Significance
Hematopoiesis is a tightly regulated process by which hematopoietic stem cells (HSCs) give rise to mature cells. The balance between the propensity of HSCs to remain quiescent, to divide and generate more HSCs (self-renewal), or to divide and give rise to mature cells (differentiation) is essential for the long-term maintenance of blood cell formation. Mechanisms underlying cell fate decisions of HSCs are not completely understood. We observed that deletion of the B-myb gene leads to depletion of the HSC pool and losses of mature cells. Our further studies strongly suggest that these effects are due to defects in HSC proliferation and differentiation. We therefore identified B-myb as a critical component of the mechanism that balances self-renewal and differentiation of HSCs.
Keywords: myeloipiesis, myelodisplastic syndrome
Abstract
The B-myb (MYBL2) gene is a member of the MYB family of transcription factors and is involved in cell cycle regulation, DNA replication, and maintenance of genomic integrity. However, its function during adult development and hematopoiesis is unknown. We show here that conditional inactivation of B-myb in vivo results in depletion of the hematopoietic stem cell (HSC) pool, leading to profound reductions in mature lymphoid, erythroid, and myeloid cells. This defect is autonomous to the bone marrow and is first evident in stem cells, which accumulate in the S and G2/M phases. B-myb inactivation also causes defects in the myeloid progenitor compartment, consisting of depletion of common myeloid progenitors but relative sparing of granulocyte–macrophage progenitors. Microarray studies indicate that B-myb–null LSK+ cells differentially express genes that direct myeloid lineage development and commitment, suggesting that B-myb is a key player in controlling cell fate. Collectively, these studies demonstrate that B-myb is essential for HSC and progenitor maintenance and survival during hematopoiesis.
Hematopoiesis is maintained by the renewal of multipotent hematopoietic stem cells (HSCs) that give rise to lineage-committed cells. HSCs are maintained in constant numbers in the bone marrow (BM), where they reside in a quiescent state (1, 2). According to the stochastic model of hematopoiesis, long-term HSCs (LT-HSCs), which can undergo extensive self-renewal, begin to differentiate into short-term HSCs (ST-HSCs) with limited self-renewing potential. ST-HSCs further differentiate into multipotent progenitors (MPPs) that unlike HSCs do not self-renew, but retain the ability to commit to multiple lineages. Lineage commitment begins to occur at the level of the common lymphoid and myeloid progenitors (CLPs and CMPs), which are thought to arise from MPPs. Whereas CLPs give rise to lymphoid cells, CMPs further differentiate into megakaryocyte–erythroid progenitors (MEPs) and granulocyte–monocyte progenitors (GMPs) (1, 2). More recent studies have identified additional intermediates, such as lymphoid-primed MPPs (LMPPs), in this developmental pathway (3).
The MYB family of transcription factors has three members: A-myb (MYBL1), B-myb (MYBL2), and c-myb (MYB). Disruption of the c-myb locus in mice results in embryonic lethality at embryonic day 15, primarily due to defective erythropoiesis in the fetal liver (4). A role for c-myb in adult hematopoiesis has been shown more recently using several conditional knockout (KO) and mutant mouse models. In adult thymocytes and B lymphocytes, disruption of c-myb blocks development at the DN3 and prepro B stages, respectively (5–8). c-myb expression is also required for erythropoiesis, myelopoiesis, and the development and maintenance of HSCs (9–12), underscoring the importance of this gene within the entire hematopoietic compartment.
B-myb–null mutant mice die in utero at days 4–6.5 of gestation due to defects in inner cell mass formation (13), which precludes the study of this gene in hematopoiesis. In this paper, we describe the effects of conditional disruption of B-myb in adult hematopoietic cells. Loss of B-myb leads to depletion of the HSC pool, resulting in dramatic losses of mature cells in multiple lineages. The effect of B-myb deficiency is autonomous and is associated with defects in HSC cell cycle progression and increased levels of cell death in the myeloid progenitor compartment. Gene expression analysis indicates that B-myb–null LSK+ cells differentially express genes that regulate myeloid lineage development and commitment, suggesting that B-myb plays a critical role in regulating HSC and progenitor cell maintenance.
Results
Loss of B-myb Expression Results in Defective Hematopoiesis.
