biomolecular templates (38–41). However, in all of these cases, the template was functionally passive, providing either only a locale for the attachment of Pt2+/4+ ions or displaying a peptide sequence motif that can promote the growth of Pt nanocrystals through binding of specific crystal planes (42, 43). Because the biological role of cyt cb562 (the parent of RIDC3) is electron transfer via its heme center, we surmised that heme groups housed periodically within RIDC3 assemblies can direct the controlled reduction of surface-attached Pt2+ into PtNPs.
We first set out to establish that RIDC3 assemblies could support reductive PtNP growth on their surfaces and whether this process indeed required the presence of the supramolecular template (Fig. 4). Briefly, the 1D nanotubes (10 µM total RIDC3 concentration) and 2D arrays (50 µM total RIDC3) were incubated with 5 or 10 mM Pt2+, respectively, at room temperature for 24 h to allow coordination to surface Asp and Glu residues. Subsequent addition of 50 mM ascorbate resulted in a gradual color change of the suspensions from red (due to heme absorption) to black over the course of ∼4 h. TEM images of unstained samples showed that the crystalline 1D and 2D RIDC3 architectures were unaffected by this chemical treatment (Fig. S7A) and revealed the presence of surface-attached dendritic nanoparticles 33 ± 8 and 23 ± 8 nm in diameter for 1D nanotubes and 2D arrays, respectively, as well as some single nanoparticles (Fig. 4 C and E). High-resolution TEM images revealed that the nanodendrites are crystalline and composed of single particles 3 ± 0.5 nm in diameter and that many of these single particles are aligned throughout the dendritic clusters (Fig. S7B).
Fig. 4.
Templated formation of Pt nanoparticles on the surfaces of 1D and 2D RIDC3 assemblies. (A and B) TEM images of unstained samples prepared from solutions containing 1D nanotubes or 2D arrays. (C–F) TEM images of unstained samples prepared after Pt2+ reduction on the surface of the arrays. Reduction of Pt2+ at room temperature led to the growth of dendritic Pt0 nanoparticles (C and E), whereas reduction at 99 °C (D and F) favored the growth of mostly single nanoparticles.
During these experiments, we found that the high thermostability of the 2D RIDC3 arrays is further improved on overnight Pt2+ incubation, likely due to cross-linking of surface residues. Therefore, we were able to carry out templated PtNP growth experiments also at 99 °C. In line with previous findings (44), the heat treatment resulted in a considerable decrease in the size of PtNPs, yielding in near uniformity single particles with a narrow size distribution (2.2 ± 0.7 nm in diameter; Fig. 4 D and F).
When the PtNP growth experiments were carried out in the absence of RIDC3 arrays at room temperature, mostly large amorphous Pt aggregates formed (Fig. S8A). Similar aggregates were formed at 99 °C in the absence of a template, although to a lesser extent than at room temperature (Fig. S8A). Omission of the overnight incubation step with Pt2+ before heating resulted in an increase in background reduction and sparse PtNP coverage of the RIDC3 templates (Fig. S8B). Incubation of preformed particles with RIDC3 arrays at room temperature or after a 10-min period at 99 °C resulted in only disordered aggregates (Fig. S8B). Finally, when RIDC3 monomers were used rather than their assemblies, we observed a significant decrease in PtNP formation. A possible explanation is that any growing Pt seeds on an RIDC3 monomer can be capped by other free RIDC3 monomers, thus preventing their growth into larger nanoparticles. Alternatively, the nucleation and growth of PtNPs may be taking place primarily at interfacial coordination sites in the RIDC3 arrays, which are not present in the monomers. In any case, our control experiments clearly show that the assembly of RIDC3 into supramolecular arrays is necessary for the templated growth of PtNPs and that these supramolecular arrays can operate under conditions not typically tolerable by most biomolecular assemblies.
Light-Activated Growth of Platinum Nanoparticles on RIDC3 Assemblies via Zn-Substituted Heme Cofactors.
