Significance
This paper uses a single-molecule imaging approach based on energy transfer to study how telomeric DNA is protected against the DNA damage-signaling protein replication protein A (RPA). Telomeres terminate with a single-stranded overhang, which is protected by protection of telomere (POT1) and POT1-interacting protein 1 (TPP1) against RPA. Telomeric overhangs have a guanine-rich sequence, forming a four-stranded G-quadruplex structure. Using model telomeric DNA, we studied the competition between POT1/TPP1 and RPA to access telomeric G-quadruplexes in vitro. We showed that G-quadruplex formation of telomeres significantly enhances the ability of POT1/TPP1 to block RPA’s access to telomeres. The results suggest that secondary structure of the telomeric overhang plays an important role in suppressing the DNA damage signals in telomeres.
Keywords: telomere protection, DNA damage response, single molecule imaging
Abstract
Human telomeres terminate with a single-stranded 3′ G overhang, which can be recognized as a DNA damage site by replication protein A (RPA). The protection of telomeres (POT1)/POT1-interacting protein 1 (TPP1) heterodimer binds specifically to single-stranded telomeric DNA (ssTEL) and protects G overhangs against RPA binding. The G overhang spontaneously folds into various G-quadruplex (GQ) conformations. It remains unclear whether GQ formation affects the ability of POT1/TPP1 to compete against RPA to access ssTEL. Using single-molecule Förster resonance energy transfer, we showed that POT1 stably loads to a minimal DNA sequence adjacent to a folded GQ. At 150 mM K+, POT1 loading unfolds the antiparallel GQ, as the parallel conformation remains folded. POT1/TPP1 loading blocks RPA’s access to both folded and unfolded telomeres by two orders of magnitude. This protection is not observed at 150 mM Na+, in which ssTEL forms only a less-stable antiparallel GQ. These results suggest that GQ formation of telomeric overhangs may contribute to suppression of DNA damage signals.
Human telomeres consist of 2,000–30,000 bp of double-stranded TTAGGG repeats and terminate with a 50- to 200-nt-long, single-stranded 3′ G overhang (1). Telomeric termini need to be protected against DNA damage signals to ensure genome integrity. Replication protein A (RPA) nonspecifically binds ssDNA, is highly abundant in eukaryotes, and plays a role in DNA replication and repair (2). RPA binding to ssDNA, including telomeric overhangs, activates the ataxia telangiectasia and Rad3-related checkpoint (2, 3). The protection of telomeres (POT1)/telomere protection protein (TPP1) subunit of the shelterin complex contributes to telomere protection by specifically binding to the G overhang (4, 5). RPA is 1,000-fold more abundant than POT1/TPP1 and has a similar affinity for single-stranded telomeric DNA (ssTEL) (6, 7). Therefore, POT1/TPP1 alone may not be able to effectively compete against RPA binding (8, 9). Efficient protection of ssTEL against RPA binding may require association of POT1/TPP1 to the rest of the shelterin complex along double-stranded telomeric tracts or other telomere-associated proteins (6, 9).
Another potentially significant factor that could influence the competition between RPA and POT1/TPP1 is the ability of ssTEL to form G-quadruplex (GQ) structures (10, 11). Recent studies have shown that human telomeres form GQ structures in vivo (12, 13) and in cell extracts (14). At physiologically relevant ionic conditions (∼150 mM K+), GQs are thermodynamically more stable than competing Watson–Crick pairing (15). These structures were often regarded as obstacles for recruitment of telomerase (16) and translocation of the DNA replication machinery (17), and their unfolding requires helicase activity (17–19) or ssDNA binding proteins (20, 21). It remains unclear whether GQ formation of ssTEL plays any role in protection of telomeres.
ssTEL sequences fold into parallel, antiparallel, and hybrid GQ conformations under physiological salt and pH (22–26). Because of the heterogeneity of these structures, GQ formation was either prevented or its stability was significantly reduced to obtain a homogeneous ssTEL template in previous biochemical studies (6, 9). Understanding the molecular basis of how POT1 interacts with distinct GQ states and competes with RPA in GQ-forming conditions requires direct observation of these interactions at a single-molecule level.
In this study, we used single-molecule Förster resonance energy transfer (smFRET) to investigate how GQ formation of model telomeric DNA affects the ability of POT1/TPP1 to block RPA binding in a steady state. At physiologically relevant ionic conditions (150 mM K+), we observed at least two distinct FRET populations, consistent with the parallel and antiparallel GQ conformations (27). POT1 unfolds the majority of antiparallel GQs, whereas parallel GQs remain stably folded. POT1/TPP1 blocks RPA’s access by loading adjacent to a folded GQ and by coating the unfolded ssTEL. Such protection is not observed with the same GQ-forming sequence terminating with a short overhang, which is not long enough to accommodate POT1 loading. In 150 mM Na+, ssTEL only folds into an antiparallel GQ, and POT1-mediated protection against RPA was not observed. On the basis of these results, we propose a model to describe how GQ formation aids POT1/TPP1’s ability to suppress RPA binding to telomeric overhangs.
Results
POT1 Preferentially Unfolds Antiparallel GQ.
