Significance
Exposure to extremely stressful conditions is common, and the effect of such exposure on neuropsychiatric function is well-documented with posttraumatic stress disorder (PTSD). Epidemiological studies reveal a higher risk for cardiovascular conditions among individuals exposed to traumatic events. However, the underlying molecular mechanism for ailments associated with stress exposure is yet to be fully understood. Our study with animal models revealed genetically associated stress-induced tissue injuries on peripheral organs, including the heart. Longitudinal transcriptomics studies uncovered detailed molecular events involved in stress-related heart damage followed immediately by tissue-repairing processes; whether this injury and repairing process causes long-term effects is uncertain. Our findings on heart injury in a PTSD mouse model clearly indicate physiological changes arising from stress.
Keywords: systems biology, transcriptome, microRNA
Abstract
Posttraumatic stress disorder (PTSD) is a common condition induced by life-threatening stress, such as that experienced by soldiers under battlefield conditions. Other than the commonly recognized behavioral and psychological dysfunction, epidemiological studies have also revealed that PTSD patients have a higher risk of other diseases, such as cardiovascular disorders. Using a PTSD mouse model, we investigated the longitudinal transcriptomic changes in heart tissues after the exposure to stress through intimidation. Our results revealed acute heart injury associated with the traumatic experience, reflecting the underlying biological injury processes of the immune response, extracellular matrix remodeling, epithelial-to-mesenchymal cell transitions, and cell proliferation. Whether this type of injury has any long-term effects on heart function is yet to be determined. The differing responses to stress leading to acute heart injury in different inbred strains of mice also suggest that this response has a genetic as well as an environmental component. Accordingly, the results from this study suggest a molecular basis for the observed higher risk of cardiovascular disorders in PTSD patients, which raises the likelihood of cardiac dysfunction induced by long-term stress exposures.
In a modern society, the overall lifetime probability of an individual being exposed to a traumatic event, such as a traffic accident, physical assault, or combat situation, is 40–90% in the general population (1). However, the overall lifetime chance of an individual suffering posttraumatic stress disorder (PTSD) is lower, about 7–12% (1, 2). At any given time, about 7% of the population in the United States suffers from some form of PTSD (3). Hence, PTSD is perhaps the most common psychological disorder. The fact that not all individuals develop PTSD after experiencing one or more traumatic events suggests there may also be a genetic contribution to the development of the disease.
Patients with PTSD frequently show sociobehavioral problems and have a higher risk of drug or alcohol abuse. In addition, patients with PTSD often display structural changes in the prefrontal cortex, the amygdala, and the hippocampus and biochemical changes, such as a higher ratio of norepinephrine/cortisol in urine, higher total cholesterol levels in the blood, and elevated norepinephrine levels in the cerebrospinal fluid (4–8). These findings indicate a disruption of the normal hypothalamic–pituitary–adrenal function and the autonomic nervous system in PTSD patients (9). A systems approach to the study of the molecular changes associated with a stress-induced, PTSD-like mouse model will allow us to gain a better understanding of the disease. It also allows us to evaluate the effects of PTSD-like stress exposure on organs other than the brain. This systems approach is embedded in the concept that disease arises as a consequence of disease-perturbed networks in relevant tissues initiated by either genetic and/or environmental stimuli—and the fact that we can study dynamically, from onset to the final stage of the disease, the behavior of these disease-perturbed networks through -omic analyses in relevant tissues from animal models and subsequently, relate these disease-perturbed dynamical networks to the pathophysiology of the disease (10, 11). This approach may lead to more informative diagnostic markers for identifying the disease early, provide information as to which organs are disease-involved, and provide insights into therapeutic approaches for reversing the progression of the disease.
Individuals with PTSD also have a higher risk of cardiovascular conditions, with an increased basal heart rate and blood pressure, higher risk for hypertension and stroke, altered platelet activity, and elevated blood cholesterol and triglyceride levels (12–15). An 1871 report noted that serious cardiac disorders (cardiomyopathies, heart failure, heart pain, etc.) were a consequence of extended stress exposures in soldiers from the US Civil War (16). This report led to our interest in examining the effects of short-term stress exposure on heart tissues in the PTSD mouse model.
We adapted an aggressor-exposed social stress mouse model for PTSD by pairing aggressor mice (SJL strain) with different strains of subservient mice. This model generated a simple and reproducible set of PTSD-associated conditions in the subservient mice, such as avoidance behaviors (17). The reporting of myocarditis in short-term stressed subservient mice suggests that stress-induced PTSD may also cause transitory heart injury. Through a detailed longitudinal transcriptomic study [mRNAs and microRNAs (miRNAs)] of the heart tissues in the PTSD mouse, we observed the involvement of key molecular processes, including an inflammatory response, extracellular remodeling and, epithelial-to-mesenchymal transitions (EMTs), in PTSD-induced heart injury. These findings may provide possible molecular explanations for the higher risk of cardiovascular diseases in patients with PTSD.