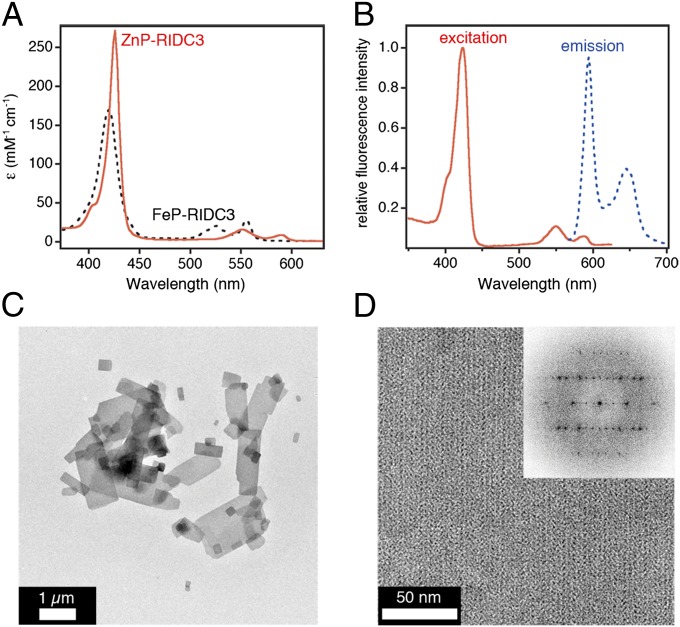
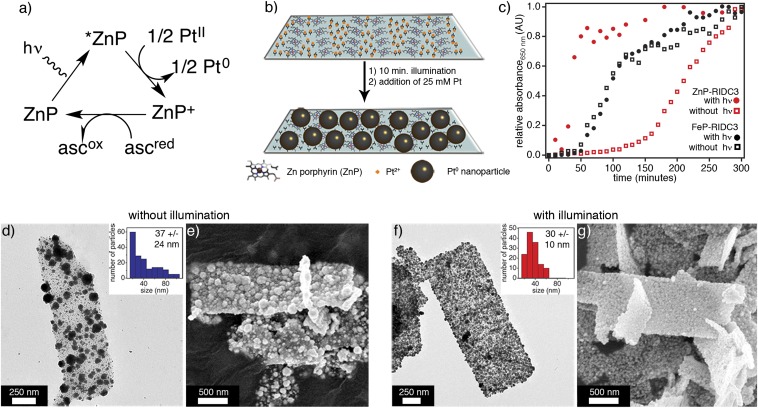
To investigate whether the heme centers within the RIDC3 arrays are actively involved in the reductive growth of PtNPs, we synthesized ZnP-RIDC3, in which the heme Fe center was replaced by Zn2+. The particular advantage of the Zn-porphyrin (ZnP) cofactor in the context of our experiments is that it is redox-inactive in its ground state but becomes a strong reductant/oxidant on UV-visible (UV-vis) excitation (45, 46). ZnP-RIDC3 was prepared in high purity according to established protocols for other cytochromes (47) and displayed the expected absorption and emission features for a ZnP species (Soret maximum at 426 nm, emission maxima at 594 and 645 nm; Fig. 5 A and B). It also formed the same Zn-mediated crystalline 2D arrays with near-identical lattice constants (a = 37 nm, b = 136 nm, α = 90°; Fig. 5 C and D) as the FeP counterpart, verifying that metal substitution did not affect the structure of the RIDC scaffold and its self-assembly.
Fig. 5.
Characterization of ZnP-RIDC3. UV-vis (A) and fluorescence (B) spectra indicate the substitution of heme Fe centers with Zn. Low- (C) and high- (D) magnification TEM images show that ZnP-RIDC3 forms 2D arrays similar to those obtained with FeP-RIDC3 (D and Inset).
2D ZnP-RIDC3 arrays were incubated with 5 mM Pt2+ overnight, after which unbound Pt and unassembled protein were removed by centrifugation. Pt2+-bound ZnP-RIDC3 arrays were then resuspended in a buffer solution that contained 50 mM ascorbate (now intended as a sacrificial electron donor in the photocatalytic scheme; Fig. 6A), but no additional Pt2+, and illuminated with white light for 10 min to create Pt0 seeds for nanoparticle growth at room temperature (Fig. 6B): 25 mM Pt2+ was then added to this suspension, and the growth of PtNPs was followed by an increase in absorbance at 650 nm (Fig. 6C) and by TEM (Fig. S9). The same procedure was carried out in parallel for a ZnP-RIDC3 sample that was not exposed to light and two FeP-RIDC3 samples that were either illuminated or not (Fig. 6C and Fig. S9).