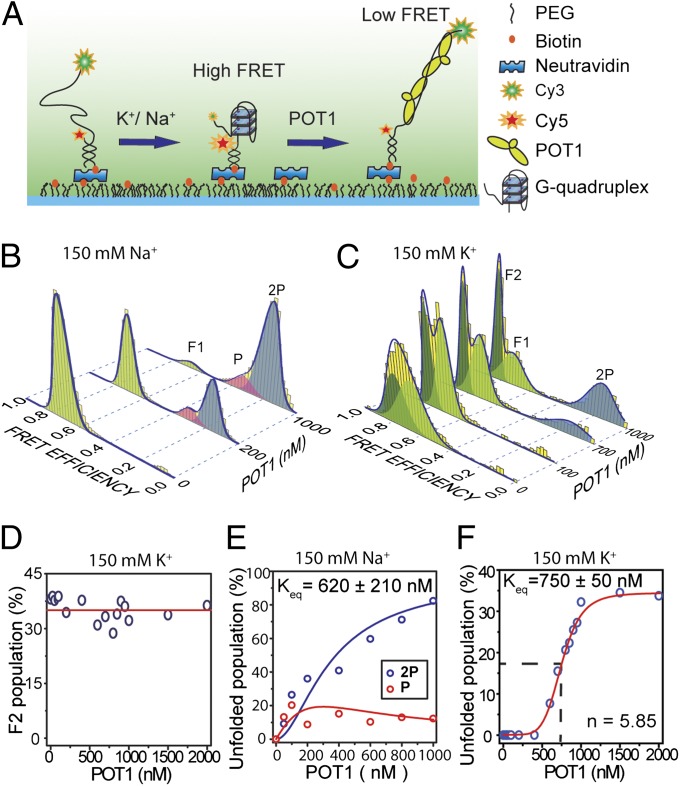
We performed smFRET measurements using total internal reflection fluorescence (TIRF) microscopy to monitor the conformational states of individual DNA molecules. Partial duplex DNA (pdDNA) constructs were immobilized on a PEG-coated surface via neutravidin–biotin linker. The single-stranded portion of the pdDNA [ssTEL4, TA(GGGTTA)4G] contains four telomeric repeats forming a single GQ and terminates with a TTAG-3′ overhang. Cy3 (donor) and Cy5 (acceptor) FRET pairs were placed as shown in Fig. 1A.
Fig. 1.
Steady-state equilibrium of POT1-mediated GQ unfolding in the presence of either 150 mM K+ or 150 mM Na+. (A) Schematic of smFRET assay under TIRF illumination to monitor the folding/unfolding of a model telomeric GQ. (B) smFRET data (yellow bars) showing unfolding of GQ at varying concentrations of POT1 in 150 mM Na+. A multi-Gaussian fitting (blue curve) determines three distinct FRET peaks (F1, P, and 2P). F1 (EF = 0.70, light green) represents antiparallel GQ. P (EF = 0.32 pink) and 2P (EF = 0.16, blue) correspond to one and two POT1-bound unfolded ssTEL4, respectively. (C) smFRET data showing unfolding of GQ at varying concentrations of POT1 in 150 mM K+. Unlike Na+, two folded populations, F1 (EF = 0.70, light green) and F2 (EF = 0.80, dark green), and one unfolded population (2P, EF = 0.16, blue) were observed. F2 is consistent with the parallel GQ conformation. (D) In 150 mM K+, POT1 is unable to unfold the F2 population, which remains nearly constant (red line) from 0 to 2,000 nM POT1. (E) The percentage of P and 2P at 150 mM Na+ as a function of POT1 concentration. Global fitting of the data to fractional occupancy of two distinguishable sites as a function of substrate concentration yields Keq = 620 ± 210 nM and cooperativity of −1.5 kBT. Increasing the parameters of the fit for the cooperative model significantly improves the fit (F test, F = 11.2, α = 0.05), and hence is justifiable. (F) At 150 mM K+, the percentage of the 2P population (blue circles) increases as a function of POT1 concentration, owing to the unfolding of the F1 conformation. The 2P population is fitted to a Hill equation (red line).
The GQ structure is stabilized by addition of a monovalent cation, Na+ or K+. In 150 mM Na+, nearly all of the pdDNA molecules (n = 3,405) formed stable GQs (Fig. 1B). The conversion of various secondary structures into GQ is evidenced by a transition from intermediate-FRET to high-FRET states upon salt addition (Fig. S1). Gaussian fitting to the FRET histogram in Fig. 1B revealed a single population (referred to as F1), with a mean FRET efficiency (EF) of 0.71 ± 0.05. We assume that the F1 peak represents the antiparallel GQ, evidenced by CD spectrum of ssTEL4 (Fig. S2) and NMR studies on similar telomeric sequences (22) in the presence of Na+.
To monitor how POT1 affects ssTEL4 structures, we measured FRET efficiencies of prefolded GQ molecules at varied concentrations of POT1 in 150 mM Na+. Upon addition of 200 nM–1 μM POT1, two new FRET peaks were observed at lower FRET efficiencies (0.16 ± 0.06 and 0.32 ± 0.06, Fig. 1B). To identify the stoichiometry of POT1 binding corresponding to these FRET states, we repeated the smFRET measurements with a mutant ssTEL4 sequence (ssTEL4ΔGQ, Table S1), which can accommodate up to two POT1s but is unable to form GQ (Fig. S3). Under the same ionic conditions, addition of 20 nM POT1 resulted in two distinct FRET peaks (EF = 0.32 ± 0.06 and EF = 0.16 ± 0.05) located at the same positions of the peaks observed for ssTEL4. The population of EF = 0.16 peak increases compared with the EF = 0.32 peak as POT1 concentration is increased. We interpret these peaks as one POT1-bound (EF = 0.32, referred to as P) and two POT1-bound (EF = 0.16, referred to as 2P) unfolded states, respectively. Change in P and 2P populations in ssTEL4ΔGQ as a function of [POT1] fits well to fractional occupancy of two indistinguishable sites. The global fit of the data results in a Kd = 26 ± 4 nM (Fig. S3). POT1 binding to unfolded ssTEL4 was noncooperative. The results are consistent with a recent smFRET study that showed binding of one or two POT1 molecules to four telomeric repeats in 100 mM Na+ (28).
