Abstract
The extensive set of NMR doublings exhibited by the immunophilin FKBP12 (FK506-binding protein 12) arose from a slow transition to the cis-peptide configuration at Gly89 near the tip of the 80′s loop, the site for numerous protein-recognition interactions for both FKBP12 and other FKBP domain proteins. The 80′s loop also exhibited linebroadening, indicative of microsecond to millisecond conformational dynamics, but only in the trans-peptide state. The G89A variant shifted the trans–cis peptide equilibrium from 88:12 to 33:67, whereas a proline residue substitution induced fully the cis-peptide configuration. The 80′s loop conformation in the G89P crystal structure at 1.50 Å resolution differed from wild-type FKBP12 primarily at residues 88, 89 and 90, and it closely resembled that reported for FKBP52. Structure-based chemical-shift predictions indicated that the microsecond to millisecond dynamics in the 80′s loop probably arose from a concerted main chain (ψ88 and ϕ89) torsion angle transition. The indole side chain of Trp59 at the base of the active-site cleft was reoriented ~90o and the adjacent backbone was shifted in the G89P crystal structure. NOE analysis of wild-type FKBP12 demonstrated that this indole populates the perpendicular orientation at 20%. The 15N relaxation analysis was consistent with the indole reorientation occurring in the nanosecond timeframe. Recollection of the G89P crystal data at 1.20 Å resolution revealed a weaker wild-type-like orientation for the indole ring. Differences in the residues that underlie the Trp59 indole ring and altered interactions linking the 50′s loop to the active site suggested that reorientation of this ring may be disfavoured in the other six members of the FKBP domain family that bear this active-site tryptophan residue.
Keywords: conformational dynamics, FK506-binding protein 12 (FKBP12), linebroadening analysis, NMR, slow exchange, X-ray structure
Abbreviations: FKBP, FK506-binding protein; mTOR, mammalian target of rapamycin; RyR, ryanodine receptor
Short abstract
Extensive resonance doubling arises from a cis–trans peptide transition at Gly89, whereas linebroadening appears due to a concerted shift in the neighbouring torsion angles. The active site Trp59 ring adopts a perpendicular orientation at a population of 20%.
INTRODUCTION
The peptide prolyl isomerase FKBP12 (FK506-binding protein 12) was initially characterized for its role in binding the immunosuppressants FK506 and rapamycin. The ternary complex of FKBP12 and FK506 with calcineurin inhibits the enzymatic activity of this protein phosphatase, blocking a key T-cell-activation pathway involved in tissue transplant rejection [1]. The binding of FKBP12 and rapamycin to mTOR (mammalian target of rapamycin) inhibits its role in regulating cell growth and cancer progression [2]. Despite considerable effort directed towards developing active-site inhibitors of mTOR, the only two clinically approved therapeutics for this protein kinase are allosteric inhibitors derived from rapamycin [3].
The challenge of FKBP12 pharmacology is increased markedly by the presence of 22 homologous FKBP domains within the human genome [4]. In addition to the closely homologous FKBP12.6 and the more evolutionarily divergent FKBP13, there are 19 different FKBP domains that are modules within larger proteins. In addition to structural similarities, there is also some degree of functional overlap among these FKBP domain proteins. The mTOR interactions of FKBP12 can be functionally replaced in cellular model systems by the larger FKBP domain proteins FKBP51 and FKBP52 [5]. In a similar fashion, siRNA studies indicate that FKBP12 contributes to ~60% of the FK506-mediated inhibition of calcineurin, whereas the rest of the inhibitory effect is contributed nearly equally by FKBP12.6 and FKBP51 [6].
Although the pharmacological utility of the bacterial macrolides FK506 and rapamycin is well established, the physiological relevance of their interactions is less clear. Despite more than two decades of research, no endogenous small-molecule ligand has been shown to mediate protein–protein interactions for the FKBP domains [7]. Both biochemical [8,9] and genetic [10,11] evidence support the importance of the binding of FKBP12 and FKBP12.6 to the RyR (ryanodine receptor) Ca2+ channels in cardiac and skeletal muscle in the regulation of contraction, as well as binding to the RyR channels of pancreatic islet cells for the regulation of insulin secretion [12] and to RyR channels in the central nervous system that are involved in memory processing [13] and in stress-induced cognitive dysfunctions [14]. FKBP12 has also been implicated in the regulation of a number of membrane-bound hormone receptors [15].
The prolyl isomerization activity of FKBP12 has not been shown to participate in any of its protein signalling interactions, and a similar lack of dependence on catalytic activity has been observed for the well-studied glucocorticoid receptor interactions of the closely homologous FKBP domains of FKBP51 and FKBP52 [16]. On the other hand, prolyl isomerization activity does appear to play a role in the association of FKBP12 with aggregated α-synuclein in the Lewy body deposits of patients with Parkinson's disease [17], with the conformationally disordered tau protein in the neurofibrillary tangles of Alzheimer's disease [18] and in the binding to the amyloid precursor protein [19]. FKBP12 has been shown to accelerate the aggregation of α-synuclein in vitro [20] and in vivo [21].
Recently, we characterized a previously unreported slow conformational transition for FKBP12 in which a minor state population of 12% interchanges with the major conformational state at a rate of ~0.05 s−1 at 25°C [22]. Although the primary site of the conformational transition appears to lie at the tip of the 80′s loop, which spans between the last two strands of the central β-sheet, the doubling of the backbone amide resonances that arises from the two slowly interchanging conformations extends not only to residues lining the active-site cleft, but beyond to a number of residues in and surrounding the 50′s loop on the opposite side of the protein. Interestingly, the major state, but not the minor state, of this slow transition also exhibits linebroadening of the amide resonances in the 80′s loop, indicative of conformational exchange in the microsecond to millisecond timeframe. Evidence for conformational flexibility within the 80′s loop gains particular relevance due to the fact that this loop provides a major proportion of the interprotein interactions for each of the four distinct protein–protein complexes that have been structurally determined for FKBP12 [23–26]. The homologous loop has also been shown to provide critical protein-recognition interactions in other FKBP domain proteins [16].
In the present study we have analysed the structural basis of the slow resonance doubling transition of FKBP12 and the more rapid conformational linebroadening transition in the 80′s loop to gain insight into how these effects are propagated through the protein structure. In turn, these studies have led to the characterization of a well-populated conformational transition within the active-site cleft.
EXPERIMENTAL
Protein preparation
Genes for the wild-type and G89A and G89P variants of the human FK506-binding protein FKBP12 were synthesized chemically (Genscript) from the wild-type gene sequence, with codon optimization for expression in Escherichia coli cells. The genes were cloned into the expression vector pET11a and then transformed into the BL21(DE3) strain (Novagen) for expression. The protein expression and purification procedure for the FKBP12 proteins were carried out as described previously [22,27].
All isotopically labelled samples were prepared via protein expression in minimal medium containing 0.1% 15NH4Cl as a nitrogen source [28]. For U-13C,15N-enriched samples, 0.2% [U-13C]glucose (Cambridge Isotopes) was substituted for the unlabelled glucose used for preparing the U-15N samples. For the selective [13C]methyl-labelled samples, 85 mg/l [3-2H,4-13C]α-oxoisovalerate and 50 mg/l [3-2H2,4-13C]α-oxobutyrate [29] were supplemented into a medium for U-2H,15N-enriched sample growth as described previously [28].
For the protein samples expressed in 2H2O medium, 1 mM tris(2-carboxyethyl)phosphine and solid Tris base was added to a solution of the purified protein to obtain a pH value above 9, and the samples were incubated at 25°C for 3 h and then neutralized with solid monobasic sodium phosphate. All protein samples were concentrated via centrifugal ultrafiltration (1000 g for 30 min at 4°C) and then equilibrated into a pH 6.50 buffer containing 25 mM sodium phosphate, 2 mM DTT and 2 mM tris(2-carboxyethyl)phosphine by a series of centrifugal concentration steps (1000 g for 30 min at 4°C). For the crystallization trials, the protein sample was neutralized and then equilibrated into 5 mM sodium chloride and concentrated by centrifugal ultrafiltration (1000 g for 30 min at 4°C).
X-ray crystallography
Crystals of the C22V,G89P variant of FKBP12 were grown at room temperature (22°C) in hanging drops by microseeding from a crushed crystal of the FKBP C22V,H87V variant [22]. Before microseeding, 2 μl of protein solution at 22 mg/ml concentration was mixed with an equal volume of reservoir solution containing 64% saturated ammonium sulfate, 0.1 M Hepes (pH 7.5) and 4% methyl-2,4-pentanediol, and equilibrated overnight. The crystals belonged to space group C2 and had the cell parameters a=71.37 Å, b=35.86 Å, c=41.24 Å and β=96.65°. There was one molecule per asymmetric unit with a crystal solvent content of 48%. Before data collection, crystals were gradually transferred into a reservoir solution containing a higher concentration of ammonium sulfate (up to 80%) at 5% per step, and then flash-cooled under a nitrogen stream at 100 K and stored in liquid nitrogen. Diffraction data were collected at 100 K using an RAxisIV++ detector and an in-house Rigaku microfocus Micromax-007 X-ray generator. These diffraction data were processed and scaled using CrystalClear 1.3.6 (Rigaku). Subsequently, higher-resolution diffraction data were collected at 100 K using beamline X25 of the National Synchrotron Light Source (Brookhaven National Laboratory). These data were processed and scaled using HKL2000 [30]. With the high-resolution structure of FKBP12 [31] (PDB code 2PPN) used as a search model, clear solutions were found with the PHASER molecular replacement program within the PHENIX suite [32]. Structural refinement was carried out using PHENIX and SHELX-97 [33]. Model rebuilding was carried out using Coot [34]. Figures of crystallographic structures were generated using the Chimera software [35].
NMR spectroscopy
NMR data for the G89A and G89P variants of FKBP12 were collected on a Bruker Avance III 600 MHz and 900 MHz spectrometers at 25°C. Backbone resonance assignments were carried out using standard HNCO [36], HN(CA)CO [36], HNCACB [37] and HN(CO)CACB [38] experiments. The resonance assignments for the wild-type protein have been reported previously [22] (Biological Magnetic Resonance Data Bank entries 19240 and 19241). The cis-glycine linkage analysis and the indole ring reorientation analysis were carried out using 3D 13C-13C-1H NOESY and 13C-15N-1H NOESY [39] experiments respectively. FELIX software (Felix NMR) was used for the NMR data processing. Relaxation analysis of the 15N indole resonance of Trp59 utilized a CSA (chemical shift anisotropy) value of −126 p.p.m. [40].