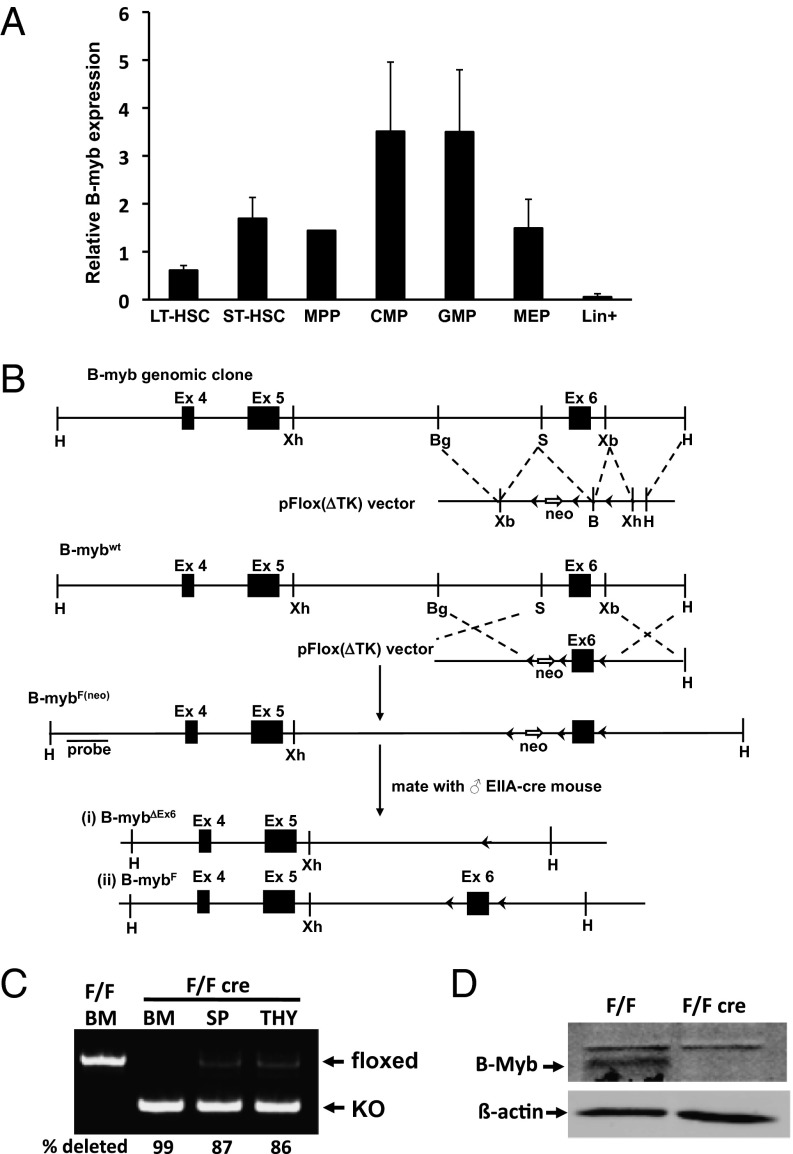
In BM, B-myb mRNA is expressed at appreciable levels in the HSC and myeloid progenitor compartments, with the highest levels in the CMP and GMP populations. These levels are in stark contrast to those of mature, lineage+ (Lin+) cells (Fig. 1A). To determine the role of B-myb in adult hematopoiesis, we generated a conditional B-myb floxed (B-mybF/F) mouse model (Fig. 1 B–D) and crossed this mouse with the Mx1-cre strain (14). Administration of polyinosinic:polycytidylic acid (pIpC) to B-mybF/F-Cre mice (designated “F/Fcre”) resulted in a dramatic loss of cellularity in the BM and spleen (Fig. 2 A and B) compared with identically treated littermate control (F/F or +/F) animals. Notably, we observed significant decreases in the number of B- and T-lymphoid, erythroid, and myeloid lineage cells, with the myeloid compartment being most profoundly affected in both tissues (Fig. 2 C and D). Statistically significant reductions in the number of platelets, total white blood cells, lymphocytes, and neutrophils were also observed in the peripheral blood (PB) of B-myb–deficient F/Fcre animals (Fig. S1). These differences resulted in changes in the frequencies of cell types in the spleen and BM (Fig. S2), as the percentages of B, erythroid, and myeloid cells were altered in the absence of B-myb expression. Although the number of T-lineage (CD3+) cells were also decreased in both the BM and spleen, thymocytes and the CD3+ cells in the thymus were not significantly depleted (Fig. 2 C–F).
Fig. 1.
Murine B-myb genomic structure and target production. (A) Expression of B-myb in HSCs and progenitor cells. (All mRNA levels shown are normalized to that of β-actin.) All values represent mean ± SEM. (B) Schematic representation of the mouse B-myb genomic clone. (Note: not drawn to scale.) LoxP sites are depicted as black arrowheads. Homologous recombination will generate the B-mybF(neo) allele that will be used for subsequent recombination by Cre recombinase. ES cells with either a type I or II deletion (B-mybΔEx6 and B-mybF) will be produced following Cre expression. The probe that can be used to determine homologous recombination is indicated. Restriction enzyme sites: B, BamHI; Bg, BglII: H, HindIII; S, SmaI; Xb, XbaI; Xh, XhoI. Deletion of exon 6 produces a B-MYB protein without a DNA-binding domain and a shift in reading frame. (C) Representative semiquantitative PCR analysis of genomic DNA isolated from pIpC-treated control and B-myb floxed mice showing the presence of floxed and deleted alleles in the BM, spleen (SP) and thymus (THY). The percentage of deletion is shown. (D) Western blot analysis of B-MYB protein (arrow) in sorted Lin− BM cells derived from pIpC-treated control (F/F) and floxed/Mx1Cre+ (F/Fcre) mice. β-Actin is shown as a loading control.