To gain insight on POT1-mediated GQ unfolding at physiologically relevant ionic conditions, we repeated the smFRET measurements in 150 mM K+ (Fig. 1C). The FRET histogram revealed two distinct folded populations centered at EF = 0.66 ± 0.10 (matches the F1 peak observed in 150 mM Na+) and EF = 0.75 ± 0.06 (referred to as F2). The existence of at least two distinct GQ conformations agrees well with the CD measurements (Fig. S2). Even though the GQ unfolding is not observed at low POT1 levels, F1 and F2 peaks become more distinct as the POT1 concentration is increased from 0 to 200 nM (Fig. S4). The gamma parameter, obtained from a ratio of the intensity change in acceptor and donor intensities upon acceptor photobleaching, revealed the underlying mechanism of the shifting of the F1 and F2 peaks as POT1 is titrated in the 0–200 nM range (Fig. S5) (29). We interpret the changes in the gamma parameter to be due to POT1 loading to the TTAG overhang, which spaces the donor away from the GQ. Furthermore, in the presence of 1 µM POT1, replacement of 150 mM Na+ with 150 mM K+ in a sample chamber shifts the FRET distribution from that of Na+ to K+ (Fig. S6), suggesting that steady-state conformations are interchangeable in a cation-dependent manner.
The F1 and F2 peaks are consistent with antiparallel and parallel GQ conformations, respectively. In agreement with our interpretation, a recent computational study showed that antiparallel conformation has a lower FRET efficiency than the parallel conformation (27). High-resolution NMR and EPR studies showed that, in K+, human telomeric sequence folds into various hybrid GQ conformations, in addition to the parallel and antiparallel conformations (14, 25, 30, 31). The most stable telomeric GQ conformation strongly depends on the overhang sequence, pH, and ionic conditions (14). Therefore, we cannot rule out the possibility that the F2 peak may contain both parallel and hybrid conformations with similar distance between the FRET probes. We will refer to F2 as the parallel GQ state for simplification, and precise conformation in this population requires future investigation.
As POT1 concentration increased from 200 nM to 2 µM in 150 mM K+, the F1 population consistently decreased from ∼63% of total population to ∼30% (Fig. 1C). Remarkably, F2 population is unaffected by increased POT1 concentration, even at 2 µM (Fig. 1D). The results suggest that POT1 specifically unfolds the antiparallel GQ, whereas the parallel conformation remains stably folded. A single low FRET peak arises at EF = 0.16 ± 0.08, which matches the 2P peak observed in 150 mM Na+. Unlike in the 150 mM Na+ dataset, the P peak was not observed.
Our results suggest two POT1 molecules may be required to stabilize the unfolded state in 150 mM K+, whereas a single POT1 is sufficient in 150 mM Na+. To test this hypothesis, we performed smFRET measurements on ssTEL41P that forms stable GQ but accommodates only one POT1 (Fig. S7 and Table S1). We preincubated ssTEL41P with 500 nM POT1 in the absence of salt to facilitate POT1 binding to the unfolded DNA. The sample chamber was then washed with 150 mM Na+ or K+ while maintaining the same POT1 concentration. Consistent with our hypothesis, POT1 was able to maintain a significant fraction of the unfolded conformation after addition of 150 mM Na+, but not after 150 mM K+. The unfolded fraction in ssTEL41P (EF = 0.34 ± 0.13) matches with the P population (EF = 0.32 ± 0.06) of ssTEL4 in Na+, supporting our interpretation of the P population as the one-POT1 bound state.
We calculated the equilibrium constant (Keq) for unfolded populations in the presence of Na+ and K+. In 150 mM Na+, 94% of the GQ molecules were unfolded at 1 µM POT1. Because our model assumes the existence of both one and two POT1 binding states in 150 mM Na+, we calculated fractional occupancy of two sites as a function of POT1 concentration. Global fitting of P and 2P populations resulted in Keq = 620 ± 210 nM and positive cooperativity of −1.5 kBT (Fig. 1E). In 150 mM K+, only 34% of the DNA molecules were unfolded at 1 µM POT1. Because the P intermediate was not observed in this case, we used the Hill equation to calculate Keq (750 ± 50 nM, Fig. 1F). Our Keq values are nearly two orders of magnitude higher than the previously reported Kd for POT1 (1–70 nM) (6, 7, 32), because Keq represents both POT1-mediated GQ unfolding and POT1 binding to the unfolded DNA. Similarly, for ssTEL4ΔGQ, Kd = 26 ± 4 nM (Fig. S3) was comparable to Kd = 21 nM from our gel shift assays (Fig. S8).
TPP1 Significantly Enhances POT1’s Ability to Unfold GQ in Na+ but Not in K+.