The initial rate of NOE build-up between an isolated proton pair is proportional to:
| (1) |
where ω is the 1H Larmor frequency and Jij(X) are the com-ponents of the spectral density function. If the correlated internal motion of the two nuclei dissipates rapidly as compared with the rate of global molecular tumbling, the internal motion correlation function can be factored out as a generalized order parameter (S) [41] which for a three state methyl jump model can be represented as:
| (2) |
where rj is the distance between the methyl proton j and a distal proton and θij is the angle between the vectors formed between a distal proton and the two methyl protons i and j.
RESULTS AND DISCUSSION
Mutational variation of the slow exchange conformational equilibrium in FKBP12
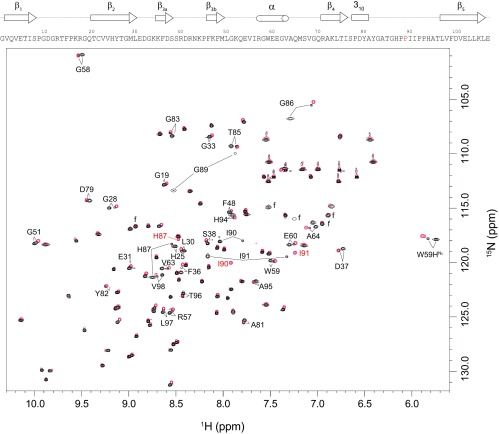
The largest chemical-shift differences for the doubled amide resonances of FKBP12 are seen for residues in the 80′s loop [22]. The resonance doubling extends into the first two residues of the β5-strand and the structurally contiguous residues of the adjacent β2- and β3α-strands (Figure 1). The doubling also extends backwards along the chain into the 310 turn and into the nearby α-helix which bears the Trp59 indole side chain that forms the base of the active-site cleft. The residues exhibiting resonance doubling extend beyond the active site to the beginning of the 50′s loop that abuts the C-terminal end of the α-helix.
Figure 1. 1H–15N 2D NMR correlation spectra of wild-type and the G89P variant of FKBP12.
Cross-peaks from the G89P variant are indicated in red. Resonances that are doubled in the wild-type spectrum are identified. The Ala84 resonances were not observed in this spectrum due to severe linebroadening presumably because of the rapid amide hydrogen exchange as observed in FKBP12 [27]. f, folded side chain resonances.
We reported previously the crystal structure and 15N relaxation analysis of an H87V variant of FKBP12. Despite the close similarity of the H87V structure [31] (0.25 Å Cα RMSD with respect to the 0.92 Å wild-type structure), the slow resonance doubling transition is completely quenched throughout the protein [22]. Chemical shift-based prediction of the backbone torsion angles for the minor slow exchange state yielded a negative ϕ torsion angle for Gly89 as compared with a value of +103° reported in the 0.92 Å resolution structure of FKBP12 [31] (PDB code 2PPN).
In the present study, the G89A mutation was introduced so as to shift the conformational equilibrium towards a negative ϕ value for this residue. The 2D 1H,15N HSQC spectrum of the G89A variant yielded a pattern of amide resonance doublings with chemical-shift values quite similar to that observed for the wild-type protein [22], although the relative intensity of cross-peaks from the two slow exchange conformational states is markedly different (Supplementary Figure S1 at http://www.biochemj.org/bj/458/bj4580525add.htm). In the wild-type spectrum the minor slow exchange state represented only 12% of the total population, whereas in the G89A variant the analogous set of cross-peaks yielded 67% of the protein signal. This change in population corresponded to a 6.7 kJ/mol difference in the relative stability of the two conformations that was induced by the alanine residue substitution.
To examine further how enforcing a negative ϕ torsion angle at residue 89 affects the structure and dynamics of FKBP12, the G89P variant was analysed by NMR. The proline residue substitution eliminates peak doubling for all of the amide resonances (Figure 1). The similarity in chemical-shift behaviour for the G89P variant and the minor slow exchange conformation of the wild-type protein indicated strongly a corresponding similarity in structure. The only marked differences in the superimposition of these two sets of resonances occurred at residue 89, for which the G89P variant lacked an amide resonance, and Ile90 for which the 15N of the G89P was shifted downfield as anticipated from the inductive effects resulting from the side chain substitution for the preceding residue [42,43].
Crystal structure for a G89P variant of FKBP12 at 1.50 Å resolution
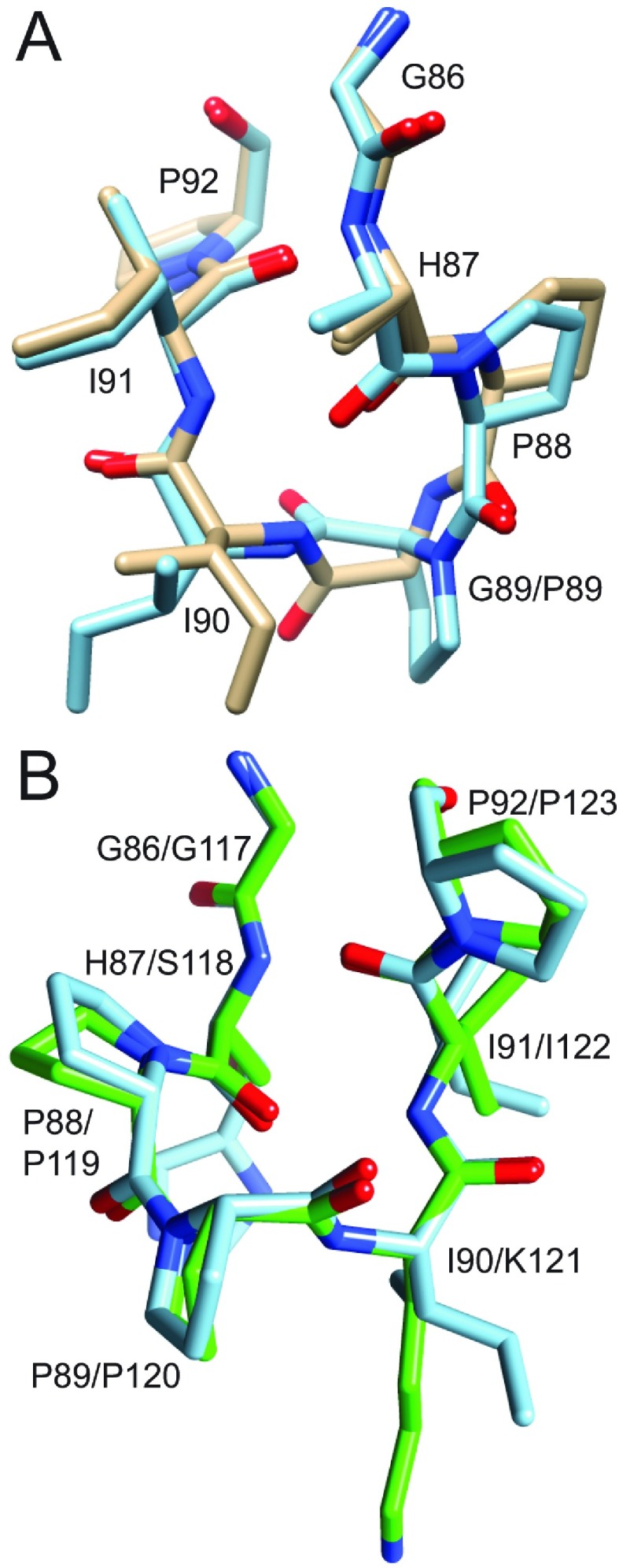
Taking advantage of the close structural similarity between the wild-type and the evolutionarily conservative C22V variant of FKBP12 and the increased stability of the cysteine residue-free variant [22], the G89P mutation was introduced into the C22V background for crystallization trials. Crystals were grown in the C2 space group using microseeding from crystals of the C22V,H87V variant [22]. Diffraction data were collected to 1.50 Å resolution using an in-house diffractometer. Following refinement, the backbone ϕ and ψ torsion angles for Pro89 were found to be (−92 and −12) with the ϕ value being nearly 180° from that of Gly89 in the wild-type structure. Furthermore, this crystal structure adopted a cis-peptide conformation for the linkage between Pro88 and Pro89. Superimposition with the high-resolution wild-type structure indicated that, although the conformation of residues 89 and 90 differed markedly, the residues preceding Pro88 and those following Ile90 aligned closely (Figure 2A).
Figure 2. Superimposition of region surrounding the Pro88–Pro89 peptide bond in the crystal structure of the G89P variant (blue) as compared with the wild-type FKBP12 (gold) and to the homologous segment from the first FKBP domain of FKBP52 (green).
(A) In the comparison between the two FKBP12 structures, the His87 side chain is truncated at Cβ to facilitate visualization of the backbone conformations. The cis-peptide linkage in the G89P structure perturbs the backbone of residues 88–90, whereas the preceding and following residues superimpose fairly closely. (B) As viewed from the opposite face of the loop, the structural changes induced by the cis-peptide linkage in the G89P variant yields a backbone conformation that closely follows that of the first FKBP domain of FKBP52 (PDB code 1N1A).
The sequence at the tip of the 80′s loop differed by only one residue between the first FKBP domain of FKBP52 (PPKIPP) and the G89P variant of FKBP12 (PPIIPP). A cis-peptide linkage at the position homologous with Pro89 has been reported in crystal structures of this FKBP52 domain (e.g. PDB codes 1N1A [44] and 4LAV [45]). This region of the 80′s loop in the G89P structure superimposed closely with that of the PDB code 1N1A structure for FKBP52 (Figure 2B).