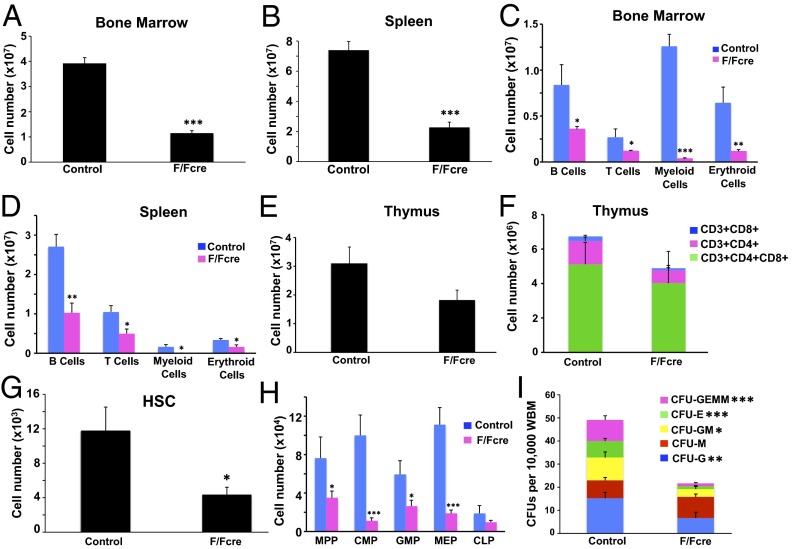
Fig. 2.
B-myb is required for hematopoiesis. B-myb–deficient (F/Fcre; Mx1-cre-B-mybF/F) and control (B-myb+/F or B-mybF/F) mice were treated with pIpC every other day over a 5-d period. Tissues were harvested on day 21 posttreatment. Total number of cells isolated from BM (A) and spleen (B) of control and B-myb–deficient mice. Analysis of mature lineage cells present in the BM (C) and spleen (D) of control and B-myb–deficient mice. Erythroid, B-, T-, and myeloid lineage cells are defined as TER119+, B220+, CD3+, and CD11b+Gr1+, respectively. Total number of thymocytes (E) and mature populations (F) in the thymuses of control and B-myb F/Fcre mice. (G) Total number of HSCs in the BM of control and B-myb–deficient mice. (H) Total number of MPPs, myeloid progenitors (CMPs, GMPs, and MEPs), and lymphoid progenitors (CLPs) in the BM of control and B-myb–deficient mice. (I) Colony-forming assay showing the frequency of whole BM cells that formed megakaryocyte (GEMM), erythroid (E), granulocyte–monocyte (GM), monocyte (M), or granulocyte (G) colonies in culture. All values represent mean ± SEM. n ≥ 3 per population for each genotype. *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005.
A more detailed examination of the HSC and progenitor compartments showed that B-myb–deficient mice have marked decreases in the absolute number of HSCs (Lin−Sca1+cKit+CD150+CD48−), MPPs (Lin−Sca1+cKit+CD150−CD48−) (15) as well as CMPs (Lin−Sca1−cKit+CD34+CD16/32[FcγR]mid), GMPs (Lin−Sca1−cKit+CD34+CD16/32[FcγR]hi) and MEPs (Lin−Sca1−cKit+CD34loCD16/32[FcγR]lo) (Fig. 2 G and H) (16). Although we also observed a decrease in the number of CLPs (Lin−/IL7Rα+/Sca1lo/cKitlo) (Fig. 2H) (17), this difference was not significant. Consistent with the reductions in myeloid progenitors, B-myb–deficient BM also formed fewer colonies in in vitro colony-forming unit (cfu) assays (Fig. 2I). All of the myeloid lineage colonies that were scored, with the exception of CFU-M, were reduced in number in cultures derived from B-myb–deficient BM. The increased number of CFU-M colonies, although not statistically significant, mirrors the CD11b+Gr1lo monocyte-containing population whose frequency is increased in B-myb F/Fcre mice (Fig. S2A). Taken together, these results show that B-myb expression is required for adult hematopoiesis and for the maintenance of HSCs and myeloid progenitor cells.
B-myb–Mediated Defects in the BM Are Autonomous.
We next performed noncompetitive BM transplantation assays to determine whether the defect in hematopoiesis was intrinsic to BM. Whole BM was isolated from CD45.2+ control or B-myb–deficient mice and transplanted into lethally irradiated CD45.1+ recipients (Fig. S3A). pIpC was administered once engraftment was stable (8–10 wk posttransplant) and ∼90% of the PB in the recipients was reconstituted from CD45.2+ donor-derived cells (Fig. 3A). Mice transplanted with B-myb–deficient BM and treated with pIpC became pancytopenic (Fig. 3 B and C) and showed profound decreases in the numbers of LT-HSCs and myeloid progenitors (Fig. 3 D and E) as well as reduced PB counts (Fig. S4). On the other hand, reciprocal, noncompetitive BM transplants, whereby wild-type BM (CD45.1+) was transplanted into lethally irradiated control or B-myb F/Fcre (CD45.2+) mice, failed to produce a phenotype (Fig. S5). These observations confirm that loss of B-myb expression impairs hematopoiesis in a cell-autonomous manner.
Fig. 3.