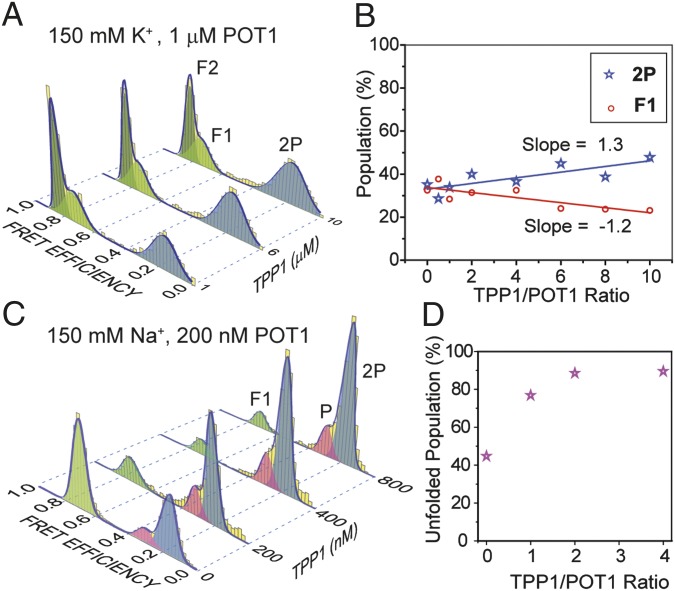
POT1 forms a heterodimer with TPP1 (33), and the POT1/TPP1 complex has higher affinity to ssTEL than POT1 alone (7). We confirmed that TPP1 is unable to unfold GQ without POT1 (Fig. S9). To explore how TPP1 affects the interactions of POT1 with telomeric GQs, we performed smFRET measurements with ssTEL4 in 150 mM K+ (Fig. 2 A and B) or 150 mM Na+ (Fig. 2 C and D) at 1–10 μM TPP1, while keeping POT1 concentration at 1 µM. The resulting low FRET peaks (EF = 0.32 ± 0.04 and EF = 0.16 ± 0.03) were at the same locations as P and 2P observed in the absence of TPP1, suggesting that POT1–TPP1 interaction does not induce further conformational changes on the POT1-bound ssTEL4. A small increase of the 2P population was observed as TPP1 concentration was increased (Fig. 2B). The rate of decrease in F1 population (−1.2% per μM of TPP1) was very similar to the rate of increase in 2P population (1.3% per μM of TPP1), suggesting that TPP1 has a slight effect on POT1-mediated antiparallel GQ unfolding. The F2 population remains unaltered under all conditions. We next repeated TPP1 titrations in 150 mM Na+, and used lower (200 nM) POT1 concentration to maintain ∼50% GQ population. In contrast to the 150 mM K+ case, a substantial increase (from 45 to 89%) in the unfolded population (P + 2P) was observed as the TPP1 population was increased from 0 to 800 nM (Fig. 2 C and D).
Fig. 2.
The effect of TPP1 on POT1-mediated GQ unfolding in the presence of either 150 mM K+ or 150 Na+. (A) Unfolding of telomeric GQ as a function of TPP1 concentration in 150 mM K+ and 1 µM POT1. (B) In 150 mM K+, the 2P population increases 1.3% per μM of TPP1 (blue line) and the F1 population decreases at a similar rate (1.2% per μM of TPP1, red line). (C) Unfolding of telomeric GQ as a function of TPP1 concentration in the presence of 150 mM Na+ and 200 nM POT1. (D) In 150 mM Na+, the percentage of unfolded populations (P + 2P) increases by a factor of 2 when TPP1:POT1 ratio is increased from 1:1 to 2:1 at 200 nM POT1.
The results show that TPP1 alone neither unfolds telomeric GQs nor binds to the unfolded ssTEL4. However, TPP1 enhances the POT1-mediated GQ unfolding depending on ionic conditions.
POT1/TPP1 Protects Telomeric GQ Against RPA-Mediated Unfolding.
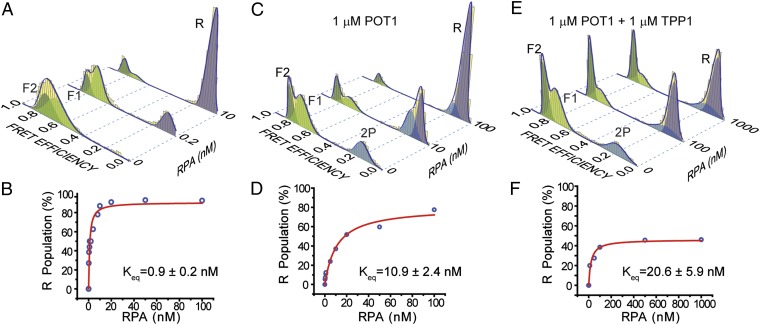
We next investigated whether GQ formation of ssTEL4 plays a role in the competition between POT1/TPP1 and RPA to access telomeres. In 150 mM K+, RPA unfolds a majority of ssTEL4 GQs in the absence of POT1 at two orders of magnitude lower concentrations than its physiological levels (Fig. 3 A and B), in agreement with an earlier study on a similar GQ construct (21). FRET value of the RPA-bound unfolded state (R, EF = 0.10 ± 0.03) is lower than 2P (EF = 0.16 ± 0.03), allowing us to distinguish between POT1-bound and RPA-bound unfolded states (Fig. 3). We measured the percentage of R population as a function of RPA concentration in RPA only, RPA with 1 μM POT1, and RPA with 1 μM POT1/TPP1 cases. The data of these three cases were fitted by a Langmuir binding isotherm of the form y = α[RPA]/([RPA] + Keq). In this expression y is the R population, α represents the R population at saturating RPA concentration, and Keq is the equilibrium constant.
