ABSTRACT
The interferon (IFN)-inducible antiviral state is mediated in part by the 2′,5′-oligoadenylate (2-5A) synthetase (OAS)/RNase L system. 2-5A, produced from ATP by OAS proteins in response to viral double-stranded RNA, binds to and activates RNase L. RNase L restricts viral infections by degrading viral and cellular RNA, inducing autophagy and apoptosis, and producing RNA degradation products that amplify production of type I interferons (IFNs) through RIG-I-like receptors. However, the effects of the OAS/RNase L pathway on IFN induction in different cell types that vary in basal levels of these proteins have not been previously reported. Here we report higher basal expression of both RNase L and OAS in mouse macrophages in comparison to mouse embryonic fibroblasts (MEFs). In MEFs, RNase L gene knockout decreased induction of IFN-β by encephalomyocarditis virus infection or poly(rI):poly(rC) (pIC) transfection. In contrast, in macrophages, RNase L deletion increased (rather than decreased) induction of IFN-β by virus or pIC. RNA damage from RNase L in virus-infected macrophages is likely responsible for reducing IFN-β production. Similarly, direct activation of RNase L by transfection with 2-5A induced IFN-β in MEFs but not in macrophages. Also, viral infection or pIC transfection caused RNase L-dependent apoptosis of macrophages but not of MEFs. Our results suggest that cell-type-specific differences in basal levels of OAS and RNase L are determinants of IFN-β induction that could affect tissue protection and survival during viral infections.
IMPORTANCE
Type I interferons (IFNs) such as IFN-β are essential antiviral cytokines that are often required for animal survival following infections by highly pathogenic viruses. Therefore, host factors that regulate type I IFN production are critically important for animal and human health. Previously we reported that the OAS/RNase L pathway amplifies antiviral innate immunity by enhancing IFN-β production in mouse embryonic fibroblasts and in virus-infected mice. Here we report that high basal levels of OAS/RNase L in macrophages reduce, rather than increase, virus induction of IFN-β. RNA damage and apoptosis caused by RNase L were the likely reasons for the decreased IFN-β production in virus-infected macrophages. Our studies suggest that during viral infections, the OAS/RNase L pathway can either enhance or suppress IFN production, depending on the cell type. IFN regulation by RNase L is suggested to contribute to tissue protection and survival during viral infections.
Observation
The 2′,5′-oligoadenylate (2-5A) synthetase (OAS)/RNase L system is a uniquely regulated innate immune pathway that restricts viral infections (reviewed in reference 1). The interferon (IFN) inducible OAS family of proteins include pathogen recognition receptors for viral double-stranded RNA (dsRNA) that synthesize 2-5A from ATP. In mice, the enzymatically active species of OAS include OAS1a, OAS2, and OAS3 (reviewed in reference 2). 2-5A binds with high affinity and specificity to the latent cytoplasmic endoribonuclease, RNase L, causing its dimerization and activation (3). RNase L then cleaves both viral and cellular single-stranded regions of RNA, predominantly at UpU and UpA dinucleotides (4), leading to inhibition of viral replication in vivo (5).
The RNase L antiviral mechanism varies, depending on the types of RNA molecules that are cleaved (reviewed in reference 1). If the RNA substrate for RNase L is viral genomic single-stranded RNA, even a single cleavage event per viral genome might prevent replication. Cleavage of viral mRNA is a mechanism that could potentially apply to RNA and DNA viruses with different replication strategies. In addition, degradation of cellular RNA, including rRNA in intact ribosomes, could damage host cell machinery required for protein synthesis (6, 7). Also, RNase L restricts viral infections by causing apoptosis through an unknown mechanism involving Jun N-terminal kinase (JNK) (5, 6, 8). Furthermore, RNase L activation results in autophagy in a pathway involving JNK and protein kinase R (PKR) that can either enhance or decrease viral replication (9, 10). Finally, RNase L indirectly controls viral infections by enhancing viral induction of IFN-β induction in some cell types and in animals (11).
