Abstract
The detailed understanding of adult tissue stem cells has significance for both regenerative medicine and oncology. This perspective will discuss how major advances in our ability to identify and monitor these cells, which include genetic lineage tracing, FACS purification, and robust in vitro clonogenic assays, have changed our view of their roles in many organs. Label retention and quiescence are no longer considered obligatory stem cell features. Furthermore, some tissues have more than one type of stem cell, each used in only particular situations of regenerative stress. Thus, there is no “one size fits all” adult tissue stem cell paradigm.
Spurred by the excitement surrounding the potential of pluripotent stem cells in regenerative medicine (Takahashi et al., 2007), interest in tissue-specific adult stem cells also increased significantly since the inception of this journal. A detailed understanding of how organs maintain and repair themselves in the postnatal organism is obviously required in order to properly utilize the derivatives of pluripotent cells in therapy. In addition, it is conceivable that the ability to control and manipulate tissue stem cells in situ may obviate the need for exogenous cell transplantation altogether. There is much evidence that tissue-resident stem cells may be the origin of malignant tumors (Barker et al., 2009). Therefore, knowing the mechanisms that turn stem cells into cancer stem cells or understanding how cancers derived from differentiated cells acquire stem cell characteristics would probably yield targets for therapy. Although it would be difficult to comprehensively cover all of the advances recently made in our understanding of the many different kinds of tissue stem cells, there are some common themes that apply broadly and these will be discussed herein.
Stem Cells versus Progenitors: Not Simply a Semantic Distinction
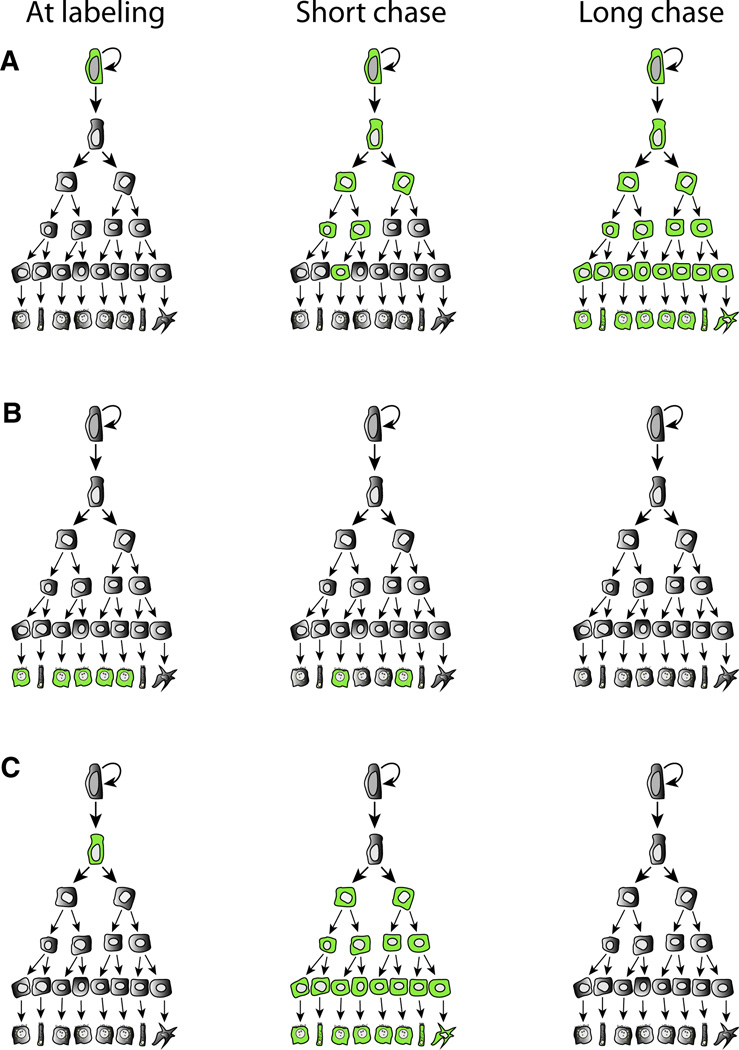
Perhaps because of its popularity with the lay public and funding agencies, the term “stem cell” is used liberally and often indiscriminately. However, despite much progress, the existence of true stem cells has to date been shown for only few adult tissues when the strict criterion of extensive clonal self-renewal (van der Kooy and Weiss, 2000; Weissman et al., 2001) is used. This widely accepted stem cell definition was derived from the hematopoietic field where in vivo transplantation assays are available to assess replicative potential. Similar in vivo reconstitution assays do not yet exist for many adult tissues and thus it is not possible to clearly prove self-renewal in those organs by this classic approach. In some rapidly regenerating tissues—most notably the intestine—clonal marking techniques can prove life-long self-renewal in the absence of transplantation and firmly establish the identity of stem cells (Figure 1). However, in many tissues the existence of true stem cells remains yet to be fully proven and the term “progenitor” is more appropriate to describe the cells that have been found to date. Like true stem cells, progenitors often are multipotential and may give rise to several different mature lineages. However, they can be distinguished from true stem cells by the fact that clonal labeling is transient and does not last for the life of the animal (Figure 1C). In contrast, true stem cells will produce labeled offspring permanently (Figure 1A). Extensive self-renewal in culture may represent an in vitro artifact and does not conclusively prove that the cell in question truly is a stem cell in vivo. The distinction is not just semantic because the limited self-renewal of progenitors may signify restricted regenerative capacity of a particular tissue, particularly in the setting of chronic injury or aging. Although the existence of self-renewing stem cells in highly regenerative organs has been established for some time now (Mackenzie and Bickenbach, 1985; Potten and Hendry, 1975), the precise identity, molecular regulation, and anatomic location of these cells in organs such as colon (Barker et al., 2007), small intestine (Barker et al., 2007), stomach (Barker et al., 2010), breast (Shackleton et al., 2006; Stingl et al., 2006), and skin (Snippert et al., 2010a) were described only fairly recently.