Fig. 3.
Competition between RPA versus POT1 or POT1/TPP1 to bind ssTEL4 in 150 mM K+. (A) FRET histograms display unfolding of GQ and binding of RPA to the unfolded ssTEL4 (R peak at EF = 0.10, lavender); ∼90% of GQ molecules are unfolded at 10 nM RPA. (B) Langmuir binding isotherm analysis (red curve) of RPA-mediated unfolding for the RPA-only case. (C) Unfolding of GQ and binding of RPA to the unfolded ssDNA in the presence of 1 µM POT1; ∼90% of GQ molecules are unfolded at 100 nM RPA concentration. (D) Langmuir binding isotherm analysis (red curve) of RPA-mediated unfolding in the presence of 1 µM POT1. (E) Unfolding of GQ by RPA in the presence of 1 µM POT1/TPP1; ∼47% of GQ molecules are unfolded at 1 µM RPA. Peak position of R (0.10) is distinct from 2P (0.16). Peak positions of F1 and F2 are similar to the POT1-only case. (F) Langmuir binding isotherm analysis (red curve) of RPA-mediated unfolding in the presence of 1 µM POT1/TPP1.
In the absence of POT1, RPA efficiently unfolded both F1 and F2 conformations with α = 90% and Keq = 0.9 ± 0.2 nM RPA. In 1 μM POT1, the R peak was populated at a much lower rate compared with the RPA-only case with α = 90% and Keq = 10.9 ± 2.4 nM RPA (Fig. 3 C and D). The 12-fold increase in Keq suggests that POT1 provides a significant protection of ssTEL4 at lower RPA concentrations. For comparison, in the absence of POT1 R population is 87% of the total population at 10 nM RPA. The corresponding R is 25% at 10 nM RPA when 1 μM POT1 is in the environment. Saturating RPA concentrations displaces POT1 from ssTEL4 with α reaching a value similar to that of the RPA-only case.
To test whether TPP1 can enhance the ability of POT1 to block RPA’s access to ssTEL4, we incubated folded ssTEL4 GQ molecules with 1 μM POT1 and 1 μM TPP1 in 150 mM K+ before titrating RPA (Fig. 3 E and F). The protection of ssTEL4 was even more dramatic in this case with α = 47% and Keq = 20.6 ± 5.9 nM RPA. Compared with the RPA-only case, Keq increased by a factor of 23 and α decreased by a factor of 2. In particular, the reduction in α suggests that about half of ssTEL4 molecules are protected against physiological RPA levels (1 μM) by equimolar POT1/TPP1.
Another outcome of this analysis is the variation in the 2P population before and after RPA is introduced to the chamber. This variation represents the fraction of bound POT1 or POT1/TPP1 molecules that are displaced from ssTEL4 by RPA. At 1 μM POT1, 34% of folded ssTEL4 are unfolded and bound by two POT1 molecules in the absence of RPA. Upon introducing 100 nM RPA to the chamber, the 2P population reduces to 14%, suggesting that RPA displaces 59% of bound POT1 molecules from ssTEL4. Similarly, introduction of 1 μM RPA displaces 42% of bound POT1/TPP1 molecules at 1 μM POT1/TPP1. Therefore, TPP1 increases the binding stability of POT1 to unfolded ssTEL4, resulting in a smaller fraction of displacement by RPA.
POT1 and POT1/TPP1 selectively protected the F2 population against RPA-mediated GQ unfolding. At 1 μM POT1, F1 population was reduced from 34 to 0%, and F2 population was reduced from 32 to 8% as RPA concentration was increased from 0 to 100 nM (Fig. 3D). At 1 μM POT1/TPP1, the F2 population was reduced from 33 to 26%, whereas F1 population was reduced from 48 to 14% as RPA concentration was increased from 0 to 1 μM (Fig. 3F). Therefore, POT1 alone is not adequate to protect F1 or F2 conformations at high (>100 nM) RPA concentrations, whereas significant fractions of the GQ population remain stably folded even at 1 μM RPA in the presence of POT1/TPP1.
We next examined whether POT1 could protect ssTEL4 against RPA binding in 150 mM Na+ (Fig. 4). Similar to the 150 mM K+ case, we compared α and Keq for RPA titration with or without incubating with POT1. The sample was incubated with a lower concentration of POT1 (200 nM), compared with 1 µM POT1/TPP1 used in the 150 mM K+ case, to maintain a significant fraction of folded GQ population. Addition of POT1 did not lead to significant reduction in RPA’s efficiency to unfold the GQ (Fig. 4). At low (∼2 nM) RPA, 70% of the GQs were unfolded and all of the P population was displaced by R, whereas 80% of 2P remained unaltered. Increasing the RPA concentration to 10 nM unfolded almost all of the GQs and displaced 70% of the 2P population (Fig. 4B). Therefore, POT1 is unable to protect ssTEL4 against RPA binding in 150 mM Na+. These results demonstrate the significance of using physiologically relevant ionic conditions in in vitro studies while interpreting the in vivo outcomes of protein–GQ interactions.
Fig. 4.