RNase L amplifies IFN-β induction by producing small RNA cleavage products from cellular or viral RNA that interact with RIG-I and MDA5 (11, 12). Accordingly, Rnasel−/− mice produce less IFN-β, as measured in serum, after infection with encephalomyocarditis virus (EMCV) (intraperitoneally) or Sendai virus (intranasally) than do wild-type (WT) mice (11). The small RNA cleavage products produced by RNase L that enhance IFN-β production can be either cellular (self) or viral (nonself) and typically have some double-strand regions, a 5′-hydroxyl, and a phosphate at the 2′,3′-cyclic terminus (11, 12). Previously we showed in human hepatoma Huh7 cells that RNase L released a small RNA product from the NS5B region of hepatitis C virus (HCV) genomic RNA that bound to RIG-I, causing displacement of its repressor domain, stimulating its ATPase function, and propagating signaling through the mitochondrial antiviral signaling protein (MAVS) to the IFN-β gene (12). The small RNA from HCV RNA was also shown to induce a hepatic innate immune response when injected into mice. Similarly, an mRNA of parainfluenza virus 5 activated type I IFN production, independently of the encoded protein, by a pathway involving RNase L and MDA5 (13). Also RNase L was shown to be a factor in type I IFN induction in response to herpes simplex virus 2 infection (14). Recently, we reported large differences (up to 100-fold) in basal expression of different OAS isoforms between different types of primary mouse cells (15). Notably, bone marrow-derived macrophages (BMMs), as well as primary microglia, brain resident macrophages, expressed the highest levels of different OAS isoforms compared with several other primary cell types. In a separate study, peritoneal macrophages (p-Macs) were observed to have high basal expression of OAS (16). In addition, we previously compared basal levels of RNase L in 9 rodent and 11 human cell lines (17). The mouse and human macrophage-like cell lines RAW264.7 and U937, respectively, were among the cell lines expressing the highest levels of RNase L protein. In the same study, different mouse fibroblast cell lines (NIH 3T3, SVT2, and L929) were shown to express relatively low levels of RNase L protein. However, the effect of RNase L on IFN induction might vary, depending on where the IFN is measured, the subtype of IFN being measured, the type of virus and its route of infection, and the type of cells that are infected. Here we have investigated the last of these variables, namely cell-type-specific effects of RNase L on IFN induction.