If the minor slow exchange state of FKBP12 corresponds to the transition of the Gly89 peptide linkage to a cis configuration, the crystal structure of the G89P variant provides a ready explanation for how the H87V substitution for FKBP12 might suppress this transition. In the crystal structure of the H87V variant, the Cγ2 atom of that valine residue is 3.7 Å from the Cδ1 atom of the evolutionarily invariant Tyr82 side chain [22]. When a valine residue side chain was modelled on to His87 of the G89P crystal structure, the separation between the valine Cγ2 atom and the Cδ1 atom of Tyr82 was reduced to 2.9 Å, suggestive of a substantial steric hindrance to such a conformational transition.
Glycine cis-peptide configuration at residue 89 in wild-type FKBP12
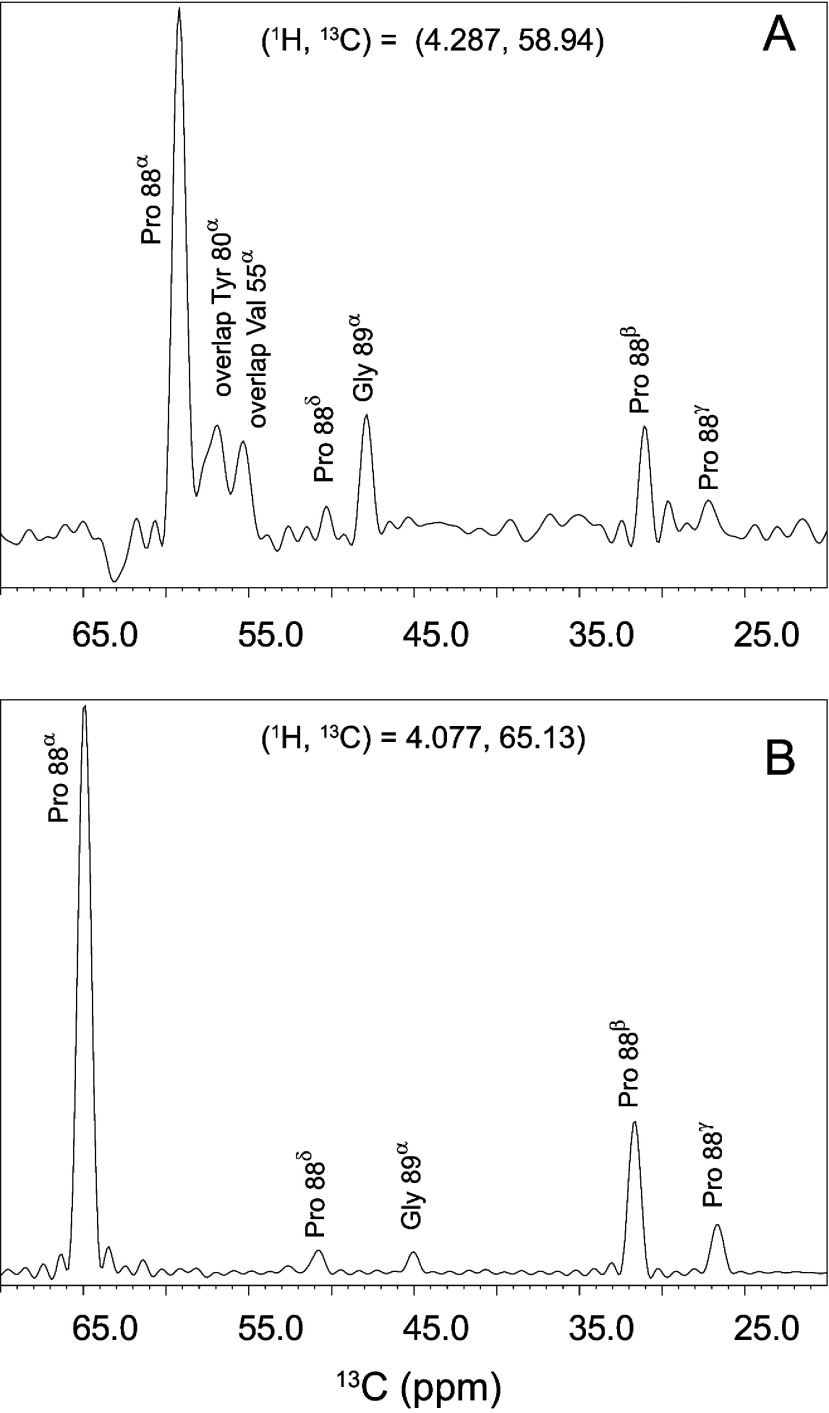
If the peptide linkage of Gly89 in wild-type FKBP12 assumes a cis-peptide linkage in the minor state of the slow exchange transition, this implies that this critical protein-recognition loop of FKBP12 adopts a FKBP52-like conformation in a substantial fraction of the conformational distribution. One indirect line of support for this possibility is the 70 kJ/mol activation energy that was determined for the slow exchange process in FKBP12 [22], which is typical for cis–trans isomerization of peptides. On the other hand, non-proline residue cis-peptide linkages are quite rare in protein crystal structures, occurring at a frequency of only 0.03% [46]. To demonstrate directly whether the minor slow exchange state of FKBP12 adopts a cis-peptide linkage at this residue, 3D 13C-13C-1H NOESY measurements were carried out to determine the presence of a short range distance between the Hα atoms of Pro88 and Gly89 in the minor conformation. For the more common trans-peptide linkage, the distance between sequential Hα atoms is ~4.5 Å, nearly independent of the intervening ψi and ϕi+1 torsion angles [47]. On the other hand, assuming that the minor slow exchange form of the wild-type protein adopts the geometry of this linkage, as found in the G89P crystal structure, the distances from the Pro88 Hα to the two Gly89 Hα atoms were 2.0 and 3.4 Å.
For the minor state resonances of wild-type FKBP12, the intensity of the sequential NOE transfer from the 1Hα resonances of Gly89 to the 1Hα of Pro88 was more intense than the intraresidue NOE transfer to Pro88 1Hα from its own 1Hβ resonances (Figure 3A). In contrast, for the major slow exchange state the sequential NOE transfer from the 1Hα resonances of Gly89 to the 1Hα of Pro88 was much weaker than the intraresidue transfer from the 1Hβ resonances (Figure 3B). A similar pattern of NOE cross-peak intensities were observed for transfer in the reverse direction from the 1Hα of Pro88 to the 1Hα resonances of Gly89 for both the major and minor slow exchange states.
Figure 3. 1D slices from the 3D 13C-13C-1H NOESY spectrum of wild-type FKBP12.
(A) At the 1H and 13C frequencies for the Cα of Pro88 in the minor slow exchange state the NOE cross-peak correlating the sequential Cα of Gly89 is more intense than the corresponding NOE cross-peak correlating the intraresidue Pro88 Cβ interaction. (B) In contrast, the corresponding sequential NOE connectivity correlating the Cα nuclei of Pro88 and Gly89 in the major slow exchange state is comparatively far less intense than the intraresidue NOE to the Pro88 Cβ.
NMR chemical-shift analysis of the backbone conformations for the 80′s loop
Having established that the extensive resonance doubling of FKBP12 arises from the slow cis–trans isomerization of the peptide linkage between Pro88 and Gly89, we then examined the structural basis for the much more rapid linebroadening conformational dynamics of the 80′s loop. The transition from the wild-type FKBP12 crystal structure to a conformation resembling the G89P crystal involved not only the change to the Gly89 cis-peptide configuration. The ϕ torsion angle of Gly89 is +103° in the high-resolution crystal structure of wild-type FKBP12 [31], whereas the five-membered ring of the homologous Pro120 in the FKBP52 structures forces the ϕ angle to be negative. To examine the plausibility that Gly89 of FKBP12 might undergo a relatively rapid transition between positive and negative ϕ angle values while remaining in the predominant trans-peptide configuration, the other crystal structures of FKBP52 were considered. As noted above, two crystal structures of FKBP52 (PDB codes 1N1A [44] and 4LAV [45]) exhibit a cis-peptide linkage between Pro119 and Pro120. However, there are two other crystal structures of FKBP52 (PDB codes 1Q1C [48] and 4LAW [45]) for which this peptide linkage is in a trans configuration. These latter two crystal structures of FKBP52 can provide models for the conformation of the 80′s loop of FKBP12 in which Gly89 switches to an ϕ angle<0, whereas its peptide linkage remains in a trans configuration.
T2 transverse relaxation experiments serve to measure the 15N linebroadening effects which arise from the difference in chemical shift for a given nucleus that interchanges between conformational states in the microsecond to millisecond timeframe. A number of empirical algorithms have been developed for predicting NMR chemical shifts from protein structures [49–52]. One widely used algorithm is the SPARTA+ program of Shen and Bax [49] which has applied an artificial neural network analysis against a database of 580 proteins to derive its parameterization. Consistent with the results from other such chemical-shift prediction algorithms, SPARTA+ yielded an S.D. of 2.45 p.p.m. in its predictions of backbone 15N chemical shifts for a set of 11 validation proteins, an appreciably lower precision than was obtained for the 13C′, 13Cα, 13Cβ, 1Hα and 1HN chemical shifts. In modelling the linebroadening for the 80′s loop resonances of FKBP12, the 15N chemical shift predictions were only applied in a differential mode in anticipation of an increased precision in the resultant analysis.
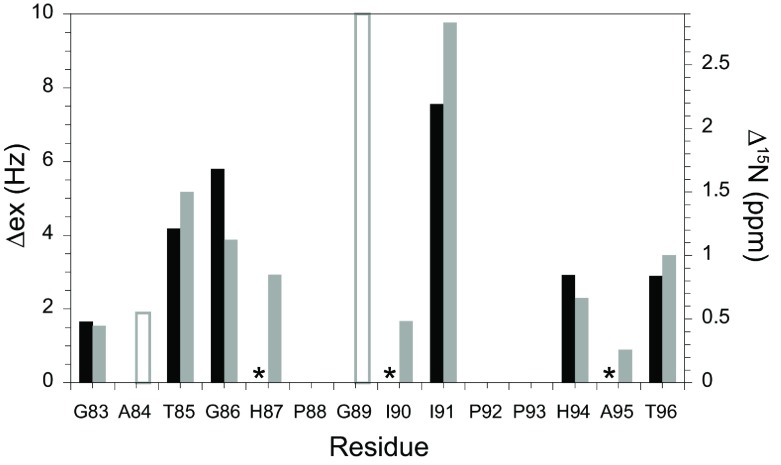
Exploiting the fact that these chemical shift predictions are dominated generally by the local backbone geometry, the crystal structures for the 80′s loop in FKBP12 [31] (PDB code 2PPN) and the first FKBP domains of FKBP52 (PDB codes 1Q1C [48] and 4LAW molecules A and B [45]) were transformed to a common sequence by converting the disparate residues into alanine, except for the site of the cis-peptide transition for which the FKBP52 structures were converted into Gly120. The predicted 15N chemical-shift values from the FKBP12-derived structure were compared against those from the three FKBP52-derived structures, both individually and as a simple average over those three structures. In comparing these predicted differences in 15N chemical shift with the experimentally observed 15N linebroadening effects [22], averaging the predictions over the three FKBP52-derived structures provided the strongest correlation (Figure 4).