Results
We designed our initial study (study I) with four experimental conditions, in which we varied the length of time that subservient mice were exposed to aggressor mice as well as the length of rest time after the exposure [number of days of trauma exposure (T); number of days of rest after exposure (R)]. The conditions included were short exposure–short rest (T5R1), short exposure–long rest (T5R10), long exposure–short rest (T10R1), and long exposure–long rest (T10R28) (Table 1). SJL mice were used as the aggressor strain, and C57BL/6j, DBA/2j, and BALB/cj mice were used as subservient strains. Multiple subservient strains were used to identify the shared features of this stress exposure and be able to subtract away the strain-specific (biological or genetic) features—an important aspect of using a systems approach to reduce biological noise associated with the strain-specific features of the response. We were also interested in whether the different strains exhibited different responses to stress under similar environmental conditions—thus implying a genetic contribution. The social stress mouse model produced some of the behavioral and physiological changes commonly observed in human PTSD patients, including avoidance behavior, increased body weight, decreased memory, and decreased prefrontal cortex volume (17). Evaluating the cardiac histopathology data, the C57BL/6j strain showed the strongest pathological effects among the three subservient strains, suggesting a genetic contribution to the susceptibility to develop PTSD-associated heart conditions. Except for myocarditis and vasculitis associated with the heart tissues (Fig. S1), there was no additional observable damage in other major organs in subservient C57BL/6j mice (or indeed, the organs of the other strains) during pathological examination. Therefore, we conducted a detailed transcriptome study, including miRNA and mRNA analyses, on the C57BL/6j heart tissues. We also conducted limited transcriptome studies on tissues from the other two strains.
Table 1.
Summary of experimental conditions
| Experimental group | Exposure (d) | Rest (d) |
| Study group I* | ||
| T5R1 | 5 | 1 |
| T5R10 | 5 | 10 |
| T10R1 | 10 | 1 |
| T10R28 | 10 | 28 |
| Study group II† | ||
| T1R1 | 1 | 1 |
| T2R1 | 2 | 1 |
| T3R1 | 3 | 1 |
Three subservient mouse strains were used: C57BL/6j, DBA/2j, and BALB/Cj.
C57BL/6j strain was used for subservient mice.
Perturbation of Heart Transcriptome Associated with Stressed Mice.
Agilent microarrays were used to analyze the heart transcriptomes of control and stressed mice, and significant changes in a number of gene expression levels were found in the short-term rest groups. We observed significant changes in the expression level of 455 differentially expressed genes (DEGs) from the T5R1 group and 40 genes from the T10R1 group, with 31 shared transcripts, 29 of which were up-regulated [based on greater than ±1.5-fold changes with false discovery rate (FDR) < 0.1]. The expression-level changes for eight genes were verified by quantitative PCR (Fig. S2). No genes showing statistically significant changes from animals in the two long rest groups (T5R10 and T10R28) were identified, which suggests an active ongoing tissue repair process in the heart that is complete by 10 d. The T10R1 group showed fewer changes between the control and stressed mice compared with the T5R1 group, which indicates that a similar repair process seems to be initiated early and is functioning even under the stress environment (from 455 DEGs to 40 DEGs). This contention is supported by the fact that the changing expression patterns of the DEGs identified in the T5R1 group are similar to those patterns seen in the T10R1 group, despite the observation that most of them did not show greater than ±1.5-fold changes and had an FDR < 0.1 (hence, missing our selection criteria for DEGs; only 31 of 455 genes were classified as DEGs in T10R1). Enrichment analyses of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and gene ontology (GO) terms with 377 up-regulated genes identified between the two groups (of 464 DEGs total) revealed a strong association of cell cycle and ECM remodeling activities—consistent with active tissue repair processes. The full list of enriched GO terms is given in Tables S1 and S2. No significantly enriched pathways or GO terms were associated with 87 down-regulated genes identified from either the T5R1 or T10R1 group. The 31 shared DEGs between T5R1 and T10R1 groups showed enrichment in ECM-related functions, including cell adhesion and collagen fibril organization. No group-specific enrichment analysis was performed, because the entire list of DEGs showed similar changing expression profiles between the two groups, despite the fact that the majority of them did not show as DEGs in the T10R1 group based on our criteria.
The levels of a number of ECM-related genes, such as various collagen transcripts (Col1a2, Col4a1, Col4a2, and others), fibronectins (Fn1 and Fndc1), the matrix metalloprotease gene (Mmp2), and its inhibitor (Timp1), were increased in the heart tissues from the T5R1 and T10R1 stressed mice, supporting the idea that an active ECM remodeling process is occurring. Several immune response-related genes, including chemokines, chemokine receptors (Ccl8, Cxcl10, Cx3cr1, and Ccrl1), and integrins (Itga11 and Itgbl1), were also increased under the short-term rest condition (Fig. S3).
miRNA analysis on the heart tissues was performed, and we identified three miRNAs (miR-29b, miR-302a, and let-7d) that showed a statistically significant concentration decrease (greater than ±1.5-fold changes with FDR < 0.1) in the short exposure and short rest group (T5R1). No significant change in miRNA expression levels was observed in the other experimental conditions. The miR-29 family has three known members in mice (miR-29a, miR-29b, and miR-29c), and all of them were decreased in T5R1, although only miR-29b was identified as differentially expressed based on our selection criteria (Fig. S4). The miRNAs that are highly enriched in heart tissues, such as miR-208a, miR-208b, and miR-499, did not show any significant concentration changes.
We could not find any observable pathological changes in the other two mouse strains used in the experiment (DBA/2j and BALB/cj) compared with C57BL/6j (17). Influence on the degree of phenotypic changes by genetic background has been reported in the literature; for example, C57BL/6j is more susceptible to diet-induced atherosclerosis than the BALB/cj or DBA/2j strain (18). We did not observe any significant pathological changes associated with BALB/cj, but a number of DEGs identified in C57BL/6j heart tissues also showed similar changing patterns in BALB/cj heart tissues, although the magnitude of changes did not rise to the criteria for DEGs (Fig. S5). Similar to the C57BL/6j strain, most of the changes were observed in the short rest groups (T5R1 and T10R1). This observation implies that some genetic factors may also affect the susceptibility to stress-related tissue injuries in the heart.