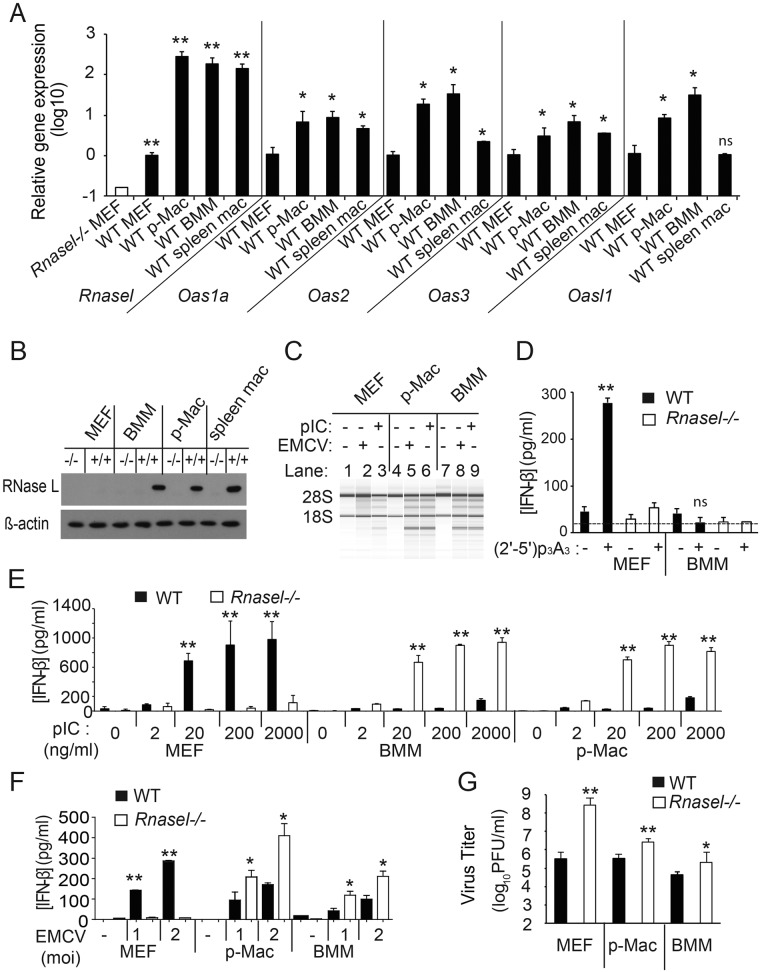
Levels of mRNAs for Rnasel and different Oas genes were monitored by quantitative reverse transcription-PCR (qRT-PCR) in MEF cell lines, BMMs, peritoneal macrophages (p-Macs), and freshly isolated splenic macrophages (Fig. 1A). Compared to MEFs, BMMs had elevated levels of mRNAs for Rnasel (200-fold), Oas1a (9-fold), Oas2 (34-fold), Oas3 (7-fold), and Oasl1 (32-fold) (Fig. 1A). Oas1a, Oas2, and Oas3 encode OAS isoforms that are enzymatically active, whereas Oasl1 encodes a protein that lacks the ability to synthesize 2-5A and is a negative regulator of type I IFN synthesis (18). Similarly, compared to MEFs, p-Macs had increased levels of mRNAs for Rnasel (260-fold), Oas1a (7-fold), Oas2 (19-fold), Oas3 (3-fold), and Oasl1 (8-fold). To control for possible activation of macrophages in vitro, splenic macrophages were isolated by fluorescence-activated cell sorter (FACS) and immediately processed for RNA isolation followed by qRT-PCR. Splenic macrophages also had increased basal levels of mRNAs for Rnasel (142-fold), Oas1a (4.6-fold), Oas2 (2.2-fold), and Oas3 (3.5-fold); however, Oasl1 mRNA levels were unchanged. These findings show enhanced basal mRNA expression of Rnasel and most enzymatically active isoforms of OAS in macrophages compared with MEFs. Western blot assays were done to monitor differences in RNase L protein levels. Whereas RNase L was observed in wild-type BMMs, p-MACs, and splenic macrophages, RNase L was not observed in wild-type MEFs (Fig. 1B). However, in WT MEFs, low levels of Rnasel mRNA were detected by qRT-PCR, and low levels of RNase L protein were apparent by monitoring characteristic, discrete rRNA cleavage products during poly(rI):poly(rC) (pIC) transfection or encephalomyocarditis virus (EMCV) infection (Fig. 1A and C, lanes 2 and 3, respectively). The rRNA fragments were, however, produced at much higher levels in p-Macs and BMMs than in MEFs (Fig. 1C, lanes 5, 6, 8, and 9). Consistent with our prior results (11), 2-5A transfection induced IFN-β in WT MEFs but not in Rnasel−/− MEFs, as determined by enzyme-linked immunosorbent assay (ELISA) (Fig. 1D). In contrast, 2-5A transfection failed to induce IFN-β in either WT or Rnasel−/− BMMs (Fig. 1D). We verified uptake of 2-5A and activation of RNase L in BMMs with rRNA cleavage assays (data not shown).
FIG 1 .