Fig. 6.
Redox-controlled growth of Pt nanoparticles on RIDC3 surfaces via integrated heme centers. (A) Photocatalytic cycle for reduction of Pt2+ to Pt0 by ZnP. (B) Scheme for the photoredox-mediated growth of PtNPs on ZnP-RIDC3 arrays. (C) Growth kinetics of PtNPs on ZnP-RIDC3 arrays (red symbols) or FeP-RIDC3 arrays (black symbols) in the presence (solid symbols) or absence of light exposure (open symbols). (D) TEM or (E) SEM images of nonirradiated ZnP-RIDC3 arrays reveal a nonuniform coverage with relatively polydisperse PtNPs (D, Inset). (F and G) Images of irradiated arrays display uniform coverage with PtNPs that have a narrower size distribution (F, Inset).
The various one-to-one comparisons between these four samples in terms of their Pt0-nanoparticle growth kinetics are highly revealing (Fig. 6C). In the irradiated ZnP-RIDC3 sample, the PtNP growth commenced immediately and was nearly complete by ∼1 h, whereas in the nonirradiated ZnP sample, there was nearly a 2-h lag phase before growth. For the FeP-RIDC3 samples, light exposure did not have any effect on the PtNP growth as expected. However, for these samples, the growth kinetics were significantly faster compared with those of the nonirradiated ZnP-RIDC3 templates, but slower than those of the irradiated ZnP-RIDC3 sample. In addition, PtNPs on nonirradiated ZnP-RIDC3 arrays were highly irregular with a broad size distribution (37 ± 24 nm, with some particles exceeding 100 nm in diameter) and unevenly distributed over the array surfaces (Fig. 6 D and E). In contrast, the light-exposed ZnP-RIDC3 arrays supported the growth of smaller PtNPs with a tighter size distribution (30 ± 10 nm), and the surface coverage was uniform (Fig. 6 F and G). These observations are consistent with the following two conclusions:
-
i)
The built-in FeP cofactor of RIDC3, which has an Fe3+/2+ reduction potential of 0.18 V (24), acts as a redox conduit between ascorbate (E° ∼ 0.1 V at pH 5.5) (48) and the surface-bound Pt2+ ions, thus giving rise to more efficient and uniform nucleation/growth of PtNPs compared with that observed with the ground-state, redox-inactive ZnP-RIDC3 arrays. In the latter case, the nucleation and growth of PtNPs by necessity occurs through the bimolecular reduction of Pt2+ ions by ascorbate, which offers far less spatiotemporal control compared with the intramolecular reduction of surface-immobilized Pt2+ by integrated FeP cofactors, leading to a less uniform spatial and size distribution of PtNPs on their surfaces.
-
ii)
The triplet excited state of ZnP (3ZnP, E° ∼ –0.8 V) (49) is a significantly more potent reductant than FeP, which results in a more efficient/rapid generation of Pt0 seeds on the array surface and the growth of PtNPs without a detectable induction period (Fig. 6C).
Undoubtedly, a firm establishment of these conclusions will require a detailed examination of the various possible electron transfer pathways operative during templated PtNP nucleation and growth. Nevertheless, our findings clearly show the RIDC3 arrays are actively involved in the redox chemistry of these processes through their integrated heme cofactors.
Conclusions.