The dynamical mRNA and miRNA profiling results on the four experimental groups from study I (Table 1) suggested that the tissue injury observed in the hearts probably occurred shortly after exposure to stress. To determine when the injury occurred, we conducted a series of shorter stress exposures with C57BL/6j subservient mice (study II) with 1, 2, and 3 d of exposure and 1 d of rest for all of the exposure conditions (i.e., T1R1, T2R1, and T3R1) (Table 1). The transcriptomic analysis of the short exposure study II heart samples revealed that 494 genes were significantly affected (based on greater than ±twofold changes with FDR < 0.1): 422 genes increased, and 72 genes decreased. Although only 19 (7 up- and 12 down-regulated) DEGs were identified in the T1R1 group, 250 (198 up- and 52 down-regulated) and 466 (405 up- and 61 down-regulated) genes were significantly changed in the T2R1 and T3R1 groups, respectively (Fig. S6A). Most of the DEGs identified in the T2R1 group were also seen in the T3R1 group (227 of 250) (Fig. S6B). This finding indicates that the stressful condition immediately affects the expression levels of some of the genes and that the number of perturbed genes increases linearly with the duration of stress exposure. This finding also indicates that some of the early DEGs are also seen in the longer trauma exposures.
In study II, we did not find any significantly enriched pathways or GO terms associated with 72 down-regulated genes. For 422 up-regulated DEGs identified in study II, a number of pathways associated with the immune response and tissue remodeling emerged, including chemokine signaling pathway, Fc-γ receptor-mediated phagocytosis, phagosome, ECM–receptor interaction, and focal adhesion. In addition, cell proliferation-related pathways, such as the cell cycle and DNA replication pathways, also showed strong associations. The results of GO term enrichment analysis identified biological processes that are similar to the pathway enrichment analysis, which includes terms related with immune response, cell cycle, and ECM remodeling. In summary, inflammatory response, chemotaxis, cell division, collagen fibril organization, and ECM organization were among the most significantly enriched biological processes in these PTSD mouse model studies (Tables S3 and S4).
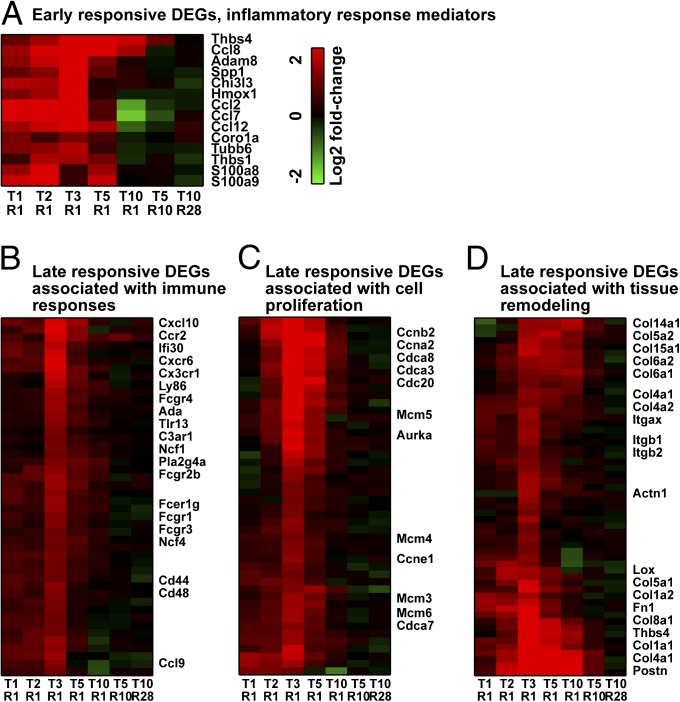
The increase of several inflammatory response mediators, including chemokines and S100 protein coding genes in the T1R1 group, indicates that a strong inflammatory signal is associated with the initial tissue injury (Fig. 1A). At later time points (from T2R1 to T5R1), genes related to cell cycle and tissue remodeling processes in addition to the immune response dominate the set of up-regulated DEGs (Fig. 1B). Although the changes representing immune system responses diminished immediately after T3R1, those changes related with cell cycle (proliferation) and tissue remodeling were sustained through T5R1 and T10R1, respectively (Fig. 1 C and D). Interestingly, most of the gene expression changes had disappeared in the longer resting groups (T5R10 and T10R28). These observations again illustrate that there is an active tissue repair process.
Fig. 1.
Heat maps of DEGs representing different biological processes. DEGs represent early [(A) inflammatory response mediators] and late responses [(B) immune response, (C) cell cycle, and (D) tissue remodeling across different lengths of stress exposure]. Red and green indicate higher and lower expression levels, respectively. The gene identities are listed on the right, and the experimental conditions are indicated at the bottom. Ccn, cyclin; Cdc, cell division cycle; Col, collagen.
miR-29 and E2F Gene Family Members Are the Key Regulators Involved in PTSD Heart Pathology.