Competition between RPA versus POT1 to bind ssTEL in 150 mM Na+. (A) FRET histograms display unfolding of GQ and binding of RPA to the unfolded ssTEL4. All GQ molecules are unfolded at 10 nM RPA. (B) Langmuir binding isotherm analysis (red curve) of RPA-mediated unfolding for the RPA-only case. (C) Unfolding of GQ and binding of RPA to the unfolded ssDNA in the presence of 200 nM POT1. All GQ molecules are unfolded at 10 nM RPA. (D) Langmuir binding isotherm analysis (red curve) of RPA-mediated unfolding in the presence of 200 nM POT1. The difference between α and Keq values obtained from the fit to those obtained in the RPA-only case is within the experimental error, suggesting that the GQ is not protected by POT1 in 150 mM Na+.
POT1 and RPA Compete for the Free 3′-Terminal Sequence.
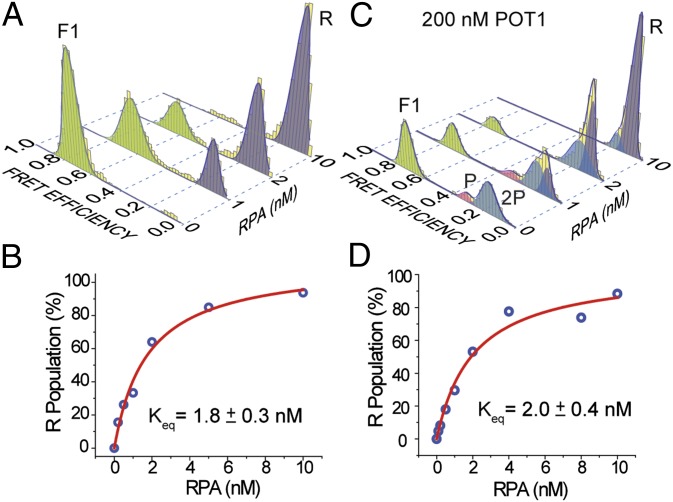
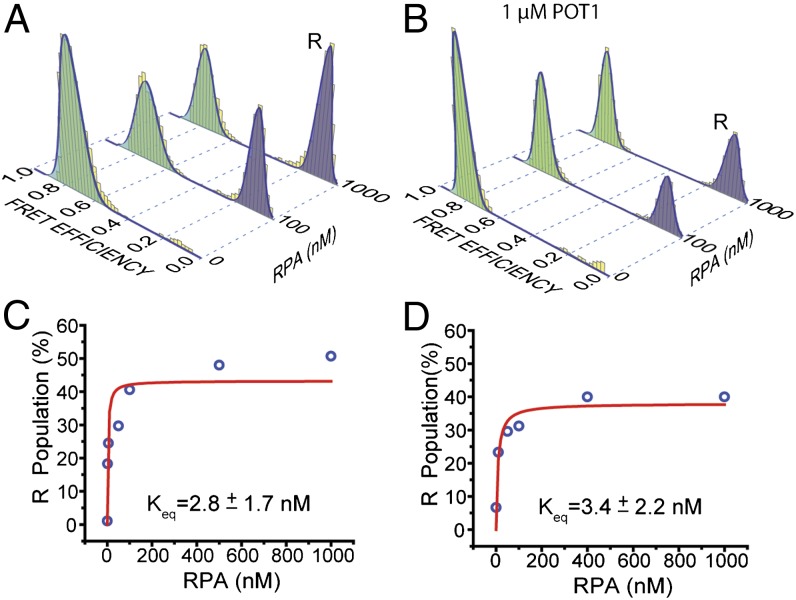
RPA consists of four active DNA-binding domains (DBDs), the most active of which (DBD-A or DBD-B) has a footprint of three nucleotides (34). POT1 has two oligosaccharide-binding (OB) domains, and OB2 interacts with T7-G10 of the minimal binding sequence 5′-TTAGGGTTAG (corresponding to the TTAG overhang of ssTEL4) (5). The TTAG overhang is long enough to serve as a substrate for RPA loading and interaction with a folded GQ. POT1 loading to this site may reduce RPA’s ability to unfold the GQ. To test this idea, we repeated RPA and POT1 competition assays in 150 mM K+ using the telomeric GQ construct terminating with a shorter (TT) overhang (ssTEL4TT), which is too short to accommodate loading of POT1 (Fig. 5). This construct has a CD spectrum similar to that of ssTEL4, and hence similar folding conformations are expected (21). RPA was less efficient in unfolding the GQ with a TT overhang, demonstrating that RPA also loads to the overhang for unfolding the GQ (Fig. 5A). Only 45% of GQ molecules were unfolded by 1 µM RPA, compared with 85% unfolding of the GQ with a TTAG overhang by 10 nM RPA. Remarkably, 1 μM POT1 was not able to unfold any of the GQ molecules with a TT overhang (Fig. 5B), highlighting the significance of the overhang in the context of POT1–GQ interactions. Incubating the prefolded GQ with 1 μM POT1 did not significantly increase Keq of RPA binding (from 2.8 ± 1.7 nM to 3.3 ± 2.2 nM). The fraction of unfolded GQ molecules at saturating RPA concentration only decreased ∼10% in the presence of 1 μM POT1, compared with the RPA-only case (Fig. 5 C and D). These results show that GQ is not protected against RPA when POT1 is unable to load onto the overhang in the vicinity of folded GQ.
Fig. 5.