We described here the improved or emergent physical and functional properties that are derived from the designed, metal-directed supramolecular assembly of a monomeric redox protein, cyt cb562, into ordered, nanoscale 1D and 2D architectures. Notably, the conversion of cyt cb562 into these functional architectures is achieved by only 13 structurally innocuous surface mutations, which amount to just 11% of the amino acid sequence. The uniform 1D and 2D RIDC3 arrays can be reproducibly generated in high yields by a simple adjustment of solution conditions. Also, the 1D RIDC3 nanotubes can be cleanly converted into the thermodynamically more stable 2D sheets by heat, demonstrating stimuli responsiveness that is characteristic of evolved protein assemblies. The metal-mediated cooperative connectivity of Zn-RIDC3 assemblies not only leads to their stability as a whole but also the dramatic stabilization of their individual components that is crucial for bio/nano-technological applications that center on the specific activity of the protein building blocks. In the present case, the native electron transfer function of the RIDC3 building blocks was exploited to spatiotemporally control the growth of dense arrays of PtNPs, which also could be triggered by light. The redox-controlled growth of PtNPs absolutely required the supramolecular assembly of RIDC3, thus demonstrating an emergent functional property. Given that electron transfer is the simplest (but also the most fundamental) chemical reaction, it is exciting to contemplate the functional scope and applications of synthetic superprotein assemblies constructed from protein building blocks with more sophisticated functions.
Materials and Methods
The expression and purification of RIDC3 were carried out as previously described (22). For the preparation of ZnP-RIDC3, protocols previously used for Zn substitution into the cytochrome c heme center were adapted (47). For a more detailed description of experimental methods on ZnP-RIDC3 preparation, Zn-mediated self-assembly, nanoparticle growth, and physical characterization (EM, AFM, UV-vis, and circular dichroism spectroscopy), see SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the US Department of Energy Division of Materials Sciences, Office of Basic Energy Sciences, Award DE-FG02-10ER46677 (to F.A.T.). The electron microscopy facilities used in this work are supported by funding to Prof. Timothy S. Baker from the National Institutes of Health (Grant R37 GM-033050), the Agouron Foundation, and University of California, San Diego.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319866111/-/DCSupplemental.
References
- 1.Liu XF, Theil EC. Ferritins: Dynamic management of biological iron and oxygen chemistry. Acc Chem Res. 2005;38(3):167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 2.Schröder HC, et al. Silicatein: Nanobiotechnological and biomedical applications. Prog Mol Subcell Biol. 2009;47:251–273. doi: 10.1007/978-3-540-88552-8_11. [DOI] [PubMed] [Google Scholar]
- 3.Shimizu K, Cha J, Stucky GD, Morse DE. Silicatein alpha: Cathepsin L-like protein in sponge biosilica. Proc Natl Acad Sci USA. 1998;95(11):6234–6238. doi: 10.1073/pnas.95.11.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleytr UB, Messner P, Pum D, Sara M. Crystalline bacterial cell surface layers (S layers): From supramolecular cell structure to biomimetics and nanotechnology. Angew Chem Int Ed. 1999;38(8):1035–1054. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1034::AID-ANIE1034>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Flenniken ML, et al. A library of protein cage architectures as nanomaterials. Curr Top Microbiol Immunol. 2009;327:71–93. doi: 10.1007/978-3-540-69379-6_4. [DOI] [PubMed] [Google Scholar]
- 6.Mann S. Life as a nanoscale phenomenon. Angew Chem Int Ed Engl. 2008;47(29):5306–5320. doi: 10.1002/anie.200705538. [DOI] [PubMed] [Google Scholar]