Comparing 494 DEGs identified from study II (i.e., early responsive) with 464 DEGs observed in study I (i.e., late responsive), we found 245 genes in common. We did three different functional enrichment analyses, including the study I-specific, study II-specific, and common DEGs between the two studies analyses. Genes involved in immune responses and cell cycle/proliferation processes were strongly enriched in the common DEGs. Study II-specific DEGs (early response genes) showed a strong association with immune response terms, whereas study I-specific DEGs (late response genes) showed a moderately enriched GO term representing ECM organization (Table S5).
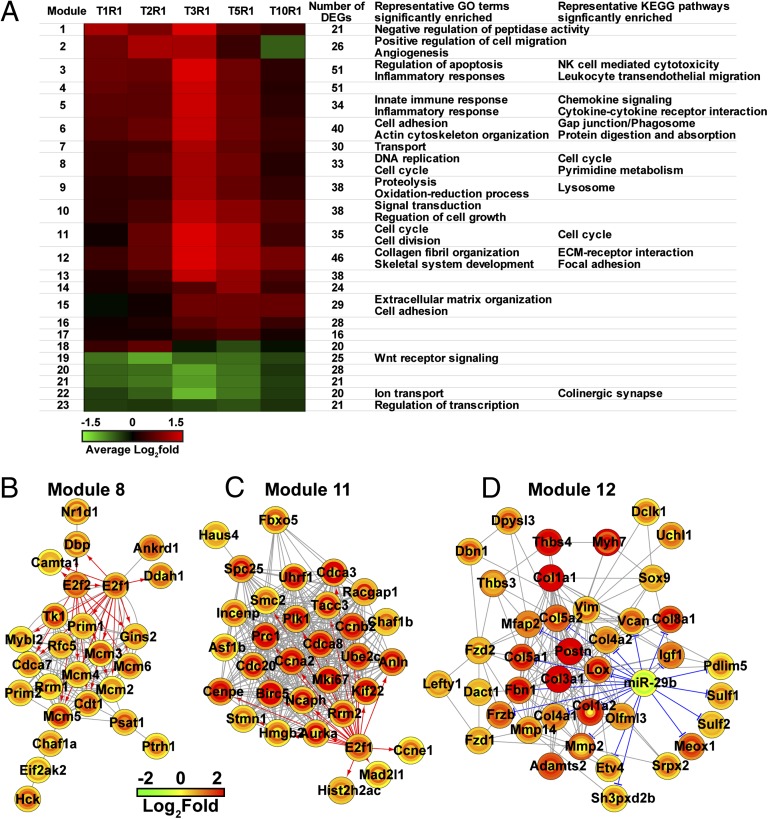
Merging the two sets of DEGs, we identified a total of 713 unique DEGs that might be involved in stress-induced acute heart injury. To gain a global view of biological effects associated with 713 DEGs, we used semantic similarity-integrated approach for modularization that integrated expression profiles, protein–protein interactions, and GO terms to construct gene modules involved in acute heart injury (19). This process produced 23 functional gene modules covering all major biological activities described earlier, including immune responses, cell proliferation, and tissue remodeling. The modules related to inflammatory responses (for example, modules 3 and 5) were composed of up-regulated DEGs at early stages (from T1R1 to T3R1), whereas modules associated with tissue remodeling (modules 12 and 15) were mostly comprised of up-regulated DEGs at later stages (from T3R1 to T10R1) (Fig. 2A). DEGs associated with cell proliferation (modules 8 and 11) showed an expression pattern similar to inflammatory response-related modules. This overall picture revealed that a typical wound-healing process occurred in the heart tissues of stressed mice over time: the initial insult induces inflammatory responses followed by cell proliferation and ECM organization-related processes.
Fig. 2.
Using the semantic similarity-integrated approach for modularization approach, 713 DEGs associated with stress-induced heart injury were grouped into functional modules. (A) The mean fold change profiles of the modules are depicted by a heat map, and the biological processes associated with each module are listed next to each heat map. NK, natural killer. We also constructed hypothetical regulatory networks for three selected modules regulated by (B and C) TFs and (D) miRNA. Expression-level changes of individual genes across different experimental conditions are expressed in concentric circles, where the innermost circles represent the fold changes between stressed and control samples in the shortest exposure group (T1R1) and the outermost circles represent the longest exposure group (T10R1).
Using databases containing predicted and validated interactions between genes and their transcriptional regulators [transcription factors (TFs) and miRNAs], we identified 49 putative regulators for 713 DEGs identified in either study I or II. The 49 putative regulators include 37 TFs and 12 miRNAs. Among them, only E2F transcription factor 1 and 2 (E2f1 and E2f2) (gradual increase in their levels from T1R1 to T5R1) and miR-29b (a significant decrease in its level in T5R1) showed statistically significant dynamical concentration changes in our data. Based on the predicted interactions in the databases, gene module 8 (enriched and representative of DNA replication and cell cycle-related GO terms and KEGG pathways) is regulated by both E2f1 and E2f2 (Fig. 2B), gene module 11 (representing cell cycle and cell division) is regulated by E2f1 (Fig. 2C), and gene module 12 (enriched with ECM remodeling-related genes) is regulated by miR-29b (Fig. 2D). Several E2F TF family members are known to be involved in the EMT process, a process in which cells lose epithelial traits and acquire mesenchymal characteristics, such as fibroblastic morphology and enhanced motility, a key process involved in tissue repair.