Cell-autonomous role for B-myb in hematopoiesis. Wild-type mice (CD45.1) were transplanted with control or B-mybF/Fcre (CD45.2) whole BM (WBM). (A) Animals transplanted with BM of both genotypes showed nearly equal donor cell contribution at 8–10 wk posttransplant, before pIpC administration. (B) Total number of control and B-myb–deficient CD45.2+ cells in the WBM of recipient animals following treatment with pIpC. (C) Total number of erythroid (TER119+), B- (B220+), T- (CD3+), and myeloid lineage (CD11b+Gr1+) CD45.2+ cells in the BM of recipients transplanted with control and B-myb–deficient BM following pIpC treatment. Total number of control and B-myb–deficient CD45.2+ HSCs (D) and myeloid progenitors (CMPs, GMPs and MEPs) (E) in the BM of recipient animals following treatment with pIpC. All values represent mean ± SEM. n ≥ 3 per population for each genotype. ***P ≤ 0.0005.
B-myb–Deficient HSCs Fail to Long-Term Repopulate.
Because loss of B-myb caused a dramatic decrease in the number of HSCs, we performed competitive repopulation studies to determine whether B-myb KO HSCs were capable of long-term reconstitution in vivo. Whole BM was isolated from pIpC-treated control and B-myb F/Fcre animals (CD45.2+), mixed in a 1:1 ratio with competitor wild-type (CD45.1+) BM and transplanted into lethally irradiated recipient (CD45.1+) mice (Fig. S3B). Engraftment was monitored every 4 wk to determine the contribution of CD45.1+ and CD45.2+ cells. The control CD45.2+ cells reconstituted the myeloid and B- and T-lineage cells in the PB at levels comparable to the competitor for the 16-wk duration of the experiment (Fig. S6). In contrast, multilineage reconstitution by B-myb–deficient BM was strongly impaired at 4 wk posttransplant, and decreased progressively over time, demonstrating that B-myb is required for the maintenance and self-renewal of HSCs.
B-myb Disruption Leads to Aberrant Cell Cycle Progression of HSCs.
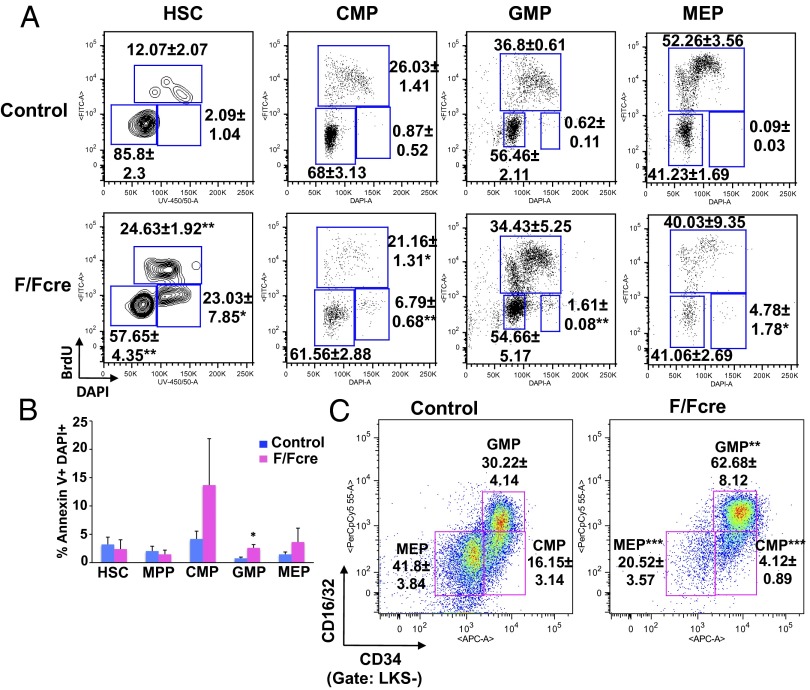
Loss of HSC renewal is often associated with a defect in proliferative control. In their steady-state, HSCs are largely quiescent and enter the cell cycle in response to stimuli that trigger differentiation and lineage commitment (18). B-myb has been shown to play a role in the cell cycle progression of multiple cell types and is involved in regulating progression through the S and G2/M phases (19–25). To determine whether loss of B-myb expression led to altered cell cycle kinetics in HSCs, pIpC-treated control and B-myb F/Fcre mice were injected with BrdU and BM was harvested 2 h posttreatment. BrdU incorporation was significantly higher in B-myb–deficient HSCs compared with those of the control, reflecting increases in the fraction of cells in the S and G2/M phases (Fig. 4A). In the myeloid compartments, more subtle but statistically significant increases in the frequencies of cells in the G2/M phase were observed as a consequence of B-myb deletion. These studies indicate that loss of B-myb expression alters cell cycle progression predominantly in HSCs, and to a lesser extent, in myeloid progenitors.
Fig. 4.