Basal levels of OAS and RNase L regulate induction of IFN-β by virus or pIC. (A) Basal levels of Rnasel and Oas1a, Oas2, Oas3, and Oasl1 mRNAs were determined by qRT-PCR with total RNA isolated with mirVANA RNA kits (LifeTechnology) with Oas primers described previously (15) and with Rnasel forward primer: 5′ TAGGCGAACACATCAATGAGGA 3′ and reverse primer 5′ CTGCCTCTGGAACGCTGAG 3′. RNA expression relative to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was expressed as 2−ΔCT, where ΔCT represents the threshold cycle (CT) of the gene of interest − the CT of GAPDH. The open bar represents the baseline determined with RNA from Rnasel−/− MEFs. The data are shown as the means ± standard deviation (SD) calculated from a minimum of 3 (up to 6) biological replicates. MEFs immortalized with simian virus 40 (SV40) T antigen were cultured in RPMI 1640 with 10% fetal bovine serum (FBS) (5). Isolation and culture conditions for BMMs and thioglycolate-elicited p-Macs were as described previously (24, 25). Splenic macrophages were isolated by FACS with fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse CD11b IgG2b (BD Pharmingen; catalog no. 557396, clone M1/70) and allophycocyanin-conjugated rat anti-mouse F4/80 IgG2b (Bio-Rad/Abd Serotec; product code MCA497APC) as described previously (25). Experiments involving mice were performed under an approved IACUC protocol from the Cleveland Clinic. (B) Western blots of the different cell-type extracts were performed with rabbit polyclonal antibody against murine RNase L (B. K. Jha, B. Dong, and R. H. Silverman, unpublished data) and antibody to β-actin (Sigma-Aldrich). (C) Cells were either infected with EMCV (a gift from Ian M. Kerr, London, United Kingdom) (MOI of 0.1) for 16 h or were Lipofectamine 2000 (Life Technologies) transfected with pIC (Sigma-Aldrich) at 2 µg/ml for 6 h. Total RNA was isolated with Trizol (Life Technologies), and rRNA degradation was monitored in RNA chips with an Agilent 2100 bioanalyzer. (D) Transfections of (2′-5′) p3A3 (10 µM) with Lipofectamine 2000 (Life Technologies) and mock transfections were done for 18 h. IFN-β levels in the cell supernatant were determined by ELISA (PBL Assay Science). The lower limit of detection of IFN-β in the ELISA was about 15 pg/ml (dashed line). (E and F) IFN-β concentrations were measured by ELISA from media of cells (E) transfected with pIC (at the indicated concentrations) for 8 h or mock transfected or (F) infected with EMCV (MOI of 1 or 2) for 16 h. Values are means from biological triplicate assays ± SD. (G) EMCV yields following infection (MOI of 0.1) at 8 h postinfection determined from the culture supernatant by plaque assay on Ifnar−/− MEF indicator cells as described previously (5). Results are shown as the means ± SD from three biological replicates. Two-tailed t tests were done. **, P < 0.001; *, P < 0.05.
To determine effects of RNase L on IFN-β induction by dsRNA, cells were transfected with different concentration of pIC (Fig. 1E). Similar to our prior study (11), RNase L enhanced IFN-β induction to a much greater extent in WT MEFs than in Rnasel−/− MEFs (Fig. 1E). Remarkably, however, the effect of RNase L on IFN induction was reversed in both BMMs and p-MACs. IFN-β accumulated to higher levels in Rnasel−/− BMMs (6.5-fold) and p-Macs (4.5-fold) than in WT p-Macs and BMMs (in response to 2 µg per ml of pIC) (Fig. 1E). Without transfection reagent, however, there was no effect of RNase L deletion on pIC induction of IFN-β in BMMs and p-Macs, suggesting that Toll-like receptor 3 (TLR3)-mediated induction of IFN was unaffected by RNase L (19) (data not shown).
To extend these studies to viral induction of IFN-β, cells were infected with EMCV. Viral infection induced IFN-β in WT MEFs but not in Rnasel−/− MEFs (Fig. 1F). Again, there was a reversal of the effect of RNase L on IFN-β induction in the myeloid cell types. Deletion of RNase L increased IFN-β production in response to EMCV infection by about 2-fold in p-Macs and BMMs (Fig. 1F). To determine if the differences in viral induction of IFN-β were related to the antiviral effect of RNase L, after one cycle of virus growth (8 h at a multiplicity of infection [MOI] of 0.1), viral yields were measured by plaque assays on type I IFN receptor (Ifnar)−/− MEF indicator cells. Viral yields were increased 85-, 8-, and 5-fold in Rnasel−/− MEFs, p-Macs, and BMMs, respectively, compared to the corresponding WT cells (Fig. 1G). Therefore, RNase L deficiency resulted in higher viral yields in MEFs than in p-Macs and BMMs. These results are consistent with the observation that RNase L gene knockout increased IFN-β production in p-Macs and BMMs but decreased IFN-β production in MEFs. In the Rnasel−/− MEFs, the decrease in IFN-β production in combination with a loss of nuclease activity likely combined to cause an increase in EMCV titers. In Rnasel−/− BMMs, the absence of nuclease activity normally provided by RNase L overrides the increase in IFN levels, allowing virus to replicate to higher titers in Rnasel−/− than in WT macrophages.