To verify the involvement of EMT in PTSD-associated heart injury, we conducted gene expression analyses on TGF-β–treated A549 cells, a well-known in vitro EMT model system. The idea was to identify what fraction of the DEGs from the in vitro EMT model was also seen in the PTSD model. Expression analysis revealed that 629 transcripts showed temporal expression changes during TGF-β treatment (based on greater than ±1.5-fold changes with FDR < 0.1). Compared with 713 DEGs identified in the heart tissues, 52 genes (44 genes were up-regulated and 8 genes were down-regulated) (SI Materials and Methods) showed similar expression changes in DEGs in A549 cells undergoing TGF-β–induced EMT (Fig. S7). The results from GO term enrichment analysis showed a strong enrichment of EMT-related terms (FDR < 0.1), such as cell migration, positive regulation of cell migration, and angiogenesis for 52 shared DEGs.
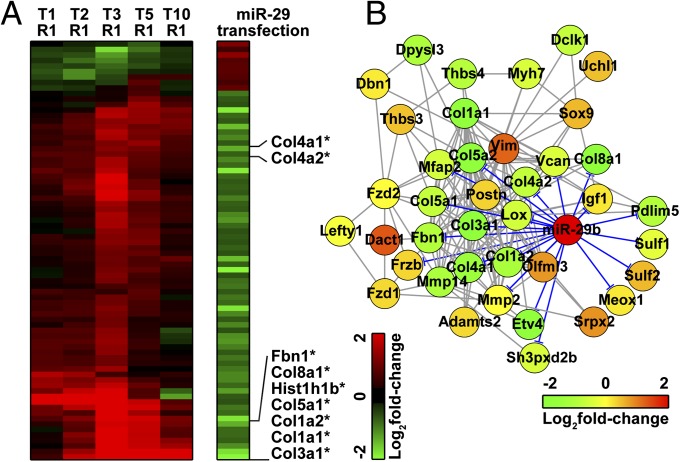
To validate the interactions of miR-29 and its predicted targets, we conducted transcriptome analyses on a mouse fibroblast cell line that was transfected with a miR-29 miRNA mimic. Among 713 DEGs identified in the heart tissues, 76 genes showed strong correlation with transcripts affected by miR-29 transfection (Fig. 3A). Pathway and GO term enrichment analyses on 76 shared DEGs revealed a strong association with tissue remodeling and wound-healing processes (Tables S6 and S7). The levels of several ECM-associated genes (grouped in module 9), including various collagen and fibronectin genes, were strongly affected by the increase of the miR-29 level in the cells (Fig. 3B).
Fig. 3.
(A) Heat map of genes that might be associated with miR-29 expression level. Among the 713 DEGs identified in heart tissues, 76 DEGs showed association with miR-29 transfection. Genes involved in module 12 are indicated by asterisks. The effect of miR-29 transfection on the gene module 12 is shown in B. Expression-level changes of individual genes across different experimental conditions are expressed in concentric circles, where the innermost circles represent the fold changes between stressed and control samples in the shortest exposure group (T1R1) and the outermost circles represent the longest exposure group (T10R1).
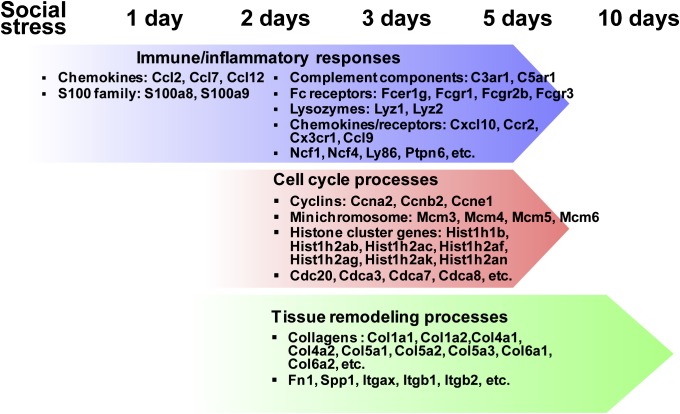
Fig. 4 summarizes the molecular processes in response to acute heart injury induced by the exposure to a traumatic event. Inflammatory signaling molecules, such as S100 proteins and chemokines, initiate the tissue repair process followed by cell proliferation, EMT, and ECM remodeling. By day 10, even under the long exposure of stress environment, most of the repair process has already been completed, which has been suggested by the return of most of the DEGs to their normal expression levels at T10R1 (Fig. 1).
Fig. 4.
A summary of biological process associated with stress-induced heart injury. Molecular processes are perturbed in response to acute heart injury in the order of inflammatory responses, cell cycle and proliferation, and tissue remodeling processes.
A Possible Systemic Response to Stress Exposure.
Because heart disorders are commonly associated with kidney conditions in human patients, we also investigated possible molecular changes in the kidney. Although we did not find statistically significant DEGs in C57BL/6j kidneys with microarray-based transcriptome profiling, we did observe a number of transcripts that showed low-level dynamical expression patterns similar to those DEGs identified in heart tissue. Examples of such DEGs are changes in the levels of some of the ECM-related transcripts in the kidney (Fig. S8). Although the magnitude of expression changes for these genes in the kidney was less than those changes in the heart, it suggests a possible system-wide response to acute stress exposure in our animal model.