Loss of B-myb results in aberrant cell cycle progression in HSCs and increased cell death of myeloid progenitors. (A) BM was harvested from pIpC-treated control and B-myb F/Fcre mice 2 h following injection with BrdU. The cells were then subjected to flow cytometric analysis to determine the percentage of cells in the indicated populations that are in the G0/G1 (lower left gate), S (upper gate), and G2/M (lower right gate) phases of the cell cycle. (B) Frequency of Annexin V+DAPI+ HSCs and myeloid progenitors of pIpC-treated control and B-myb–deficient mice. (C) Frequencies of myeloid progenitors in the BM of pIpC-treated control and B-myb–deficient mice. Representative plots are shown. All numerical values represent mean ± SEM for three independent experiments. n ≥ 3 per population for each genotype. *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005.
Loss of B-myb Increases Cell Death of Myeloid Progenitors.
We next determined whether altered cell cycle kinetics in B-myb KO mice resulted in differentiation-associated cell death, which might account for the depletion of HSCs and progenitors, by staining BM from pIpC-treated control and B-myb KO mice with Annexin V. B-myb KO mice did not have increased percentages of apoptotic (Annexin V+ DAPI+) HSCs or MPPs (Fig. 4B). However, the fraction of apoptotic CMPs, GMPs, and MEPs was higher in B-myb–deficient animals, with the difference in cell death being significant in the GMP population. Interestingly, despite the fact that B-myb–deficient mice have a profound reduction in the number of myeloid progenitors and mature myeloid cells (Fig. 2), the relative frequency of GMPs was actually increased in these animals (Fig. 4C). To determine whether the higher percentage of GMPs in the B-myb KO mice resulted from increased differentiation of CMPs and/or proliferation of GMPs, we sorted these populations from control and B-myb F/Fcre mice (16) and plated them in GM-CSF–containing medium in the presence of IFN-α to delete floxed sequences in vitro (12). Loss of B-myb resulted in lower cell numbers, reflecting an inhibition of both CMP and GMP proliferation (Fig. S7A). Furthermore, a sharp decrease in the frequency of CD11b+ and Gr1+ mature myeloid cells was observed in cultures of CMPs. The frequency of the CD11b+Gr1hi population in the B-myb F/Fcre GMP cultures was also significantly decreased (Fig. S7B), suggesting that loss of B-myb affects proliferation and differentiation along the myeloid lineage.
Disruption of B-myb Results in Altered Gene Expression in LSK+ Cells.
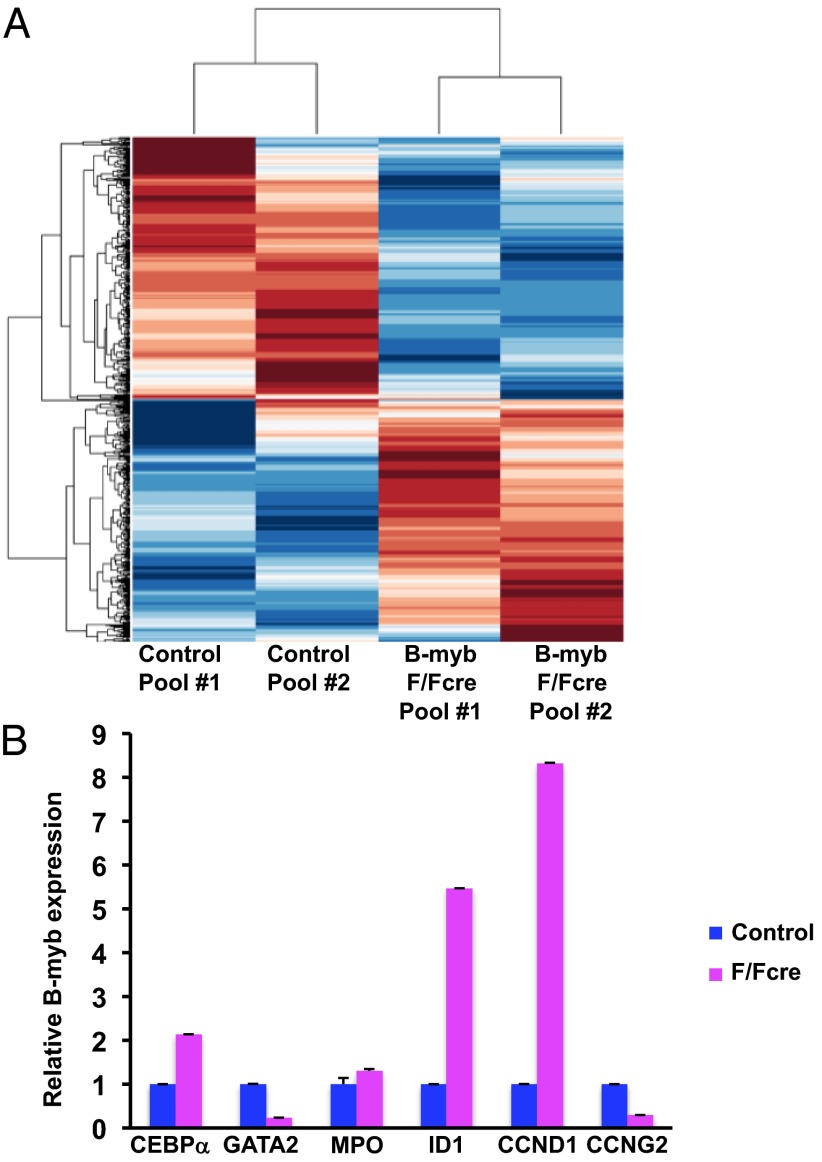
To gain mechanistic insight into the hematological defects in B-myb–deficient mice, we performed a oligonucleotide microarray analysis of the Lin−Sca1+ckit+ (LSK+) population because the defect in development originates in the HSCs. Comparative analysis and hierarchical clustering identified 438 and 470 genes that were significantly up- and down-regulated, respectively, in the B-myb KO cells (Fig. 5A). When these gene lists were further analyzed using the ChIP Enrichment Analysis (ChEA) database within ENRICHR (http://amp.pharm.mssm.edu/Enrichr) (26, 27), considerable overlap was observed with the promoters of genes that are occupied by transcription factors during the control of maintenance, development, and multipotency of HSCs and progenitor cells as well as embryonic stem cells (ESCs) (Dataset S1) (28–34). Of these transcription factors, C-MYC and E2F-1 are known to be intricately involved in B-MYB–dependent transcription (22, 25, 35–38), whereas others, such as JARID1A, RUNX1 (AML1), TAL1 (SCL), LMO1, and FLI1, are not as well defined with respect to B-MYB transcriptional activation and repression of its targets (25, 39). In addition to the transcription factors listed above, we also detected an overlap with genes that are targets of C-MYB, which is known to regulate promoters that are both distinct from and common with other members of the MYB family (40, 41).