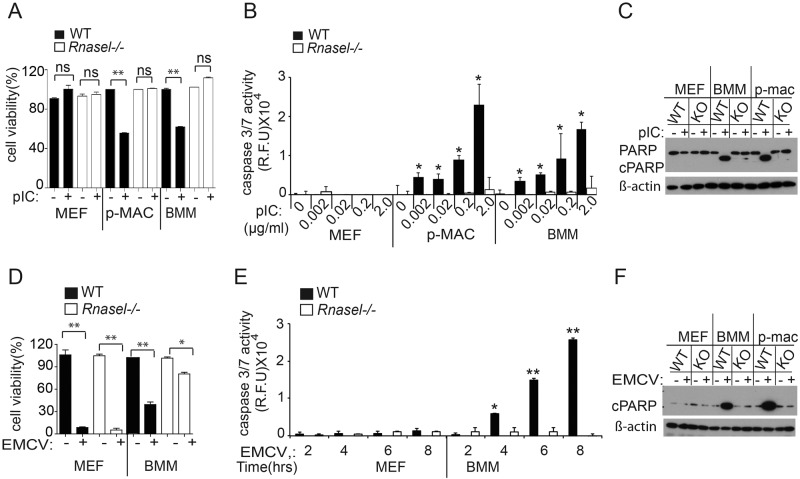
Apoptosis is known to occur in cells that sustain high levels of RNA damage due to RNase L activity (5, 6, 8). Therefore, cell viability and apoptosis were monitored in cell types expressing low and high basal levels of OAS and RNase L. There was no effect on cell viability of pIC transfection in WT MEFs or Rnasel−/− MEFs as determined by 3-(4,5-dimethyl-2-thiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) colorimetric assays (Fig. 2A). In contrast, pIC transfection reduced viability of WT p-Macs or BMMs by ~2-fold, whereas pIC transfection had no effect on viability of Rnasel−/− p-Macs or BMMs. Similarly, no apoptosis was detected in pIC-transfected WT or Rnasel−/− MEFs as determined by lack of caspase 3/7 activation (Fig. 2B). However, caspase activation assays showed that pIC transfection induced apoptosis of WT p-Macs and BMMs, but not in Rnasel−/− p-MACs and BMMs (Fig. 2B). These results were confirmed by monitoring poly(ADP-ribose) polymerase (PARP) cleavage in Western blots (Fig. 2C). Cleaved PARP was observed after pIC transfection of WT BMMs and p-Macs but not in identically treated WT MEFs. EMCV infection resulted in death of both WT and Rnasel−/− MEFs (Fig. 2D). However, while EMCV reduced viability of WT BMMs by about 2.5-fold, a smaller (1.2-fold) reduction of viability was observed in EMCV-infected Rnasel−/− BMMs (Fig. 2D). EMCV appears to have caused necrotic death of MEFs, as there was no measurable apoptosis as determined by lack of caspase 3/7 activity (Fig. 2E). In contrast, EMCV infection caused apoptosis of WT BMMs but not of Rnasel−/− BMMs, as determined by caspase 3/7 activity assays (Fig. 2E). In addition, PARP cleavage assays showed viral induction of apoptosis in WT BMMs and WT p-Macs but not in WT MEFs (Fig. 2F). These findings show that RNase L contributes to apoptosis of BMMs and p-Macs in response to either pIC transfection or EMCV infection. However, low levels of OAS isoforms and RNase L in WT MEFs are insufficient to induce apoptosis in response to either pIC transfection or EMCV infection.
FIG 2 .