Discussion
The association between PTSD and heart disorders is well-documented (12–15); however, some of these associations have been attributed to anxiety disorder-associated symptoms instead of heart tissue damage, such as found in Da Costa syndrome (soldier’s heart) (16). The mouse model used in these studies clearly displays some of the behavior and psychological characteristics of PTSD in human patients. Our findings on heart injury in a PTSD mouse model clearly indicate physiological changes in the heart arising from stress in addition to the psychological effects of PTSD. It is intriguing to observe the connection between psychical stress and tissue damage, which is probably mediated by the changes of the neuroendocrine system during the exposure of extreme stress environment. For example, the release of epinephrine significantly affects heart rate, blood vessel radius, breath pattern, and digestion.
We observed changes in the expression levels of a number of genes in the heart after the mice had been exposed to traumatic stress. Although most of the changes in the heart transcriptome occurred after 3 d of exposure (T3R1) (Fig. S6), we could clearly see some stress-induced effects, such as the up-regulation of a battery of inflammation-related genes, after as little as 1 d of exposure to stress (Fig. 1A). Most of the affected genes returned to normal levels in the longer exposure groups (T10R1 and T10R28), indicating that the repair process begins early and is underway during the longer exposures to stress. Histopathological examination of the C57BL/6j stress-treated mice also failed to identify any observable heart or other tissue damage in the T10R28 group. In addition to the heart, we also observed similar, although less extensive, molecular changes occurring in the kidney, although no gross pathological change could be identified. Whether the damage in the kidney is a direct effect caused by a stressful environment or an indirect effect induced by the short-term heart pathology is yet to be determined.
Surprisingly, longer exposure to the traumatic environment did not increase the severity of the tissue injury within our animal model. Most of the perturbed genes in shorter exposure groups (T1R1 to T5R1) reverted to normal levels in the T10R1 mice. These findings suggest that (i) traumatic stress conditions can induce acute tissue damage, (ii) the damage is reversible at the molecular level, (iii) there is an ongoing active tissue repair process even during ongoing exposure to the stressful conditions, and (iv) animals may be able to adapt to the stressful conditions at molecular levels. However, the initial heart injury followed by a quick tissue repair process may have long-term functional implications on the heart and provide some molecular bases for conditions, such as Da Costa syndrome. Indeed, the study by Da Costa (16) showed long-term and irreversible changes in the hearts of soldiers exposed to the stressful conditions of the US Civil War over periods of 1–3 y. Perhaps really long-term stress exposures lead directly to irreversible changes in the heart, which is reflected by heart pain, cardiac arrhythmias, and heart failure. In addition, our global transcriptome analyses on the entire heart tissue cannot exclude the possibility that there is small and local tissue damage in the long exposure group. Although an active tissue repair process was observed in our PTSD animal model, the likelihood of permanent and irreversible heart damage resulting from the exposure of a long-term constant or repeated stressful condition (such as soldiers with 1–3 y of combat duty in the US Civil War) could not be excluded (16). Nevertheless, the mouse model used in this study provides a good model to study the short-term physiological changes resulting from traumatic event exposure. Future studies of longer durations of rest will better separate acute and chronic responses to stress environment.
Wound healing in skin has been studied extensively, and several critical molecular processes, such as an inflammatory response, EMT, and ECM remodeling, have been identified (20, 21). Similar injury-healing processes were also observed in the heart tissues of our PTSD mouse model. As with skin, the immune mediators probably initiate the healing process, which was illustrated by the increase in expression levels of several cytokines and S100 family genes in the 1-d exposure group (T1R1) (Fig. 1A). Other than the inflammatory response, an increase in cell proliferation activities and tissue remodeling processes became evident in the T2R1 group, which was shown by the up-regulation of several cyclin, cell division cycle, and collagen genes. The activities of these wound-healing processes are gradually decreased (presumably because the healing process is finished) over time, even under the stress environment, which was evident by the decreased number of perturbed genes in the 10-d exposure group (Figs. 1 and 4).
The study identified several key gene regulators, members from the miR-29 family and E2F TF family (miR-29b, E2f1, and E2f2, respectively) that are involved in the wound-healing process (Fig. 2). The miR-29 family members have been implicated in arrhythmias, myocardial fibrosis, and other heart conditions (22–24). In addition, the miR-29 and E2F family members are known to be involved in ECM remodeling and cell proliferation, respectively (25, 26). The E2F TF family contains nine members: E2F1, E2F2, and E2F3a are transcription activators, and the rest of the members are transcriptional suppressors (27). The balance of activation and suppression activities is critical for the precise control of cell proliferation. Interrupting the balance results in deregulated cell cycle activities, which were evident in many different tumors (28, 29). Using miR-29–transfected fibroblasts, we can identify a fraction of the DEGs (76 of 713 genes) in the heart that may be directly or indirectly affected by the miR-29 levels. Most of these genes are involved in reshaping the ECM (Fig. 3A)—a picture consistent with the pathophysiology described in the hearts of the PTSD-like mice.
Using a well-known in vitro EMT model system (TGF-β–treated A549 cells), we found that a subset of DEGs identified in the heart tissues (52 of 713 genes) exhibited similar expression changes, such as observed in the EMT model. This finding shows the involvement of EMT in the heart tissue repair, which was reported for the skin. Cells from epicardium probably play a key role in the EMT process (30, 31).
Maintaining the proper ECM structure is critical to preserving the architecture and proper function of the heart (32). We observed an increase in the expression levels for a number of molecules involved in maintaining the ECM structure—serine proteases, serum protease inhibitors, matrix metalloproteinases, and metalloproteinase inhibitor—in the course of our PTSD experiments in mice. The balance of protease and protease inhibitor activities is important to maintain the integrity of the ECM structure. Unlike skin, the heart is an iteratively dynamic and constantly flexing organ, and the integrity of the ECM is critical for the function of the heart. The involvement of epicardium in EMT may provide insights as to how the heart repairs damage while maintaining its essential function of pumping blood.