Figure 4. Correlation between the 15N conformational exchange linebroadening (Δex) in the 80′s loop of FKBP12 (black) and the differential chemical-shift predictions derived from two crystal forms of FKBP52 (grey).
Model-free [41] analysis of the R1, R2 and NOE 15N relaxation measurements was used to estimate conformational exchange linebroadening [22]. The differences in 15N chemical shift were predicted with SPARTA+ [49] applied to FKBP12 (PDB code 2PPN [31]) and the first FKBP domain of FKBP52 (PDB code 1Q1C [48] and non-equivalent monomers of PDB code 4LAW [45]). An asterisk indicates not statistical significant conformational exchange linebroadening (<~0.4 Hz). The amide resonances for both Ala84 and Gly89 are severely broadened due to rapid hydrogen exchange [22], precluding a reliable determination of conformational exchange linebroadening, and the predicted chemical shift differences for these two residues are indicated as open bars.
Given the substantial challenges for quantitative structure-based predictions of conformational exchange linebroadening effects, the degree of correlation between the predicted and observed linebroadening in the 80′s loop of FKBP12 suggests strongly that Gly89 transiently adopts a negative ϕ torsion angle when that residue is in a trans-peptide configuration. On the other hand, the absence of 15N conformational exchange linebroadening for the 80′s loop when Gly89 is in the cis-peptide configuration of the minor slow exchange state [22] suggests that positive ϕ torsion angles for Gly89 are not energetically accessible from that state in a comparable population or timeframe to that observed for the trans-peptide configuration. The simplest mechanistic interpretation consistent with these data is that the two slowly interchanging Gly89 peptide configuration states, monitored by the extensive resonance doubling, interchange with each other only via a more weakly populated transient state in which the ϕ torsion angle of Gly89 is negative, whereas the peptide configuration remains trans.
As noted above, the H87V substitution suppresses all of the amide resonance doubling, presumably by its steric collision with the Tyr82 ring in the Gly89 cis-peptide configuration. This substitution also suppresses the microsecond to millisecond linebroadening transition for the resonances in the 80′s loop [22]. We propose that this linebroadening arises from a ~180° shift in the ϕ torsion angle of Gly89. The ψ angle of Pro88/119 also shifted by an average of 130° between the wild-type FKBP12 and three FKBP52 crystal structures in the trans-peptide configuration. These three crystal structures of FKBP52 can be used to examine this model for the quenching of linebroadening in the 80′s loop. Residue 118 (homologous with His87 of FKBP12) was computationally transformed into a valine for each of these three FKBP52 structures. For PDB code 1Q1C and monomer A of PDB code 4LAW, this valine residue side chain must strongly overlap with either the aromatic ring of Tyr113 (homologous with the evolutionarily invariant Tyr82) or with the backbone of Ile122. Such a steric conflict could be expected to preclude the transition of Gly89 to a negative ϕ torsion angle in the H87V variant, thus eliminating the linebroadening effect for the 80′s loop. A caveat to this detailed structural interpretation is that in the significantly differing local geometry of monomer B in the PDB code 4LAW structure of FKBP52 the introduction of the valine residue side chain at residue 118 does not appear to yield a severe steric conflict.
Both the H87V [53] and G89P [54] variants have been shown to have an equivalent peptide isomerase activity with that of wild-type FKBP12 in the standard assay using blocked tetrapeptides. The H87V variant appeared to block the resonance doubling and the conformational linebroadening of the 80′s loop by constraining Gly89 to remain in the trans-peptide configuration with a positive ϕ torsion angle. The G89P variant appeared to block the resonance doubling and the conformational linebroadening of the 80′s loop by constraining residue 89 to remain in the cis-peptide configuration with a negative ϕ torsion angle. The linebroadening transition and the much slower resonance doubling transition did not appear to participate in the peptide isomerization activity of this enzyme, at least for simple conformationally unstructured peptides. Any functional relevance of this conformational plasticity/dynamics may instead relate to a role in the protein-recognition interactions of FKBP12.
Perpendicular reorientation of the Trp59 indole at the base of the active-site cleft in a G89P variant of FKBP12
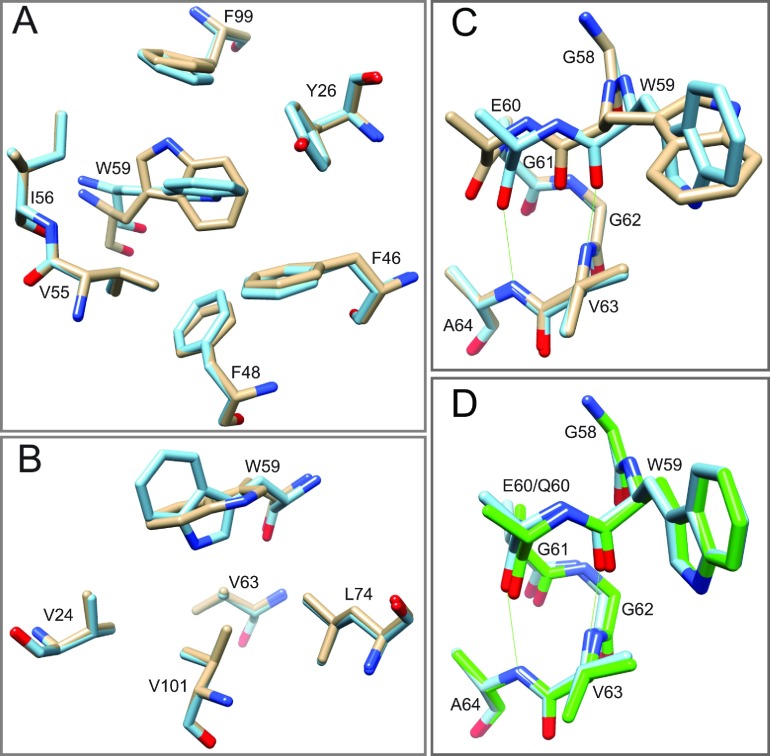
The most unexpected aspect of the G89P crystal structure was that the indole ring of Trp59 at the base of the active-site cleft was oriented perpendicular to the conformation observed in each of the more than 30 crystal structures of wild-type FKBP12 (Figure 5A). The aromatic rings of Tyr26, Phe46, Phe48 and Phe99 and the side chains of Val55 and Ile56 form the walls of the active-site cleft surrounding the Trp59 ring. The reorientation of the indole ring in the G89P crystal structure occluded much of the volume of the active-site cleft, conflicting with the binding geometry observed in the crystal structures of the FK506- and rapamycin-ligated proteins. As compared with the wild-type crystal structure, the aromatic rings of Phe48 and Phe99 in the G89P structure appeared to move slightly inward towards the reoriented indole ring. The conformation of the residues that lie behind the Trp59 indole ring was essentially identical for the wild-type and the G89P structures. A small cavity lies beneath the indole ring in the wild-type structure, and the nearby side chains of Val24, Val63, Leu74 and Val101 were essentially unperturbed by the rotation of the indole ring (Figure 5B).
Figure 5. Superimposition of the active-site region surrounding Trp59 and Glu/Gln60 from the wild-type (gold), G89P (blue) and E60Q (green) crystal structures.
(A) Above the plane of the indole ring in the PDB code 2PPN structure, the aromatic rings of Tyr26, Phe46, Phe48 and Phe99 form much of the wall of the active-site cleft. (B) Underneath the indole ring of Trp59, the Cδ1 and Nε1 of that residue can occupy a cavity formed by the side chains of Val24, Val63, Leu74 and Val101 which is unperturbed by this structural rearrangement. (C) The superimposition of the wild-type and G89P crystal structures illustrates a displacement of the backbone such that in the G89P structure canonical geometries are established for the hydrogen bonds between Glu60 O and Ala64 HN and between Trp59 O and Val 63 HN. (D) This region of the crystal structure of the E60Q variant is highly similar to that for G89P.
This perpendicular reorientation of the Trp59 aromatic ring in the G89P crystal structure was strikingly similar to that reported for the E60Q (PDB code 2PPP) and E60A (PDB code 2PPO) variants of FKBP12 [31]. In addition, the backbone of the helical residues surrounding Trp59 in the G89P crystal structure was closely similar to that observed in the E60Q (and E60A) structure (Figure 5D). In contrast, the hydrogen-bonding interactions for Trp59 O to Val63 HN and Glu60 O to Ala64 HN were significantly irregular in the crystal structure of wild-type FKBP12 with the carbonyl groups being shifted laterally by over 1 Å with respect to the amide bond vectors as compared with the canonical hydrogen-bonding geometry seen in the G89P structure (Figure 5C). Regarding the role of crystal packing interactions in the present study, it may be noted that the P21 crystal forms for the 0.94 Å resolution structure of the E60Q variant and the 1.29 Å resolution structure of the E60A variant were isomorphous with that used in the 0.92 Å resolution structure of wild-type FKBP12 [31]. The G89P variant crystallized in the C2 space group.