- 7.Huard DJE, Kane KM, Tezcan FA. Re-engineering protein interfaces yields copper-inducible ferritin cage assembly. Nat Chem Biol. 2013;9(3):169–176. doi: 10.1038/nchembio.1163. [DOI] [PubMed] [Google Scholar]
- 8.King NP, et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336(6085):1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padilla JE, Colovos C, Yeates TO. Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci USA. 2001;98(5):2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai YT, Cascio D, Yeates TO. Structure of a 16-nm cage designed by using protein oligomers. Science. 2012;336(6085):1129. doi: 10.1126/science.1219351. [DOI] [PubMed] [Google Scholar]
- 11.Ringler P, Schulz GE. Self-assembly of proteins into designed networks. Science. 2003;302(5642):106–109. doi: 10.1126/science.1088074. [DOI] [PubMed] [Google Scholar]
- 12.Kitagishi H, et al. Supramolecular hemoprotein linear assembly by successive interprotein heme-heme pocket interactions. J Am Chem Soc. 2007;129(34):10326–10327. doi: 10.1021/ja073295q. [DOI] [PubMed] [Google Scholar]
- 13.Onoda A, Kakikura Y, Uematsu T, Kuwabata S, Hayashi T. Photocurrent generation from hierarchical zinc-substituted hemoprotein assemblies immobilized on a gold electrode. Angew Chem Int Ed Engl. 2012;51(11):2628–2631. doi: 10.1002/anie.201105186. [DOI] [PubMed] [Google Scholar]
- 14.Dotan N, Arad D, Frolow F, Freeman A. Self-assembly of a tetrahedral lectin into predesigned diamondlike protein crystals. Angew Chem Int Ed Engl. 1999;38(16):2363–2366. [PubMed] [Google Scholar]
- 15.Zhang W, et al. Self-assembly of glutathione S-transferase into nanowires. Nanoscale. 2012;4(19):5847–5851. doi: 10.1039/c2nr31244a. [DOI] [PubMed] [Google Scholar]
- 16.Ge J, Lei J, Zare RN. Protein-inorganic hybrid nanoflowers. Nat Nanotechnol. 2012;7(7):428–432. doi: 10.1038/nnano.2012.80. [DOI] [PubMed] [Google Scholar]
- 17.Wang LB, et al. A new nanobiocatalytic system based on allosteric effect with dramatically enhanced enzymatic performance. J Am Chem Soc. 2013;135(4):1272–1275. doi: 10.1021/ja3120136. [DOI] [PubMed] [Google Scholar]
- 18.Miranda FF, et al. A self-assembled protein nanotube with high aspect ratio. Small. 2009;5(18):2077–2084. doi: 10.1002/smll.200900667. [DOI] [PubMed] [Google Scholar]
- 19.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci USA. 2008;105(10):3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medina-Morales A, Perez A, Brodin JD, Tezcan FA. In vitro and cellular self-assembly of a Zn-binding protein cryptand via templated disulfide bonds. J Am Chem Soc. 2013;135(32):12013–12022. doi: 10.1021/ja405318d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burazerovic S, Gradinaru J, Pierron J, Ward TR. Hierarchical self-assembly of one-dimensional streptavidin bundles as a collagen mimetic for the biomineralization of calcite. Angew Chem Int Ed Engl. 2007;46(29):5510–5514. doi: 10.1002/anie.200701080. [DOI] [PubMed] [Google Scholar]
- 22.Brodin JD, et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat Chem. 2012;4(5):375–382. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sangster J. Octanol-water partition-coefficients of simple organic-compounds. J Phys Chem Ref Data. 1989;18(3):1111–1229. [Google Scholar]
- 24.Faraone-Mennella J, Tezcan FA, Gray HB, Winkler JR. Stability and folding kinetics of structurally characterized cytochrome c-b562. Biochemistry. 2006;45(35):10504–10511. doi: 10.1021/bi060242x. [DOI] [PubMed] [Google Scholar]
- 25.Lozano P, deDiego T, Iborra JL. Influence of water-miscible aprotic solvents on alpha-chymotrypsin stability. Biotechnol Prog. 1996;12(4):488–493. [Google Scholar]
- 26.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409(6817):253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Bishop EJ, Farid RS. A de novo designed protein with properties that characterize natural hyperthermophilic proteins. J Am Chem Soc. 1997;119(4):838–839. [Google Scholar]