Adenosine, an important signaling molecule, affects an array of cardiovascular activities. Elevating the adenosine level in the heart causes vasodilation and an increase in heart rate (33). Adenosine has been implicated in the initial injury process, because it is a well-known signaling molecule for stress and tissue injury (34–36). From this study, we observed a gradual increase in the adenosine deaminase transcript level (peaking in the T3R1 group). This observation may imply a compensatory process in heart tissue to reduce the adenosine level after the tissue is in the wound-healing stage. A recent report showed that athletes competing in endurance races, such as marathons, suffered injury to the heart (particularly, the right ventricle) (37). This injury was probably caused by prolonged exposure to stressful conditions during racing, although the source of stress is different between athletes and people suffering from PTSD. The finding of acute heart injury in our PTSD animal model suggests common stress-induced heart impairment. A genetic influence on the effects of exposure to stress environment is evident based on population studies (1, 2) as well as our mouse model; the three inbred mouse strains react very differently to stress (from dramatic responses to almost no response). It would be of great interest to identify genetic factors that may be involved in stress-induced heart injury. The mouse models of the different inbred strains used in our study may provide a simple and reproducible model to study the genetic contributions to stress-induced heart injury as well as delineate the molecular mechanisms involved in this process.
The finding of heart injury in the PTSD mouse model is intriguing, because extremely stressful conditions are quite common in society. Although our findings suggest that an immediate tissue repair process after an acute injury is induced by stress, whether this injury causes any long-term effects remains to be seen. Moreover, the effects of repeated exposures to a stressful environment on the heart are still unknown—as are the effects of chronic long-term stress.
Materials and Methods
Details are described in SI Materials and Methods. Included are animals and social defeat model, pathological evaluation, tissue and RNA isolation, microarray data generation and analysis, identification of functional modules, and construction of hypothetical regulatory networks. Data analysis procedures for miR-29 transfection and EMT model studies are also represented.
Supplementary Material
Acknowledgments
We thank Bruz Marzolf, Pamela Troisch, Yue Yuan, and Sara McClarty at the Institute for Systems Biology for technical support. We also thank Linda Brennan, Julia Scheerer, and Duncan Donohue at the US Army Center for Environmental Health Research for editing the paper. This study is supported by Department of Defense Research Contracts W911NF-09-D0001 and W911SR-07-C0101 and the Institute for Systems Biology–University of Luxembourg program.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE52875).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400113111/-/DCSupplemental.
References
- 1.Breslau N. The epidemiology of posttraumatic stress disorder: What is the extent of the problem? J Clin Psychiatry. 2001;62(Suppl 17):16–22. [PubMed] [Google Scholar]
- 2.Stein MB, Jang KL, Taylor S, Vernon PA, Livesley WJ. Genetic and environmental influences on trauma exposure and posttraumatic stress disorder symptoms: A twin study. Am J Psychiatry. 2002;159(10):1675–1681. doi: 10.1176/appi.ajp.159.10.1675. [DOI] [PubMed] [Google Scholar]
- 3.Kessler RC, et al. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 4.Kroes MC, Rugg MD, Whalley MG, Brewin CR. Structural brain abnormalities common to posttraumatic stress disorder and depression. J Psychiatry Neurosci. 2011;36(4):256–265. doi: 10.1503/jpn.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawk LW, Dougall AL, Ursano RJ, Baum A. Urinary catecholamines and cortisol in recent-onset posttraumatic stress disorder after motor vehicle accidents. Psychosom Med. 2000;62(3):423–434. doi: 10.1097/00006842-200005000-00016. [DOI] [PubMed] [Google Scholar]
- 6.Solter V, Thaller V, Karlović D, Crnković D. Elevated serum lipids in veterans with combat-related chronic posttraumatic stress disorder. Croat Med J. 2002;43(6):685–689. [PubMed] [Google Scholar]
- 7.Tochigi M, et al. Serum cholesterol, uric acid and cholinesterase in victims of the Tokyo subway sarin poisoning: A relation with post-traumatic stress disorder. Neurosci Res. 2002;44(3):267–272. doi: 10.1016/s0168-0102(02)00146-3. [DOI] [PubMed] [Google Scholar]
- 8.Geracioti TD, Jr, et al. CSF norepinephrine concentrations in posttraumatic stress disorder. Am J Psychiatry. 2001;158(8):1227–1230. doi: 10.1176/appi.ajp.158.8.1227. [DOI] [PubMed] [Google Scholar]
- 9.Jones T, Moller MD. Implications of hypothalamic-pituitary-adrenal axis functioning in posttraumatic stress disorder. J Am Psychiatr Nurses Assoc. 2011;17(6):393–403. doi: 10.1177/1078390311420564. [DOI] [PubMed] [Google Scholar]
- 10.Hwang D, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin S, et al. SRM targeted proteomics in search for biomarkers of HCV-induced progression of fibrosis to cirrhosis in HALT-C patients. Proteomics. 2012;12(8):1244–1252. doi: 10.1002/pmic.201100601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kibler JL. Posttraumatic stress and cardiovascular disease risk. J Trauma Dissociation. 2009;10(2):135–150. doi: 10.1080/15299730802624577. [DOI] [PubMed] [Google Scholar]