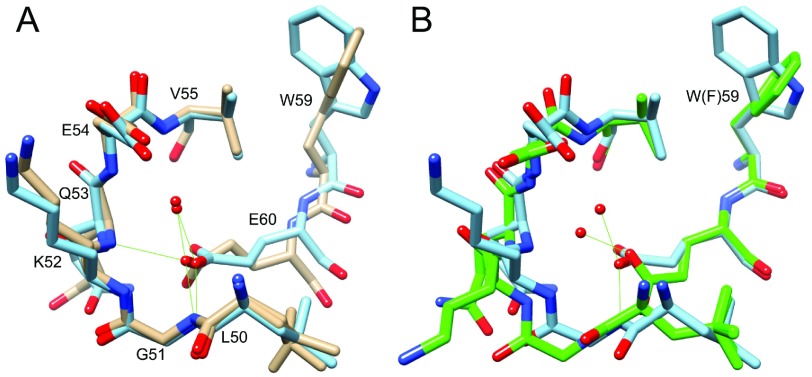
Saven and colleagues noted that the Glu60 side chain in their high-resolution structure of wild-type FKBP12 extends towards the backbone of the 50′s loop to co-ordinate the amide hydrogens of Gly51 and Gln53, as well as to a highly conserved solvent-inaccessible water molecule [31]. Introduction of glutamine at position 60 of FKBP12 results in the flipping of the peptide unit that links Lys52 and Gln53 and a shift in co-ordination for the buried water molecule. Although the backbone of the 50′s loop in the E60A variant remains similar to that of the wild-type protein, the side chain interaction of residue 60 with the buried water molecule is obviously disrupted. The authors [31] argued that these altered interactions of residue 60 with the buried water molecule results in a shift in the α-helix, which, in turn, presses the indole ring more tightly against the ring of Phe99 thus driving the reorientation of the Trp59 side chain.
The backbone of the 50′s loop and the position of the buried water molecule in the crystal structure of the G89P variant closely superimposed with that of wild-type FKBP12 (Figure 6A). The carboxy Oε1 atom of Glu60 preserved its hydrogen-bonding interactions with the amide hydrogens of Gly51 and Gln53 and with the buried water molecule. The similarity in the interactions of the Glu60 side chain with the backbone of the 50′s loop and the buried water molecule for the wild-type and G89P structures suggested that disruption of these interactions were sufficient, but not necessary, to shift the backbone of Trp59 and Glu60 into a canonical α-helical hydrogen-bonding geometry and enable the reorientation of the indole ring.
Figure 6. Interactions of Glu60 with the 50′s loop in wild-type (gold), G89P (blue) and W59F (green) FKBP12 crystal structures.
(A) The Glu60 side chain differs in the G89P structure primarily by adopting an extended trans χ1 rotamer conformation which enables the shift in the backbone into a canonical α-helical hydrogen-bonding geometry. (B) The crystal structure of the W59F variant [55] exhibits a similarly extended trans χ1 rotamer conformation for Glu60 and canonical α-helical hydrogen-bonding geometry, although the phenyl ring of Phe59 remains flat across the bottom of the active-site cleft.
The most notable difference for Glu60 between the crystal structures of wild-type FKBP12 and the G89P variant is the transition of the side chain from a gauche− to a trans χ1 torsion angle, which allows the backbone atoms of Trp59 and Glu60 to shift towards the active site indole ring without disrupting the hydrogen-bonding interactions described above. Such a shift from a gauche− to a trans χ1 torsion angle for the Glu60 side chain was reported previously in the crystal structure of the W59F variant of FKBP12 [55]. In this case, the backbone atoms of Phe59 and Glu60 underwent a similar shift to that observed in the structure of the G89P variant (Figure 6B). On the other hand, the smaller phenyl side chain at residue 59 does not become tightly pressed against the aromatic ring of Phe99 when it maintains an orientation along the bottom of the active-site cleft similar to that of the tryptophan ring in the wild-type FKBP12 structure. In comparing the crystal structures of the W59F and G89P variants, it appeared that disruption of the steric interaction between the indole ring of Trp59 and the phenyl ring of Phe99 was sufficient to enable the backbone of Trp59 and Glu60 to shift into a canonical α-helical hydrogen-bonding geometry.
Crystal structures have been reported for four of the other six human FKBP domains which contain a tryptophan ring at the base of the active-site cleft [44,56–58]. In each of these four structures, Glu60 of FKBP12 is replaced by the shorter aspartate side chain which forms similar interactions with the backbone of the 50′s loop. It is not self-evident how these interactions could be preserved while undergoing the shift in the α-helical backbone to allow for an analogous reorientation of the tryptophan ring. A further indication that the reorientation of the active site indole side chain may be energetically disfavoured in these other FKBP domains is that the unoccupied volume beneath the plane of the Trp59 ring in FKBP12 appears to be occluded in these other crystal structures. In the example of FKBP51 [56], the aliphatic side chains underlying the indole ring closely superimpose on those of FKBP12 illustrated in Figure 5(B), except Ile132 which was substituted for Val101. This substitution resulted in the isoleucine residue Cδ atom occupying the position of the void space in the FKBP12 crystal structure. Similarly, Ile94 and Ile105 of FKBP52 [45] replace the less bulky pair of Val63 and Leu74 in FKBP12, resulting in a reduced void volume beneath the tryptophan ring. If the transition of the indole ring to the perpendicular orientation is substantially more populated in FKBP12, as compared with these other FKBP domains, such a conformation might prove amenable to selective drug inhibition.
Reorientation equilibria and dynamics for the active site indole ring in wild-type FKBP12
The structural analysis of these FKBP12 variants support the possibility that there may be only a modest energy difference between the canonical and the perpendicular orientations for the indole ring of Trp59 in the wild-type protein. Potential evidence for internal mobility of the single indole ring in FKBP12 comes from the time-resolved fluorescence anisotropy measurements of Silva and Prendergast [59] in which they derived an S2 order parameter of 0.75 for the uncomplexed protein and an S2 value of ~1 for the FK506-bound complex. In contrast with the three fastest relaxation components for which the amplitudes and time constants were similar for the free and ligated forms of the protein, the time constant of 5.43 ns for the slowest component of the fluorescence anisotropy decay in the FK506-bound protein was decreased to 4.75 ns for uncomplexed FKBP12. The authors analysed these data assuming that this shift in the time constant arises from unresolved contributions from a local conformational transition of the indole ring and the global molecular tumbling [59], leading to a predicted time constant of 3 ns for the internal motion. In their molecular dynamics analysis on the role of the buried water molecule in modulating the conformational dynamics of FKBP12, Park and Saven [60] reported a simulation of the wild-type protein in which the Trp59 indole ring transitioned to a perpendicular orientation after 8 ns in a 12 ns simulation. Although no reverse transition was observed, their simulation suggested the energetic accessibility of such a conformation.
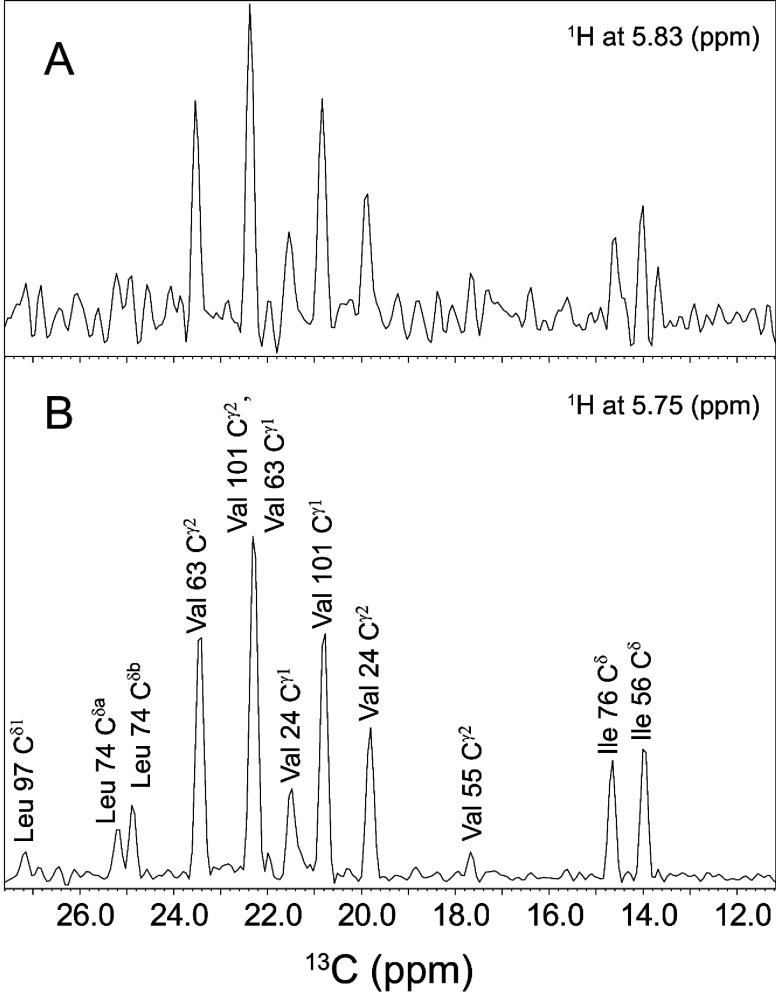
In the 3D 13C-15N-1H NOESY spectrum of U-13C,15N-labelled FKBP12 we observed substantial cross-peaks from the indole HNε1 of Trp59 to both methyl groups for Val24 and Val63 as well as to the Cγ1 methyl of Val101, despite the fact that these methyls lie ~7 Å from the indole HNε1 in the crystal structure of the wild-type protein. To examine this issue in more detail, we prepared a sample of FKBP12 with 13C enrichment for the methyl groups of valine, leucine and isoleucine residues with uniform 15N and perdeuteration at all other carbon-bound sites as described previously by Kay and colleagues [29]. For most of the moderately intense cross-peaks of the 13C-15N-1H NOESY spectra for valine, leucine and isoleucine methyl-labelled FKBP12, the NOE build-up curves were approximately linear up to a 200 ms mixing time.
A 1D 13C slice at the 1H and 15N frequencies of the Trp59 indole N–H in the major slow exchange conformational state indicates 11 peaks, most of which correspond to methyl groups that are too distant from the indole HNε1 in the high-resolution structure of wild-type FKBP12 to give rise to the observed intensities (Figure 7B). The analogous 1D slice at the 1H and 15N frequencies of the Trp59 indole N–H in the minor slow exchange conformational state yielded a qualitatively similar pattern (Figure 7A), although the lower sensitivity from this minor state precluded a more detailed analysis.
Figure 7. 1D slices from the 3D 13C-15N-1H NOESY spectrum at the 1HNε1 and 15N ε1 (117.75 p.p.m.) frequencies of Trp59.