Fig. 5.
Computational analysis of the B-myb LSK+ transcriptiome. (A) Clustering analysis of altered gene expression in microarrays of control and B-myb–depleted LSK+ cells. (B) Quantitative mRNA expression (qPCR) of differentially expressed genes in control and B-myb KO LSK+ cells. All results were normalized to β-actin expression and are graphed as the average fold-change (±SEM). PCR reactions were performed in triplicate using cDNAs isolated from at least two independent pools of sorted cells per genotype.
We next confirmed the pattern of expression of differentially expressed genes known to be expressed in HSCs that, based on the predicted level of expression in the absence of B-myb, might possibly account for the observed phenotype. Quantitative PCR (qPCR) revealed that the level of cyclin D1 (CCND1), which promotes G1/S progression, is increased by approximately eightfold in B-myb–deficient LSK+ cells (Fig. 5B). We also detected a decrease in S/G2 cyclin, cyclin G2 (CCNG2) expression after B-myb deletion. The dramatic changes in the myeloid progenitor compartment of the B-myb KO mice also prompted us to assess the expression of select genes that govern myeloid lineage development. ID1, GATA2, and CEBPα are transcription factors that regulate HSC development as well as various stages of myeloid lineage commitment (42). ID1 levels, which normally increase as cells transition to the GMP stage (43), were elevated in B-myb KO LSK+ cells (Fig. 5B), along with those of CEBPα and to a lesser extent, myeloperoxidase (MPO). Of these, CEBPα is required for the development of CMPs to GMPs (44). GATA2, on the other hand, was decreased in the absence of B-myb. We also examined the expression pattern of genes that modulate S phase progression and G2/M transition, some of which have been implicated in the proliferative defects observed in B-myb–null ESCs (22). Of the genes analyzed in Fig. S8, only PLK1 and BIRC5 (survivin) were predicted to be differentially expressed. The expression pattern of these genes does not overlap with B-myb KO ESCs, which might suggest a differential use of target genes in LSK+ cells, or, simply reflects a greater accumulation of B-myb–deficient LSK+ cells in S phase compared with ESCs (22). Given that B-myb–null HSCs display a profound engraftment defect, these studies suggest that loss of B-myb induces changes in gene expression that impair self-renewal and results in aberrant lineage commitment.