POT1 and RPA compete for the 3′ overhang of ssTEL4 in 150 mM K+. (A) The RPA-mediated GQ unfolding was significantly reduced using the ssTEL construct terminating with a short TT-3′ overhang (ssTEL4TT), rather than the TTAG-3′ overhang of ssTEL4. Even at 1,000 nM RPA, only ∼50% of the GQ molecules remain folded. (B) The RPA-mediated GQ unfolding at 1 µM POT1. (C and D) The Langmuir isotherm fits to GQ unfolding in the absence and presence of POT1, respectively. POT1 addition did not significantly affect Keq for GQ unfolding, compared with the 12-fold increase observed in ssTEL4 terminating with a TTAG overhang.
Discussion
The results of our study signify that the GQ formation not only blocks protein–DNA interactions but also regulates the competition between DNA binding proteins. Previous studies suggested that unfolding of the ssTEL GQ is an essential step for POT1 binding (16). We provided evidence that POT1 can load adjacent to a folded GQ. POT1 loading induces unfolding of the antiparallel conformation, whereas the other GQ conformation is further stabilized against RPA-mediated unfolding. RPA’s access to telomeric DNA can be reduced by at least two orders of magnitude via a synergistic activity of POT1/TPP1 and GQ formation of ssTEL4 sequence. Under these conditions, POT1/TPP1 effectively competes against equimolar RPA in accessing ssTEL. Finally, we showed that at least two POT1 molecules are required to stabilize the unfolded state of a single GQ-forming telomeric sequence in 150 mM K+, whereas a single POT1 is adequate in 150 mM Na+.
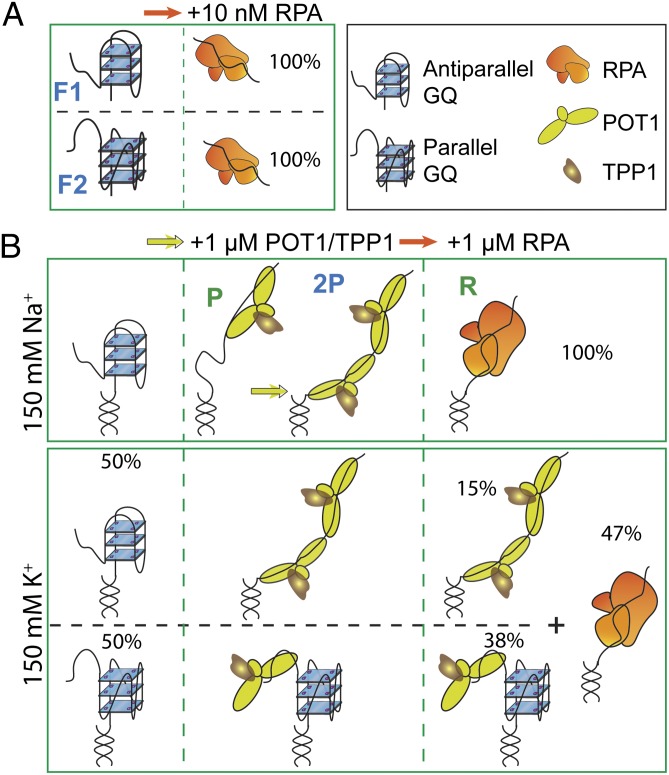
We propose a model for the molecular interactions between different GQ states, POT1, and RPA. RPA is very efficient at unfolding unprotected GQ, regardless of the folding conformation (Fig. 6A). POT1 loads onto the TTAG overhang in both conformations and unfolds the antiparallel conformation, whereas the parallel GQ remains stably folded (Fig. 6B). POT1 loading adjacent to an antiparallel GQ may destabilize it (35). As the end sequences of GQ transiently melt owing to thermal fluctuations, the GQ-forming sequences may be invaded by DNA binding domains of POT1. These transient melting events can be more likely to occur in Na+ compared with K+ because GQ is less stable in Na+ (36). Whether POT1 loading increases the stability of the parallel conformation, for example, owing to stacking of POT1 with this GQ structure as observed with synthetic ligands (37–39), or it remains bound to the overhang without influencing GQ stability requires further investigation. TPP1 increases the binding stability of POT1 to the TTAG overhang, which results in higher unfolding for the less-stable GQ in Na+ and suppresses RPA-mediated GQ unfolding in K+.
Fig. 6.
A model for protection of ssTEL4 against RPA-mediated unfolding. (A) In the absence of POT1/TPP1, RPA unfolds both antiparallel (F1) and parallel (F2) conformations at a low concentration either in Na+ or in K+. (B) In 150 mM Na+, ssTEL4 forms only antiparallel GQ. Binding of one (P) or two (2P) POT1 unfolds the antiparallel conformation. Addition of RPA displaces POT1/TPP1 from ssTEL4. In 150 mM K+, POT1/TPP1 binds to both GQ folding patterns and unfolds antiparallel GQ, whereas parallel GQ remains folded. Binding of two POT1s to ssTEL4 is required to stabilize the unfolded state. POT1/TPP1 binding effectively suppresses RPA binding to ssTEL4 even at high RPA concentrations. In particular, the majority of parallel GQ remains stably folded against RPA-mediated unfolding.
Under physiological K+ concentrations, we observed that POT1/TPP1 is able to protect a significant fraction of ssTEL4 molecules against RPA binding at equimolar concentrations (Fig. 6B). Because POT1/TPP1 protect both folded and unfolded ssTEL4 GQs against RPA, we propose that POT1/TPP1 blocks RPA’s access by coating the unfolded ssTEL4 and inhibiting access of RPA to the folded GQ. Although the model would predict all of the ssTEL to be RPA-bound at high [RPA], there are multiple reasons that could explain the plateau at 47%, including the reactions not achieving equilibrium and the existence of multiple DNA conformations.