NOE cross-peaks with 13C-enriched methyl groups in the major (B) and minor (A) slow exchange states of wild-type FKBP12. The methyl groups of Leu7 were not stereochemically assigned.
The methyl–indole HNε1 cross-peak volumes were analysed to test their consistency with FKBP12 adopting a mixture of active-site conformations corresponding to the PDB code 2PPN wild-type and G89P variant crystal structures. The familiar 1/r6 distance dependence for the NOE cross-relaxation rate typically invokes the assumption of an isolated pair of nuclear spins in the absence of internal motion.
High levels of deuteration suppress strongly spin diffusion effects thus providing a useful approximation of the isolated spin pair assumption. Furthermore, if the rapid internal motion for both of the interacting 1H nuclei is isotropic and uncorrelated (similar to the standard assumption of crystallographic B-factor analysis for all but the highest-resolution protein structures), the resultant NOE cross-relaxation rate equals that predicted for the two nuclei being fixed at their mean relative positions [61]. Although this exact cancellation of the radial and angular dispersion effects provides a useful approximation for many types of proton pairs, the 3-fold rotational jumps of methyl protons deviates strongly from spherical isotropy. As a result, NOE calculations based on a simple 1/r6 summation of distances from the methyl protons to a distal proton site give rise to severely foreshortened apparent distances in protein NOESY analyses. In the present study, methyl–indole HNε1 NOESY cross-peaks were analysed under the assumption of separability of the generalized order parameter characterizing the interaction (eqn 2) and this analysis was compared with the results derived from the more commonly used approximations of measuring NOE distances from a unified methyl group centred at either the methyl carbon or the mean position of the methyl protons.
Table 1 lists the normalized experimental volumes for the methyl–indole HNε1 NOESY cross-peaks shown in Figure 7(B). In addition to normalization against the most intense NOE resonance, the volumes for these NOE cross-peaks were normalized against the peak volumes for the corresponding methyl resonances from the 2D 1H–13C HSQC spectrum to compensate for differences in relaxation behaviour and in the levels of 13C enrichment due to incomplete biosynthetic incorporation from the labelled α-oxobutyrate and α-oxoisovalerate during protein expression. Also listed are the distances from each methyl carbon to the indole HNε1 in the PDB code 2PPN wild-type and the G89P variant crystal structures. Distances beyond 7 Å were not included in the calculations and neither were the NOE cross-peaks to the Leu74 methyls due to ambiguity in their stereochemical assignment. This residue offers minimal utility in the NOE analysis as the distance from the closest Leu74 methyl to the Trp59 HNε1 is nearly the same for both the wild-type and G89P crystal structures.
Table 1. Generalized order parameter NOE analysis of Trp59 indole reorientation in FKBP12.
Interactions for methyl–HNε1 distances >7 Å were not included. The ratio of volumes between the normalized NOEs for PDB code 2PPN wild-type/G89P was 0.28. Distance, distance between methyl carbon to the indole HNε1 in the PDB code 2PPN wild-type and the G89P variant crystal structures. NOE(exp), normalized experimental volumes for the methyl–indole HNε1 NOESY cross-peaks.
| Residue | Atom | NOE(exp) | Distance2PPN (Å) | NOE (S2) | DistanceG89P (Å) | NOE (S2) |
|---|---|---|---|---|---|---|
| Ile56 | Cδ1 | 0.795 | 4.38 | 0.903 | ||
| Ile76 | Cδ1 | 0.828 | 4.14 | 1.000 | ||
| Val55 | Cγ2 | 0.048 | 6.59 | 0.018 | ||
| Val24 | Cγ2 | 0.460 | 3.89 | 0.661 | ||
| Val101 | Cγ1 | 0.650 | 6.82 | 3.37 | 0.858 | |
| Val24 | Cγ1 | 0.308 | 4.54 | 0.173 | ||
| Val101* | Cγ2 | 1.000 | 4.98 | 0.348 | 3.32 | 1.000 |
| Val63 | Cγ2 | 0.651 | 6.88 | 0.054 | 3.60 | 0.613 |
| Leu97 | Cδ1 | 0.050 | 6.27 | 0.092 |
*This cross-peak also includes the degenerate Val63 Cγ1 which contributes only 4% of the predicted volume.
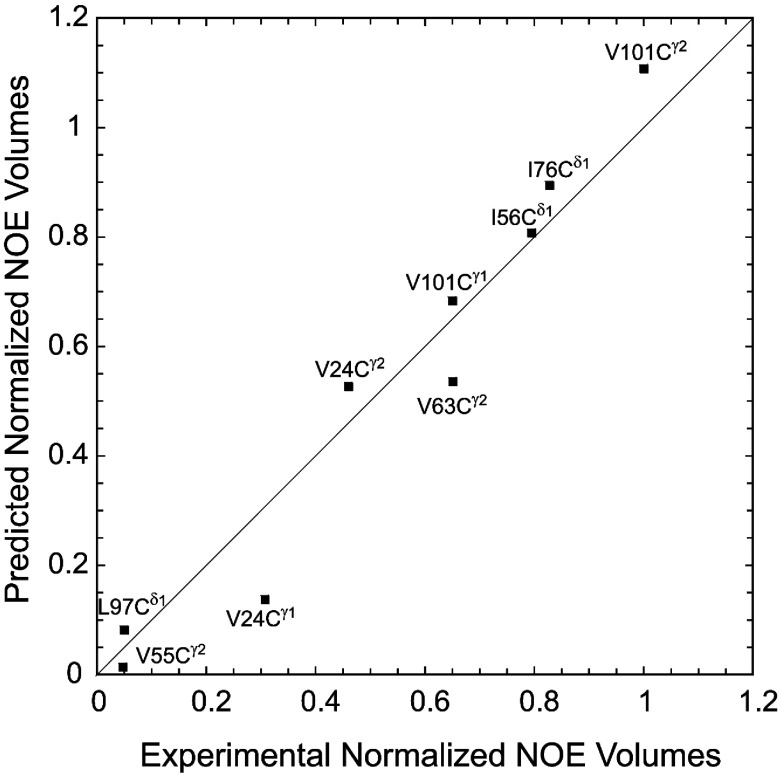
The relative NOE volumes for each of these methyl–indole HNε1 interactions were predicted from generalized order parameter calculations [41] applied to the two crystal structures. For an admixture of 80% and 20% for the PDB code 2PPN wild-type and the G89P conformations respectively, the observed NOE volumes were fitted to an RMSD of 0.086 with a correlation coefficient (r) of 0.976 (Figure 8). A 20% shift to an admixture of 84% and 16% predicts an increase in RMSD to 0.121 with an r value of 0.957, whereas, similarly, a 75–25% conformational mixture yields an RMSD of 0.108 with an r value of 0.956. When the methyl carbon position was used to model the effective methyl–indole HNε1 distance, the optimal fit was obtained for an 82–18% distribution with an RMSD of 0.134 and an r value of 0.955 (Supplementary Figure S2 and Supplementary Table S1 at http://www.biochemj.org/bj/458/bj4580525add.htm). Use of the mean methyl proton position yields an optimum distribution at 79% and 21% with an RMSD of 0.183 and an r value of 0.905 (Supplementary Figure S3 and Supplementary Table S2 at http://www.biochemj.org/bj/458/bj4580525add.htm). The markedly worse performance for the use of the mean methyl proton position reflects principally the fact that distal protons that lie along the methyl rotation axis yield elevated predictions for both the radial and angular components of the NOE interaction.
Figure 8. Predicted against observed NOE volumes for the methyl cross-peaks to Trp59 HNε1.
The experimental NOE volumes were normalized to the maximum value (Val101 Cγ2) and to the volumes of the corresponding 1H–13C cross-peaks in the 2D HSQC spectrum to compensate for relaxation and differential enrichment effects. Generalized order parameter calculations were performed on the crystal structures of the wild-type protein and G89P variants and then weighted at 80% and 20% respectively.
These results indicate significant conformational plasticity in the active site of FKBP12. As there is no apparent linebroadening for the resonances involved, the indole reorientation occurs presumably more rapidly than for the microsecond to millisecond conformational exchange regime. Indication of a much more rapid transition is drawn from the 15N relaxation analysis that we reported recently for the backbone amides of FKBP12 [22]. The R1 and R2 relaxation rates of 1.50 s−1 and 5.56 s−1 and the 15N NOE value of 0.634 at 14.1 T (600 MHz 1H) that we observed for the Trp59 Nε1 resonance are all well below the values observed for the amides in the well-ordered regions of the backbone (molecular tumbling time τc of 5.8 ns). Applying the extended model-free analysis [62] to the Trp59 Nε1 relaxation yielded an overall S2 order parameter value of 0.63 and a time constant τe of 1.5 ns for the slow component of internal motion.
For a simple jump model S2 can be calculated by eqn (3) [63,64]:
| (3) |
where pa and pb are the state populations and θ is the angle between the indole N–H bond vector orientations which is 83° for the PDB code 2PPN wild-type and G89P structures. A 20% population for the reoriented indole conformation yields an estimated S2 order parameter of 0.53. Both of the experimental and model S2 values should be regarded as approximate. In contrast with the fluorescence study cited above [59], the large number of 1H–15N bond vectors monitored in the NMR relaxation experiments facilitate the deconvolution of contributions from internal and global motion. Nevertheless, the derived order parameter values are still generally less accurate for internal motions having time constants similar to that for the global tumbling of the protein. The simple jump model does not account for the smaller scale librational motions of the 1H–15N vector which give rise to order parameter values near 0.85 for the well-ordered backbone amides and presumably for the Trp59 indole ring as well. Despite these caveats, the experimental evidence for significant conformational dynamics of the tryptophan residue indole ring in the nanosecond timeframe appears to be consistent with the ring reorientation process characterized by the NOE analysis.