Discussion
Here, we describe the effect of loss of B-myb during adult hematopoiesis. Our studies show that B-myb–deficient mice are pancytopenic, with dramatic losses in BM and splenic cellularity, have a profound cell-automous defect in HSC repopulation capacity, altered HSC (and to lesser extent of myeloid progenitor) cycling, as well increased apoptosis of myeloid progenitors. Studies have shown that cycling HSCs and those that have recently undergone cell division have profound reductions in their engraftment potential, and that long-term repopulation is dependent on the ability of these cells, or at least a portion of them, to remain in a quiescent state (18). HSC quiescence is key to maintaining the balance between long-term self-renewal and differentiation. Accordingly, genetic disruption of genes that alter cell division cause BM failure due to HSC exhaustion (45). Analysis of B-myb–deficient HSCs revealed that significant percentages of cells are in the S and G2/M phases. B-myb KO myeloid progenitors also have increased frequencies of G2/M phase cells. In addition, CMPs and GMPs in which B-myb is disrupted in vitro proliferate more slowly. These results are consistent with previous reports documenting a role for B-myb in S/G2/M progression and checkpoint control (20, 22, 25, 46, 47). A similar phenotype is also observed in the zebrafish crash&burn (crb) mutant (48), which harbors a loss of function mutation in bmyb. Cells in these embryos have delays in mitotic progression, genomic instability, and increased cell death, and it is thought that loss of bmyb during S phase impairs CYCLIN B1 expression, which is required for progression through G2 (48, 49). Although we encountered difficulties in examining the cell cycle of LT-HSCs as a distinct population in B-myb KO mice due to their extremely low numbers, our data suggest that loss of B-myb pushes HSCs out of quiescence, but arrests them in the S and G2/M phases. As loss of quiescence is associated with a failure to maintain long-term self-renewal (45), these data provide a partial explanation for HSC depletion in these animals. B-myb KO LSK+ cells express substantially higher levels of CCND1 mRNA, which, in complex with CDK4/6, drives the cell through G1 and into S-phase (50). It is interesting to note that differentiation of LT-HSCs into ST-HSCs and MPPs, but not HSC self-renewal, correlates with the induction of CCND1 mRNA (18). As the CCND1 promoter is bound by B-MYB in ChIP assays (25), this increase in cyclin D1 mRNA is likely due to the absence of repression of the CCND1 promoter by B-MYB. HSC differentiation is also accompanied by increases in CCNG2, whereas self-renewing HSCs do not exhibit such changes in gene expression (18). CCNG2 is considered an atypical cyclin in that its up-regulation is associated with cell cycle inhibition, and studies have demonstrated the involvement of this gene in the induction and/or maintenance of cell cycle arrest (51). CCNG2 mRNA levels in B-myb–deficient LSK+ cells were decreased, and it is therefore possible that B-myb KO HSCs do enter the differentiation program but ultimately fail to complete it. It is interesting to note that even though the S and G2/M checkpoints in B-myb–null ESCs are defective, these cells still progress through the cell cycle, even with a loss of genomic integrity (20, 22). Hence, defects in replication stress responses could also lead to abnormalities in HSC function and differentiation in the absence of B-myb expression.
Because multiple lineages are affected by the loss of B-myb, we also examined the level of cell death in the stem and progenitor compartments. We did not detect increases in the frequency of Annexin V+ HSCs or MPPs; however, we did observe increased cell death in the three populations of myeloid progenitors, which would additionally contribute to the depletion of mature myeloid cells. As previously stated, despite the fact that B-myb KO mice have a profound reduction in the number of myeloid progenitors and mature myeloid cells, the percentage of GMPs in these animals is actually increased compared to controls. Accordingly, the level of ID1, which is normally up-regulated as the cells transition from the LMPP to GMP stage (43), is higher in B-myb–deficient LSK+ cells. In addition, we observed higher levels of CEBPα and lower levels of GATA2 in B-myb KO LSK+ cells. Previous studies have shown that GMP development is also favored if CEBPα is up-regulated before GATA2, and these cells will continue to give rise to neutrophils and monocytes if CEBPα expression is maintained (1).
When viewed from the perspective of increased GMP frequency and its relation to pancytopenia, the B-myb KO phenotype is remarkably similar to that seen in patients with high-risk myelodisplastic syndrome (MDS) (52, 53), a group of blood disorders that are characterized by cytopenias that arise due to ineffective hematopoiesis and often progress to acute meylogenous leukemia. MYBL2 is expressed at 20–30% of normal levels in CD34+ cells of ∼65% of MDS cases, irrespective of karyotype (46, 54). Consistent with the notion that partial loss of MYBL2 expression is associated with MDS, it has recently been shown that B-myb heterozygous mice develop MDS and other myeloproliferative neoplasms at 12–24 mo of age (54). Although the hematopoietic compartment of younger animals did not show significant changes, the phenotype of a subset of the aged animals, particularly those subjected to transplant-induced replicative stress, is similar to that of B-myb KO mice immediately following pIpC administration. These data suggest that B-myb haploinsufficiency cooperates with other genetic lesions to cause disease, in contrast to that which occurs in the absence of B-myb expression or in cells expressing levels of B-myb that mirror those seen in CD34+ MDS cells (approximately 20–30% of normal) (46). A better understanding of the role of B-myb in normal hematopoietic cell development will hopefully allow us to define its role in disease states that result in defective hematopoiesis, such as MDS.
Materials and Methods
Detailed information on animals, bone marrow transplantation, and flow cytometry can be found in SI Materials and Methods. Microarray and RT-PCR analysis were performed as described in SI Materials and Methods. Primer sequences are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Stephen Cosenza and Amol Padgaonkar for assistance with animal studies and the Flow Cytometry shared resource facility (Icahn School of Medicine at Mount Sinai) for assistance with FACS. This work was supported, in part, by National Institutes of Health National Heart, Lung, and Blood Institute Grant 5R01HL085279 (to E.P.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.B. is a guest editor invited by the Editorial Board.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE53875).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315464111/-/DCSupplemental.