We believe that GQ formation of ssTEL plays an important role in cellular roles of other telomere-interacting proteins. For example, telomeric RNA and an mRNA regulator, hnRNPA1, effectively displace RPA from ssTEL and mediate POT1 binding (9). hnRNPA1 can stably bind to two telomeric repeats (40), but it only displaces RPA from ssTEL containing four or more telomeric repeats (9), which is the minimum length required for GQ formation. Indeed, hnRNPA1 unfolds telomeric GQs in vitro (41), consistent with a role for GQ formation in competition between telomere-interacting proteins.
In vivo, telomeric DNA is well protected against RPA binding even when RPA is significantly more abundant than POT1 and TPP1 (6). Therefore, effective suppression of the ATR pathway cannot be solely achieved by coating of telomeric overhangs by POT1/TPP1. We showed that GQ formation enhances the efficiency of POT1/TPP1 to compete against RPA binding to telomeres by several orders of magnitude, presenting an additional dimension to telomere maintenance. The full protection of telomeric terminus in wild-type cells may also require tethering of POT1/TPP1 to the rest of shelterin via TIN2 (6), t-loop formation (42), and other telomere-interacting proteins (9). We anticipate our single-molecule approach will serve as a platform for testing more sophisticated models on protein–protein and protein–DNA interactions for the replication and protection of telomeres. These in vitro assays must be performed under physiologically relevant ionic conditions because it proved to be a major factor determining the GQ conformation and structure.
Materials and Methods
A pdDNA with an 18-bp-long duplex stem and single-stranded tail consisting of four telomeric TTAGGG repeats terminating with a 3′ TTAG overhang was used in the smFRET studies. DNA oligomers constituting the pdDNA construct were hybridized in 10 mM Tris (pH 7.5) and 50 mM Na+. Full-length human POT1, C-terminal truncated form of human TPP1 (1–446 aa), and a p11d-tRPA construct containing the coding sequences of RPA70, RPA14, and RPA32 were expressed in Escherichia coli. Biotinylated pdDNA molecules were immobilized on PEG-coated coverslips and imaged with a prism-type TIRF microscope. Steady-state smFRET and CD measurements were performed as described (21). The smFRET histograms were constructed such that each analyzed molecule contributes equally to the FRET histogram, and the total population of the histogram is normalized to 100%. An extended description of the materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Lei (University of Michigan) and A. Sancar (University of North Carolina) for plasmids and reagents and M. D. Stone (University of California, Santa Cruz) and K. Collins (University of California, Berkeley) for carefully reading our manuscript. S.R. and M.H.Q. thank Institute for Complex and Adaptive Matter for the exchange scientist awards. Funding provided by Ellison Medical Foundation (A.Y.) and Farris Family Innovation Award (H.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321436111/-/DCSupplemental.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 3.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 4.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41(49):14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 5.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279(13):13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 6.Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell. 2011;44(4):647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445(7127):506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 8.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448(7157):1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 9.Flynn RL, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471(7339):532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334(6180):364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 11.Gomez D, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66(14):6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 12.Lam EY, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nature Communications. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5(3):182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hänsel R, Löhr F, Trantirek L, Dötsch V. High-resolution insight into G-overhang architecture. J Am Chem Soc. 2013;135(7):2816–2824. doi: 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- 15.Kumar N, Sahoo B, Varun KA, Maiti S, Maiti S. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008;36(13):4433–4442. doi: 10.1093/nar/gkn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci USA. 2005;102(31):10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38(4):1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145(5):678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30(18):3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salas TR, et al. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34(17):4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qureshi MH, Ray S, Sewell AL, Basu S, Balci H. Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J Phys Chem B. 2012;116(19):5588–5594. doi: 10.1021/jp300546u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1(4):263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 23.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417(6891):876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 24.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc Natl Acad Sci USA. 2005;102(52):18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambrus A, et al. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34(9):2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J Am Chem Soc. 2011;133(25):9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 27.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2003;100(25):14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20(11):1872–1880. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ray S, et al. RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys J. 2013;104(10):2235–2245. doi: 10.1016/j.bpj.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34(19):5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh V, Azarkh M, Drescher M, Hartig JS. Conformations of individual quadruplex units studied in the context of extended human telomeric DNA. Chem Commun (Camb) 2012;48(66):8258–8260. doi: 10.1039/c2cc32012f. [DOI] [PubMed] [Google Scholar]
- 32.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11(12):1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6(7):673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 34.Cai L, et al. Structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry. 2007;46(28):8226–8233. doi: 10.1021/bi7004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Ma L, Hao YH, Tan Z. Folding equilibrium constants of telomere G-quadruplexes in free state or associated with proteins determined by isothermal differential hybridization. Anal Chem. 2010;82(22):9469–9475. doi: 10.1021/ac102168m. [DOI] [PubMed] [Google Scholar]
- 36.Tran PL, Mergny JL, Alberti P. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011;39(8):3282–3294. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun D, et al. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol Cancer Ther. 2008;7(4):880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jena PV, et al. G-quadruplex DNA bound by a synthetic ligand is highly dynamic. J Am Chem Soc. 2009;131(35):12522–12523. doi: 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13(9):1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang QS, Manche L, Xu RM, Krainer AR. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA. 2006;12(6):1116–1128. doi: 10.1261/rna.58806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.