- 28.Korkegian A, Black ME, Baker D, Stoddard BL. Computational thermostabilization of an enzyme. Science. 2005;308(5723):857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malakauskas SM, Mayo SL. Design, structure and stability of a hyperthermophilic protein variant. Nat Struct Biol. 1998;5(6):470–475. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- 30.Eijsink VG, et al. Rational engineering of enzyme stability. J Biotechnol. 2004;113(1-3):105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Hao J, Berry A. A thermostable variant of fructose bisphosphate aldolase constructed by directed evolution also shows increased stability in organic solvents. Protein Eng Des Sel. 2004;17(9):689–697. doi: 10.1093/protein/gzh081. [DOI] [PubMed] [Google Scholar]
- 32.Lele BS, Murata H, Matyjaszewski K, Russell AJ. Synthesis of uniform protein-polymer conjugates. Biomacromolecules. 2005;6(6):3380–3387. doi: 10.1021/bm050428w. [DOI] [PubMed] [Google Scholar]
- 33.Heredia KL, Maynard HD. Synthesis of protein-polymer conjugates. Org Biomol Chem. 2007;5(1):45–53. doi: 10.1039/b612355d. [DOI] [PubMed] [Google Scholar]
- 34.Ge J, Lu DN, Liu ZX, Liu Z. Recent advances in nanostructured biocatalysts. Biochem Eng J. 2009;44(1):53–59. [Google Scholar]
- 35.Margolin AL, Navia MA. Protein crystals as novel catalytic materials. Angew Chem Int Ed Engl. 2001;40(12):2204–2222. doi: 10.1002/1521-3773(20010618)40:12<2204::aid-anie2204>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 36.Stclair NL, Navia MA. Cross-linked enzyme crystals as robust biocatalysts. J Am Chem Soc. 1992;114(18):7314–7316. [Google Scholar]
- 37.Atabekov J, Nikitin N, Arkhipenko M, Chirkov S, Karpova O. Thermal transition of native tobacco mosaic virus and RNA-free viral proteins into spherical nanoparticles. J Gen Virol. 2011;92(Pt 2):453–456. doi: 10.1099/vir.0.024356-0. [DOI] [PubMed] [Google Scholar]
- 38.Mertig M, Ciacchi LC, Seidel R, Pompe W, De Vita A. DNA as a selective metallization template. Nano Lett. 2002;2(8):841–844. [Google Scholar]
- 39.Dujardin E, Peet C, Stubbs G, Culver JN, Mann S. Organization of metallic nanoparticles using tobacco mosaic virus templates. Nano Lett. 2003;3(3):413–417. [Google Scholar]
- 40.Chen CL, Rosi NL. Peptide-based methods for the preparation of nanostructured inorganic materials. Angew Chem Int Ed Engl. 2010;49(11):1924–1942. doi: 10.1002/anie.200903572. [DOI] [PubMed] [Google Scholar]
- 41.Behrens SS. Synthesis of inorganic nanomaterials mediated by protein assemblies. J Mater Chem. 2008;18(32):3788–3798. [Google Scholar]
- 42.Nam KT, et al. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312(5775):885–888. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- 43.Li YJ, Whyburn GP, Huang Y. Specific peptide regulated synthesis of ultrasmall platinum nanocrystals. J Am Chem Soc. 2009;131(44):15998–15999. doi: 10.1021/ja907235v. [DOI] [PubMed] [Google Scholar]
- 44.Glover DJ, Giger L, Kim JR, Clark DS. Engineering protein filaments with enhanced thermostability for nanomaterials. Biotechnol J. 2013;8(2):228–236. doi: 10.1002/biot.201200009. [DOI] [PubMed] [Google Scholar]
- 45.Brugger PA, Cuendet P, Gratzel M. Ultrafine and specific catalysts affording efficient hydrogen evolution from water under visible-light illumination. J Am Chem Soc. 1981;103(11):2923–2927. [Google Scholar]
- 46.McLendon G, Miller DS. Metalloporphyrins catalyse the photo-reduction of water to H2. J Chem Soc Chem Commun. 1980;(11):533–534. [Google Scholar]
- 47.Ensign AA, Jo I, Yildirim I, Krauss TD, Bren KL. Zinc porphyrin: A fluorescent acceptor in studies of Zn-cytochrome c unfolding by fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2008;105(31):10779–10784. doi: 10.1073/pnas.0802737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borsook H, Davenport HW, Jeffreys CEP, Warner RC. The oxidation of ascorbic acid and its reduction in vitro and in vivo. J Biol Chem. 1937;117(1):237–279. [Google Scholar]
- 49.Elias H, Chou MH, Winkler JR. Electron-transfer kinetics of Zn-substituted cytochrome c and its Ru(NH3)5(Histidine33) derivative. J Am Chem Soc. 1988;110(2):429–434. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.