Dual conformers for the active site indole ring in a G89P variant of FKBP12 at 1.20 Å resolution
Given the small free-energy difference between the two perpendicular orientations that the active site indole ring of wild-type FKBP12 occupies in solution and the modest differences surrounding that ring in the wild-type and G89P variant crystal structures, we examined further whether the crystal structure of the G89P variant might also yield evidence for conformational heterogeneity at this site. Since the crystals for this variant diffracted well beyond the 1.50 Å resolution data that we had initially collected on our in-house diffractometer, we collected a second dataset to 1.20 Å resolution at the National Synchrotron Light Source (Table 2). During the later stages of refinement, dual conformers were allowed for both Trp59 and Glu60. Minor conformers for the two residues were obtained at occupancies of 0.29 and 0.34 respectively. When viewed edge-on for the major conformer of Trp59, the electron density for a perpendicular orientation of the indole ring is readily apparent (Figure 9). In the minor conformer of Glu60, the main chain carbonyl oxygen was shifted towards the 50′s loop into the position seen in the wild-type FKBP12 crystal structure. Although the Glu60 Cγ of the minor conformer was not well fitted to the electron density, the derived side chain χ1 torsion angle was shifted towards the orientation seen in the wild-type FKBP12 structure.
Table 2. Crystallographic data collection, refinement and model details (PDB code 4N19).
Values in paretheses are for the highest-resolution shell.
| Parameter | Value |
|---|---|
| Data collection | |
| Resolution range (Å) | 35–1.20 (1.3–1.20) |
| Number of unique reflections | 23973 |
| Redundancy | 5.7 (2.2) |
| Completeness (%) | 72.7 (8.5) |
| Average I/σ(I) | 48.5 (2.5) |
| Rmerge (%) | 6.0 (34) |
| Refinement | |
| Resolution limits (Å) | 35–1.20 |
| Number of reflections | 23973 |
| Rwork (%) | 12.6 |
| Rfree (%) | 18.0 |
| Non-hydrogen atoms | |
| Protein | 891 |
| SO42− | 1 |
| Water | 148 |
| Average B (Å2) | |
| All atoms | 20 |
| Solvent | 41 |
| Geometry | |
| RMSD bond length (Å) | 0.02 |
| RMSD bond angle (°) | 2.28 |
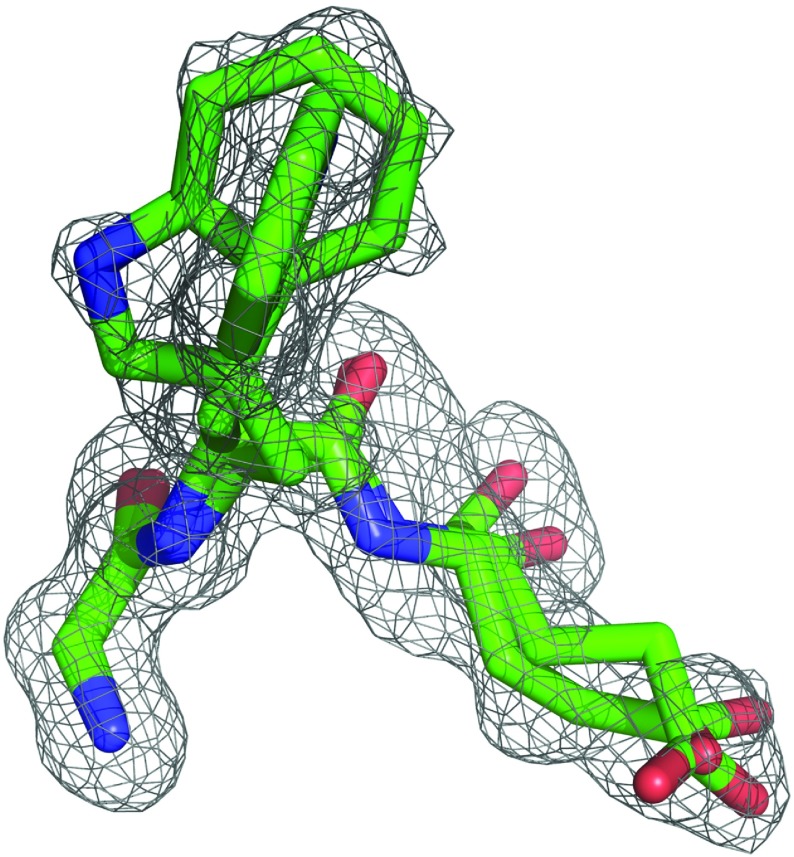
Figure 9. Electron density omit map for Gly58, Trp59 and Glu60 in the active site of the G89P variant of FKBP12 at 1.20 Å resolution.
Viewed along the plane of the indole ring for the major conformer (occupancy of 0.71), electron density for the perpendicular orientation of the minor conformer is apparent readily. The carbonyl oxygen in the minor conformer of Glu60 (occupancy of 0.34) is shifted away from the canonical α-helical hydrogen-bonding geometry seen in the major conformer and to a position close to that observed in the wild-type FKBP12 structure [31]. The contour level for the electron density grid is 0.1261 e/Å3=2σ.
The two perpendicular orientations of the Trp59 indole ring exhibited by the 1.20 Å resolution structure of the G89P variant reinforces the observation of a small difference in free energy for those two conformations in solution. The similar populations for the minor conformers of Trp59 and Glu60 in this crystal structure is consistent with a concerted shift for both the indole ring orientation and the backbone hydrogen-bonding geometry for residues 59 and 60, which, in turn, appears to be at least partially coupled to a reorientation of the Glu60 side chain.
Online data
ACKNOWLEDGEMENTS
We thank the Wadsworth Center for the use of the NMR facility, X-ray crystallography and Molecular Genetics cores, the New York Structural Biology Center for use of the NMR facility, and the National Synchrotron Light Source for use of beamline X25.
AUTHOR CONTRIBUTION
Sourajit Mustafi was involved in the design of the study, collection and analysis of the NMR data, and writing of the paper. Jing Zhang carried out the crystallization studies. Matthew Brecher was involved in the collection and analysis of the crystallographic data. Hongmin Li supervised and participated in the crystallographic studies as well as in writing the paper. David LeMaster prepared the protein samples and participated with Griselda Hernández in the design and supervision of the study, in the collection and analysis of the NMR data, and in writing of the paper.
FUNDING
This work was supported, in part, by the National Institutes of Health [grant number GM 088214].
References
- 1.Liu J., Farmer J. D., Lane J. W. S., Friedman J., Wiessman I., Schreiber S. L. Calcineurin is a common target for cyclophilin–cyclosporin A and FKBP–FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 2.Heitman J., Novva N. R., Hall M. N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 3.Sivendran S., Agarwal N., Gartrell B., Ying J., Boucher K. M., Choueiri T. K., Sonpavde G., Oh W. K., Galsky M. D. Metabolic complications with the use of mTOR inhibitors for cancer therapy. Cancer Treat. Rev. 2014;40:190–196. doi: 10.1016/j.ctrv.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galat A. Functional drift of sequence attributes in the FK506-binding proteins (FKBPs) J. Chem. Inf. Model. 2008;48:1118–1130. doi: 10.1021/ci700429n. [DOI] [PubMed] [Google Scholar]
- 5.Marz A. M., Fabian A. K., Kozany C., Bracher A., Hausch F. Large FK506-binding proteins shape the pharmacology of rapamycin. Mol. Cell. Biol. 2013;33:1357–1367. doi: 10.1128/MCB.00678-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiwad M., Edlich F., Kilka S., Erdmann F., Jarczowski F., Dorn M., Moutty M. C., Fischer G. Comparative analysis of calcineurin inhibition by complexes of immunosuppressive drugs with human FK506 binding proteins. Biochemistry. 2006;45:15776–15784. doi: 10.1021/bi061616p. [DOI] [PubMed] [Google Scholar]
- 7.Galat A. Functional diversity and pharmacological profiles of the FKBPs and their complexes with small natural ligands. Cell. Mol. Life Sci. 2013;70:3243–3275. doi: 10.1007/s00018-012-1206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timerman A. P., Ogunbumni E., Freund E., Wiederrecht G., Marks A. R., Fleischer S. The calcium release channel of sacroplasmic reticulum is modulated by FK506-binding protein. J. Biol. Chem. 1993;268:22992–22929. [PubMed] [Google Scholar]
- 9.Timerman A. P., Onoue H., Xin H. B., Barq S., Copello J., Wiederrecht G., Fleischer S. Selective binding of FKBP12.6 by the cardiac ryanodine receptor. J. Biol. Chem. 1996;271:20385–20391. doi: 10.1074/jbc.271.34.20385. [DOI] [PubMed] [Google Scholar]
- 10.Shou W., Aghdasi B., Armstrong D. L., Guo Q., Bao S., Charng M. J., Mathews L. M., Schneider M. D., Hamilton S. L., Matzuk M. M. Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature. 1998;391:489–492. doi: 10.1038/35146. [DOI] [PubMed] [Google Scholar]
- 11.Maruyama M., Li B. Y., Chen H., Xu X., Song L. S., Guatimosim S., Zhu W., Yong W., Zhang W., Bu G., et al. FKBP12 is a critical regulator of the heart rhythm and the cardiac voltage-gated sodium current in mice. Circ. Res. 2011;108:1042–1052. doi: 10.1161/CIRCRESAHA.110.237867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguchi N., Yoshikawa T., Ikeda T., Takahashi I., Shervani N. J., Uruno A., Yamauchi A., Nata K., Takasawa S., Okamoto H., Sugawara A. FKBP12.6 disruption impairs glucose-induced insulin secretion. Biochem. Biophys. Res. Commun. 2008;371:735–740. doi: 10.1016/j.bbrc.2008.04.142. [DOI] [PubMed] [Google Scholar]
- 13.Galeotti N., Quattrone A., Vivoli E., Norcini M., Bartolini A., Ghelardini C. Different involvement of type 1, 2, and 3 ryanodine receptors in memory processes. Learn. Mem. 2008;15:315–323. doi: 10.1101/lm.929008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Betzenhauser M. J., Reiken S., Meli A. C., Xie W., Chen B. X., Arancio O., Marks A. R. Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell. 2012;150:1055–1067. doi: 10.1016/j.cell.2012.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang T., Li B. Y., Danielson P. D., Shah P. C., Rockwell S., Lechleider R. J., Martin J., Manganaro T., Donahoe P. K. The immunophilin FKBP12 functions as a common inhibitor of the TGFβ family type I receptors. Cell. 1996;86:435–444. doi: 10.1016/s0092-8674(00)80116-6. [DOI] [PubMed] [Google Scholar]
- 16.Riggs D. L., Cox M. B., Tardif H. L., Hessling M., Buchner J., Smith D. F. Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Mol. Cell. Biol. 2007;27:8658–8669. doi: 10.1128/MCB.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avramut M., Achim C. L. Immunophilins and their ligands: insights into survival and growth of human neurons. Physiol. Behav. 2002;77:463–468. doi: 10.1016/s0031-9384(02)00934-4. [DOI] [PubMed] [Google Scholar]