- 13.von Känel R, et al. Non-fatal cardiovascular outcome in patients with posttraumatic stress symptoms caused by myocardial infarction. J Cardiol. 2011;58(1):61–68. doi: 10.1016/j.jjcc.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Kubzansky LD, Koenen KC. Is posttraumatic stress disorder related to development of heart disease? An update. Cleve Clin J Med. 2009;76(Suppl 2):S60–S65. doi: 10.3949/ccjm.76.s2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedersen SS, Middel B, Larsen ML. Posttraumatic stress disorder in first-time myocardial infarction patients. Heart Lung. 2003;32(5):300–307. doi: 10.1016/s0147-9563(03)00097-9. [DOI] [PubMed] [Google Scholar]
- 16.Da Costa JM. On irritable heart; a clinical study of a form of functional cardiac disorder and its consequences. Am J Med Sci. 1871;121(1):2–52. [Google Scholar]
- 17.Hammamieh R, et al. Murine model of repeated exposures to conspecific trained aggressors simulates features of post-traumatic stress disorder. Behav Brain Res. 2012;235(1):55–66. doi: 10.1016/j.bbr.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis. 1985;57(1):65–73. doi: 10.1016/0021-9150(85)90138-8. [DOI] [PubMed] [Google Scholar]
- 19.Cho JH, Wang K, Galas DJ. An integrative approach to inferring biologically meaningful gene modules. BMC Syst Biol. 2011;5:117. doi: 10.1186/1752-0509-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura M, Tokura Y. Epithelial-mesenchymal transition in the skin. J Dermatol Sci. 2011;61(1):7–13. doi: 10.1016/j.jdermsci.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Yan C, et al. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-alpha through bone morphogenic protein-2. Am J Pathol. 2010;176(5):2247–2258. doi: 10.2353/ajpath.2010.090048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W, et al. MicroRNA expression analysis: Clinical advantage of propranolol reveals key microRNAs in myocardial infarction. PLoS One. 2011;6(2):e14736. doi: 10.1371/journal.pone.0014736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Y, Perez-Polo JR, Qian J, Birnbaum Y. The role of microRNA in modulating myocardial ischemia-reperfusion injury. Physiol Genomics. 2011;43(10):534–542. doi: 10.1152/physiolgenomics.00130.2010. [DOI] [PubMed] [Google Scholar]
- 24.van Rooij E, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105(35):13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Souza SJ, et al. E2F-1 is essential for normal epidermal wound repair. J Biol Chem. 2002;277(12):10626–10632. doi: 10.1074/jbc.M111956200. [DOI] [PubMed] [Google Scholar]
- 26.Villarreal G, Jr, Oh DJ, Kang MH, Rhee DJ. Coordinated regulation of extracellular matrix synthesis by the microRNA-29 family in the trabecular meshwork. Invest Ophthalmol Vis Sci. 2011;52(6):3391–3397. doi: 10.1167/iovs.10-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierce AM, Schneider-Broussard R, Philhower JL, Johnson DG. Differential activities of E2F family members: Unique functions in regulating transcription. Mol Carcinog. 1998;22(3):190–198. doi: 10.1002/(sici)1098-2744(199807)22:3<190::aid-mc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 28.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282(4):2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 29.Rogoff HA, et al. Apoptosis associated with deregulated E2F activity is dependent on E2F1 and Atm/Nbs1/Chk2. Mol Cell Biol. 2004;24(7):2968–2977. doi: 10.1128/MCB.24.7.2968-2977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith CL, Baek ST, Sung CY, Tallquist MD. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res. 2011;108(12):e15–e26. doi: 10.1161/CIRCRESAHA.110.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gittenberger-de Groot AC, Winter EM, Poelmann RE. Epicardium-derived cells (EPDCs) in development, cardiac disease and repair of ischemia. J Cell Mol Med. 2010;14(5):1056–1060. doi: 10.1111/j.1582-4934.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spinale FG, Gunasinghe H, Sprunger PD, Baskin JM, Bradham WC. Extracellular degradative pathways in myocardial remodeling and progression to heart failure. J Card Fail. 2002;8(6) Suppl:S332–S338. doi: 10.1054/jcaf.2002.129259. [DOI] [PubMed] [Google Scholar]
- 33.Koglin J, von Scheidt W. Isolated defect of adenosine-mediated coronary vasodilation: Functional evidence for a new microangiopathic entity. J Am Coll Cardiol. 1997;30(1):103–107. doi: 10.1016/s0735-1097(97)00131-9. [DOI] [PubMed] [Google Scholar]
- 34.Bauerle JD, Grenz A, Kim JH, Lee HT, Eltzschig HK. Adenosine generation and signaling during acute kidney injury. J Am Soc Nephrol. 2011;22(1):14–20. doi: 10.1681/ASN.2009121217. [DOI] [PubMed] [Google Scholar]
- 35.Liu SQ, et al. Cardioprotective mechanisms activated in response to myocardial ischemia. Mol Cell Biomech. 2011;8(4):319–338. [PubMed] [Google Scholar]
- 36.Laubach VE, French BA, Okusa MD. Targeting of adenosine receptors in ischemia-reperfusion injury. Expert Opin Ther Targets. 2011;15(1):103–118. doi: 10.1517/14728222.2011.541441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trivax JE, et al. Acute cardiac effects of marathon running. J Appl Physiol (1985) 2010;108(5):1148–1153. doi: 10.1152/japplphysiol.01151.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.