Effects of pIC transfection or viral infection on cell viability and apoptosis. (A) Cells were transfected with pIC (2 µg/ml) for 12 h. Cell viability was quantified by MTS colorimetric assays (Promega). Results are means ± SD from three biological replicates. (B) Caspase 3/7 activation was determined with an Apo-ONE homogeneous caspase 3/7 kit (Promega) after pIC transfection (2 ng to 2 µg/ml for 8 h). Results are means ± SD from three biological replicates. (C) Western blots with antibody to uncleaved PARP and cleaved PARP (cPARP) (Cell Signaling; catalog no. 9542) or to β-actin (Sigma-Aldrich) from cells transfected with pIC (2 µg per ml) for 12 h. (D) Cells were infected with EMCV (MOI of 1.0) for 24 h, and cell viability was measured by MTS assays. (E) Activation kinetics for caspase 3/7 activity during EMCV infection (MOI of 1.0) at the indicated times postinfection. Results are means ± SD of three biological replicates. **, P < 0.0001; *, P < 0.05; ns, not significant. (F) PARP cleavage in response to EMCV infections at an MOI of 1.0 for 8 h as determined in Western blots.
Our results show that, paradoxically, RNase L can either enhance or reduce IFN-β induction by viral infection, depending on basal levels of 2-5A pathway enzymes OAS and RNase L. RNase L generates small RNA molecules from either self (cellular) or nonself (viral) RNA that activate the RIG-I or MDA5/MAVS/TBK/IRF3 axis that drives transcription of the IFN-β gene (11, 12). However, this phenomenon occurs only in cell types, such as MEFs, that have relatively low basal levels of OAS isoforms and RNase L. In cell types, such as macrophages, that have high basal levels of OAS (15) and RNase L (Fig. 1A), there is a different outcome—reduced rather than increased production of IFN-β following viral infection. RNase L is known to cleave both viral and cellular RNA molecules, including both mRNA and rRNA in intact ribosomes (7, 20). The damage to cellular RNA by RNase L, resulting in inhibition of protein synthesis (6, 21) and apoptosis (5, 6, 8), is the likely cause of reduced IFN synthesis in macrophages. The RNase L-mediated suppression of IFN production in macrophages occurs despite high expression of the IFN-stimulated gene products RIG-I and MDA5 (15) that function in signaling to the IFN-β gene. Furthermore, Mda5 expression as determined by qRT-PCR was 35-fold higher in BMMs than in MEFs (data not shown). We previously reported that induction of IFN-β in response to 2-5A transfection was partially reduced in Mda5−/− or Rig-i−/− MEFs compared with WT MEFs (11). These findings highlight the robustness of RNase L activity in myeloid cells that results in the degradation of mRNA and rRNA, thereby reducing IFN induction despite high levels of MDA5, a protein that senses both pIC and EMCV (22).
We propose that viral infections of cell types that mediate tissue integrity (such as fibroblasts) produce higher levels of type I IFNs as a result of RNase L and the small RNAs that it generates. The amplification of IFN production contributes to tissue protection (11). In virus-infected macrophages, high basal levels of OAS and RNase L result in lower levels of type I IFNs, at least in response to pIC transfection or EMCV infection. However, despite different patterns of the type I IFN response, both MEFs and macrophages contribute to host survival. In a prior study, using the murine coronavirus mouse model, we found that RNase L activity in liver sinusoidal resident macrophages, Kupffer cells, seems to prevent spread of the virus into the liver parenchyma and the consequent development of hepatitis (23). In this manner, high levels of OAS and RNase L in macrophages also contribute to tissue protection.
ACKNOWLEDGMENTS
We thank Christina Gaughan for expert technical assistance.
The studies were supported by a grant from the National Institutes of Health (NIH), National Cancer Institute (grant no. CA044059) to R.H.S. and NIH, National Institute of Allergy and Infectious Diseases (grant no. AI104887) to S.R.W. and R.H.S., and the Mal and Lea Bank Chair Fund (to R.H.S.).
Footnotes
Citation Banerjee S, Chakrabarti A, Jha BK, Weiss SR, Silverman RH. 2014. Cell-type-specific effects of RNase L on viral induction of beta interferon. mBio 5(2):e00856-14. doi:10.1128/mBio.00856-14.