- 18.Sugata H., Matsuo K., Nakagawa T., Takahashi M., Mukai H., Ono Y., Maeda K., Akiyama H., Kawamata T. A peptidyl-prolyl isomerase, FKBP12, accumulates in Alzheimer neurofibrillary tangles. Neurosci. Lett. 2009;459:96–99. doi: 10.1016/j.neulet.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 19.Liu F. L., Liu P. H., Shao H. W., Kung F. L. The intracellular domain of amyloid precursor protein interacts with FKBP12. Biochem. Biophys. Res. Commun. 2006;350:472–477. doi: 10.1016/j.bbrc.2006.09.073. [DOI] [PubMed] [Google Scholar]
- 20.Gerard M., Debyser Z., Desender L., Baert J., Brandt I., Baekelandt V., Engelborghs Y. FK506 binding protein 12 differentially accelerates fibril formation of wild-type α-synuclein and its clinical mutants A30P or A53T. J. Neurochem. 2008;106:121–133. doi: 10.1111/j.1471-4159.2008.05342.x. [DOI] [PubMed] [Google Scholar]
- 21.Gerard M., Deleersnijder A., Daniëls V., Schreurs S., Munck S., Reumers V., Pottel H., Engelborghs Y., VandenHaute C., Taymans J. M., et al. Inhibition of FK506 binding proteins reduces α-synuclein aggregation and Parkinson's disease-like pathology. J. Neuroscience. 2010;30:2454–2463. doi: 10.1523/JNEUROSCI.5983-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mustafi S. M., Chen H., Li H., LeMaster D. M., Hernández G. Analyzing the visible conformational substates of the FK506-binding protein FKBP12. Biochem. J. 2013;453:371–380. doi: 10.1042/BJ20130276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith J. P., Kim J. L., Kim E. E., Sintchak M. D., Thomson J. A., Fitzgibbon M. J., Fleming M. A., Caron P. R., Hsiao K., Navia M. A. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12–FK506 complex. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 24.Liang J., Choi J., Clardy J. Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:736–744. doi: 10.1107/s0907444998014747. [DOI] [PubMed] [Google Scholar]
- 25.Huse M., Chen Y. G., Massague J., Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF-β receptor in complex with FKBP12. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 26.Chaikuad A., Alfano I., Kerr G., Sanvitale C. E., Boegermann J. H., Triffitt J. T., von Delft F., Knapp S., Knaus P., Bullock A. N. Structure of the bone morphogenetic protein receptor ALK2 and implications for fibrodysplasia ossificans progressiva. J. Biol. Chem. 2012;287:36990–36998. doi: 10.1074/jbc.M112.365932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernández G., Anderson J. S., LeMaster D. M. Polarization and polarizability assessed by protein amide acidity. Biochemistry. 2009;48:6482–6494. doi: 10.1021/bi900526z. [DOI] [PubMed] [Google Scholar]
- 28.Hernández G., LeMaster D. M. Reduced temperature dependence of collective conformational opening in a hyperthermophile rubredoxin. Biochemistry. 2001;40:14384–14391. doi: 10.1021/bi0112560. [DOI] [PubMed] [Google Scholar]
- 29.Goto N. K., Gardner K. H., Mueller G. A., Willis R. C., Kay L. E. A robust and cost-effective method for the production of Val, Leu, Ile (δ1) methyl-protonated 15N, 13C, 2H labeled proteins. J. Biomolec. NMR. 1999;13:369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Szep S., Park S., Boder E. T., VanDuyne G. D., Saven J. G. Structural coupling between FKBP12 and buried water. Proteins. 2009;74:603–611. doi: 10.1002/prot.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheldrick G. M., Schneider T. R. SHELXL: high resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 34.Emsley P., Lohkamp B., Scott W. G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Kay L. E., Xu G. Y., Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. A. 1994;109:129–133. [Google Scholar]
- 37.Muhandiram D. R., Kay L. E. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. B. 1994;103:203–216. [Google Scholar]
- 38.Yamazaki T., Lee W., Arrowsmith C. H., Muhandiram D. R., Kay L. E. A suite of triple resonance NMR experiments for the backbone assignment of 15N, 13C, 2H labeled proteins with high sensitivity. J. Am. Chem. Soc. 1994;116:11655–11666. [Google Scholar]
- 39.Diercks T., Coles M., Kessler H. An efficient strategy for assignment of crosspeaks in 3D heteronuclear NOESY experiments. J. Biomol. NMR. 1999;15:177–180. doi: 10.1023/A:1008367912535. [DOI] [PubMed] [Google Scholar]
- 40.Hu W., Lee K. C., Cross T. A. Tryptophan in membrane proteins: indole ring orientations and functional implication in the gramicidin channel. Biochemistry. 1993;32:7035–7047. doi: 10.1021/bi00078a032. [DOI] [PubMed] [Google Scholar]
- 41.Lipari G., Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 42.Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 43.Schwarzinger S., Kroon G. J., Foss T. R., Chung J., Wright P. E., Dyson H. J. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 44.Li P., Ding Y., Wu B., Shu C., Shen B., Rao Z. Structure of the N-terminal domain of human FKBP52. Acta Crystalogr. D Biol. Crystalogr. 2003;59:16–22. doi: 10.1107/s0907444902017523. [DOI] [PubMed] [Google Scholar]
- 45.Bracher A., Kozany C., Hahle A., Wild P., Zacharias M., Hausch F. Crystal structures of the free and ligand-bound FK1–FK2 domain segment of FKBP52 reveal a flexible inter-domain hinge. J. Mol. Biol. 2013;425:4134–4144. doi: 10.1016/j.jmb.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 46.Jabs A., Weiss M. S., Hilgenfeld R. Non-proline cis peptide bonds in proteins. J. Mol. Biol. 1999;286:291–304. doi: 10.1006/jmbi.1998.2459. [DOI] [PubMed] [Google Scholar]
- 47.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: John Wiley & Sons; 1986. [Google Scholar]
- 48.Wu B., Li P., Liu Y., Ding Y., Shu C., Ye S., Bartlam M., Shen B., Rao Z. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc. Natl. Acad. Sci U.S.A. 2004;101:8348–8353. doi: 10.1073/pnas.0305969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Y., Bax A. SPARTA+: a modest improvement in empircal NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han B., Liu Y., Ginzinger S. W., Wishart D. S. SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohlhoff K. J., Robustelli P., Cavalli A., Salvatella X., Vendruscolo M. Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J. Am. Chem. Soc. 2009;131:13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- 52.Moon S., Case D. A. A new model for chemical shifts of amide hydrogens in proteins. J. Biomol. NMR. 2007;38:139–150. doi: 10.1007/s10858-007-9156-8. [DOI] [PubMed] [Google Scholar]
- 53.Itoh S., DeCenzo M. T., Livingston D. J., Pearlman D. A., Navia M. A. Conformation of FK506 in X-ray structures of its complexes with human recombinant FKBP12 mutants. Bioorg. Med. Chem. Lett. 1995;5:1983–1988. [Google Scholar]
- 54.Yang D., Rosen M. K., Schreiber S. L. A composite FKBP12–FK506 surface that contacts calcineurin. J. Am. Chem. Soc. 1993;115:819–820. [Google Scholar]
- 55.Fulton K. F., Jackson S. E., Buckle A. M. Energetic and structural analysis of the role of tryptophan 59 in FKBP12. Biochemistry. 2003;42:2364–2372. doi: 10.1021/bi020564a. [DOI] [PubMed] [Google Scholar]
- 56.Bracher A., Kozany C., Thost A. K., Hausch F. Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90. Acta Crystallogr. D Biol. Crystalogr. 2011;67:549–559. doi: 10.1107/S0907444911013862. [DOI] [PubMed] [Google Scholar]
- 57.Liang J., Hung D. T., Schreiber S. L., Clardy J. Structure of the human 25 kDa FK506 binding protein complexed with rapamycin. J. Am. Chem. Soc. 1996;118:1231–1232. [Google Scholar]
- 58.Schultz L. W., Martin P. K., Liang J., Schreiber S. L., Clardy J. Atomic structure of the immunophilin FKBP13–FK506 complex: insights into the composite binding surface for calcineurin. J. Am. Chem. Soc. 1994;116:3129–3130. [Google Scholar]
- 59.Silva N. D., Jr, Prendergast F. G. Tryptophan dynamics of the FK506 binding protein: time-resolved fluorescence and simulations. Biophys. J. 1996;70:1122–1137. doi: 10.1016/S0006-3495(96)79706-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park S., Saven J. G. Statistical and molecular dynamics studies of buried waters in globular proteins. Proteins. 2005;60:450–463. doi: 10.1002/prot.20511. [DOI] [PubMed] [Google Scholar]
- 61.LeMaster D. M., Kay L. E., Brunger A. T., Prestegard J. H. Protein dynamics and distance determination by NOE measurements. FEBS Lett. 1988;236:71–76. doi: 10.1016/0014-5793(88)80287-4. [DOI] [PubMed] [Google Scholar]
- 62.Clore G. M., Szabo A., Bax A., Kay L. E., Driscoll P. C., Gronenborn A. M. Deviations from the simple 2-parameter model-free approach to the interpretation of nitrogen 15 nuclear relaxation of proteins. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 63.Lipari G., Szabo A. Nuclear magnetic resonance relaxation in nucleic acid fragments: models for internal motion. Biochemistry. 1981;20:6250–6256. doi: 10.1021/bi00524a053. [DOI] [PubMed] [Google Scholar]
- 64.Torchia D. A. Dynamics of biomolecules from picoseconds to seconds at atomic resolution. J. Magn. Reson. 2011;212:1–10. doi: 10.1016/j.jmr.2011.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.