References
- 1.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Seita J, Weissman IL. Hematopoietic stem cell: Self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2(6):640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Mucenski ML, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65(4):677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 5.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5(7):721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 6.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci USA. 2004;101(41):14853–14858. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahl SP, Crittenden RB, Allman D, Bender TP. c-Myb is required for pro-B cell differentiation. J Immunol. 2009;183(9):5582–5592. doi: 10.4049/jimmunol.0901187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23(3):275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Vegiopoulos A, García P, Emambokus N, Frampton J. Coordination of erythropoiesis by the transcription factor c-Myb. Blood. 2006;107(12):4703–4710. doi: 10.1182/blood-2005-07-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García P, et al. Reduced c-Myb activity compromises HSCs and leads to a myeloproliferation with a novel stem cell basis. EMBO J. 2009;28(10):1492–1504. doi: 10.1038/emboj.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieu YK, Reddy EP. Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci USA. 2009;106(51):21689–21694. doi: 10.1073/pnas.0907623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieu YK, Reddy EP. Impaired adult myeloid progenitor CMP and GMP cell function in conditional c-myb-knockout mice. Cell Cycle. 2012;11(18):3504–3512. doi: 10.4161/cc.21802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka Y, Patestos NP, Maekawa T, Ishii S. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem. 1999;274(40):28067–28070. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- 14.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 15.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 16.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 17.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91(5):661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 18.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202(11):1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García P, Frampton J. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. J Cell Sci. 2006;119(Pt 8):1483–1493. doi: 10.1242/jcs.02870. [DOI] [PubMed] [Google Scholar]
- 20.Tarasov KV, et al. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS ONE. 2008;3(6):e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28(15):1737–1747. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- 22.Lorvellec M, et al. B-Myb is critical for proper DNA duplication during an unperturbed S phase in mouse embryonic stem cells. Stem Cells. 2010;28(10):1751–1759. doi: 10.1002/stem.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Down CF, Millour J, Lam EW, Watson RJ. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim Biophys Acta. 2012;1819(8):855–862. doi: 10.1016/j.bbagrm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26(5):474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan M, et al. The B-MYB transcriptional network guides cell cycle progression and fate decisions to sustain self-renewal and the identity of pluripotent stem cells. PLoS ONE. 2012;7(8):e42350. doi: 10.1371/journal.pone.0042350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen EY, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lachmann A, et al. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, et al. Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res. 2008;18(12):1177–1189. doi: 10.1038/cr.2008.309. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao L, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39(11):4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Kassouf MT, et al. Genome-wide identification of TAL1’s functional targets: Insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20(8):1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7(4):532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 35.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15(8):4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, Lucibello FC, Zwicker J, Engeland K, Müller R. Cell cycle-regulated repression of B-myb transcription: Cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24(15):2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam EW, Watson RJ. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12(7):2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joaquin M, Watson RJ. Cell cycle regulation by the B-Myb transcription factor. Cell Mol Life Sci. 2003;60(11):2389–2401. doi: 10.1007/s00018-003-3037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuzuki S, Seto M. TEL (ETV6)-AML1 (RUNX1) initiates self-renewing fetal pro-B cells in association with a transcriptional program shared with embryonic stem cells in mice. Stem Cells. 2013;31(2):236–247. doi: 10.1002/stem.1277. [DOI] [PubMed] [Google Scholar]
- 40.Golay J, et al. Redundant functions of B-Myb and c-Myb in differentiating myeloid cells. Cell Growth Differ. 1997;8(12):1305–1316. [PubMed] [Google Scholar]
- 41.Watson RJ, Robinson C, Lam EW. Transcription regulation by murine B-myb is distinct from that by c-myb. Nucleic Acids Res. 1993;21(2):267–272. doi: 10.1093/nar/21.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: Balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 43.Cochrane SW, Zhao Y, Welner RS, Sun XH. Balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions. Blood. 2009;113(5):1016–1026. doi: 10.1182/blood-2008-06-164996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21(6):853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Pietras EM, Warr MR, Passegué E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195(5):709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinrichs S, et al. MYBL2 is a sub-haploinsufficient tumor suppressor gene in myeloid malignancy. Elife. 2013;2:e00825. doi: 10.7554/eLife.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García P, Berlanga O, Watson R, Frampton J. Generation of a conditional allele of the B-myb gene. Genesis. 2005;43(4):189–195. doi: 10.1002/gene.20170. [DOI] [PubMed] [Google Scholar]
- 48.Shepard JL, et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc Natl Acad Sci USA. 2005;102(37):13194–13199. doi: 10.1073/pnas.0506583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stern HM, et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol. 2005;1(7):366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- 50.Casimiro MC, Crosariol M, Loro E, Li Z, Pestell RG. Cyclins and cell cycle control in cancer and disease. Genes Cancer. 2012;3(11-12):649–657. doi: 10.1177/1947601913479022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmermann M, et al. Elevated cyclin G2 expression intersects with DNA damage checkpoint signaling and is required for a potent G2/M checkpoint arrest response to doxorubicin. J Biol Chem. 2012;287(27):22838–22853. doi: 10.1074/jbc.M112.376855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Will B, et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 2012;120(10):2076–2086. doi: 10.1182/blood-2011-12-399683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pang WW, et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci USA. 2013;110(8):3011–3016. doi: 10.1073/pnas.1222861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarke M, et al. MYBL2 haploinsufficiency increases susceptibility to age-related haematopoietic neoplasia. Leukemia. 2013;27(3):661–670. doi: 10.1038/leu.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.