REFERENCES
- 1. Silverman RH. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720–12729. 10.1128/JVI.01471-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. 2011. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 31:41–47. 10.1089/jir.2010.0107 [DOI] [PubMed] [Google Scholar]
- 3. Dong B, Silverman RH. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 270:4133–4137. 10.1074/jbc.270.8.4133 [DOI] [PubMed] [Google Scholar]
- 4. Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. 1981. Interferon action—sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289:414–417. 10.1038/289414a0 [DOI] [PubMed] [Google Scholar]
- 5. Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355–6363. 10.1093/emboj/16.21.6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li G, Xiang Y, Sabapathy K, Silverman RH. 2004. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J. Biol. Chem. 279:1123–1131 [DOI] [PubMed] [Google Scholar]
- 7. Silverman RH, Skehel JJ, James TC, Wreschner DH, Kerr IM. 1983. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 46:1051–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castelli JC, Hassel BA, Wood KA, Li XL, Amemiya K, Dalakas MC, Torrence PF, Youle RJ. 1997. A study of the interferon antiviral mechanism: apoptosis activation by the 2-5-A system. J. Exp. Med. 186:967–972. 10.1084/jem.186.6.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. 2012. RNase L triggers autophagy in response to viral infections. J. Virol. 86:11311–11321. 10.1128/JVI.00270-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siddiqui MA, Malathi K. 2012. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J. Biol. Chem. 287:43651–43664. 10.1074/jbc.M112.399964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malathi K, Dong B, Gale M, Jr, Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816–819. 10.1038/nature06042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malathi K, Saito T, Crochet N, Barton DJ, Gale M, Jr, Silverman RH. 2010. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 16:2108–2119. 10.1261/rna.2244210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luthra P, Sun D, Silverman RH, He B. 2011. Activation of IFN-beta expression by a viral mRNA through RNase L and MDA5. Proc. Natl. Acad. Sci. U. S. A. 108:2118–2123. 10.1073/pnas.1012409108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rasmussen SB, Jensen SB, Nielsen C, Quartin E, Kato H, Chen ZJ, Silverman RH, Akira S, Paludan SR. 2009. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene-like receptors, which synergize to induce type I interferon production. J. Gen. Virol. 90:74–78. 10.1099/vir.0.005389-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao L, Birdwell LD, Wu A, Elliott R, Rose KM, Phillips JM, Li Y, Grinspan J, Silverman RH, Weiss SR. 2013. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J. Virol. 87:8408–8418. 10.1128/JVI.00769-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorgeloos F, Jha BK, Silverman RH, Michiels T. 2013. Evasion of antiviral innate immunity by Theiler’s virus L* protein through direct inhibition of RNase L. PLoS Pathog. 9:e1003474. 10.1371/journal.ppat.1003474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou A, Molinaro RJ, Malathi K, Silverman RH. 2005. Mapping of the human RNase L promoter and expression in cancer and normal cells. J. Interferon Cytokine Res. 25:595–603. 10.1089/jir.2005.25.595 [DOI] [PubMed] [Google Scholar]
- 18. Lee MS, Kim B, Oh GT, Kim YJ. 2013. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat. Immunol. 14:346–355. 10.1038/ni.2535 [DOI] [PubMed] [Google Scholar]
- 19. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- 20. Andersen JB, Mazan-Mamczarz K, Zhan M, Gorospe M, Hassel BA. 2009. Ribosomal protein mRNAs are primary targets of regulation in RNase-L-induced senescence. RNA Biol. 6:305–315. 10.4161/rna.6.3.8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams BR, Kerr IM. 1978. Inhibition of protein synthesis by 2′-5′ linked adenine oligonucleotides in intact cells. Nature 276:88–90. 10.1038/276088a0 [DOI] [PubMed] [Google Scholar]
- 22. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- 23. Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616. 10.1016/j.chom.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chakrabarti A, Sadler AJ, Kar N, Young HA, Silverman RH, Williams BR. 2008. Protein kinase R-dependent regulation of interleukin-10 in response to double-stranded RNA. J. Biol. Chem. 283:25132–25139. 10.1074/jbc.M804770200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. Chapter 14:Unit 1411. 10.1002/0471142735.im1401s83 [DOI] [PMC free article] [PubMed] [Google Scholar]