Figure 1. Lineage Tracing in Adult Tissue Stem Cell Systems.
(A) A self-renewing stem cell is labeled with a specific marker (green). All cells in the hierarchy are permanently marked in long-term follow up.
(B) Specific labeling of mature, differentiated cells. If these cells are replaced by a mature stem/progenitor, the label will disappear from the mature population with time.
(C) A progenitor, not capable of indefinite self-renewal, is labeled. After short follow-up, multiple lineages are marked. However, the marked cells disappear after longer times.
In the intestine, these breakthroughs were made by using genetic lineage tracing, a strategy originally devised by developmental biologists (Kretzschmar and Watt, 2012). Although it has been generally challenging to find highly specific markers for adult tissue stem cells, methods such as genome-wide assessment of gene expression in highly purified cell populations have suggested candidate genes (Barker et al., 2007). These loci can be used to restrict marker gene expression to the cells of interest and their progeny, thereby allowing analysis of self-renewal and potency in vivo. The results of these kinds of studies have been surprising in some cases, most notably in the small intestine, where the cell located in the crypt at the +4 position was long thought to be the true and only stem cell (Potten et al., 2009; Potten and Hendry, 1975). However, lineage tracing of cells expressing the Wnt target Lgr5 unambiguously showed that slender cells at the bottom of the crypt, which are clearly distinct from the +4 cells, also act as stem cells (Barker et al., 2007; Snippert et al., 2010b). Lgr5+ cells give rise to all of the differentiated cells types of the intestinal villus and divide hundreds of times during the life of the animal. Highly specific stem cell markers such as Lgr5 are still missing for many other tissues and their identification should be a high priority for the field. Markers specific for stem cells and progenitors in organs such as lung, heart, and liver would facilitate studies of the ontogeny of cells formed during adult organ maintenance and repair and would probably resolve some of the current controversies. Specific stem cell markers would also allow the determination of whether cancers originate from that population, as shown for Lgr5+ colon stem cells (Barker et al., 2009).
Label Retention and Quiescence: Stem Cell Markers of the Past?
Perhaps the most generally significant consequence of the discovery of the Lgr5+ intestinal stem cell is our changed view of quiescence as an essential property of tissue stem cells. Label retention was once considered a hallmark of tissue stem cells (Clarke et al., 2003; Duvillié et al., 2003; Mackenzie and Bickenbach, 1985; Morris and Potten, 1994; Terskikh et al., 2011). In order to identify label-retaining cells (LRCs), replicating DNA was labeled by administration of radioactive thymidine or BrdU for a prolonged period, followed by an even longer washout time. Tissues were then stained and cells that still retained their labels months or years after labeling were considered to be stem cells (Potten and Hendry, 1975). This paradigm was based on the notion that stem cells should persist in tissues and divide only rarely to protect their genome. Work in the epidermis had already clearly shown that LRC were not the only stem cells in skin (Claudinot et al., 2005; Li and Clevers, 2010), but the very rapid rate of division in Lgr5+ intestinal stem cells plainly demonstrated that quiescence and label retention are not inherent properties of all tissue stem cells.
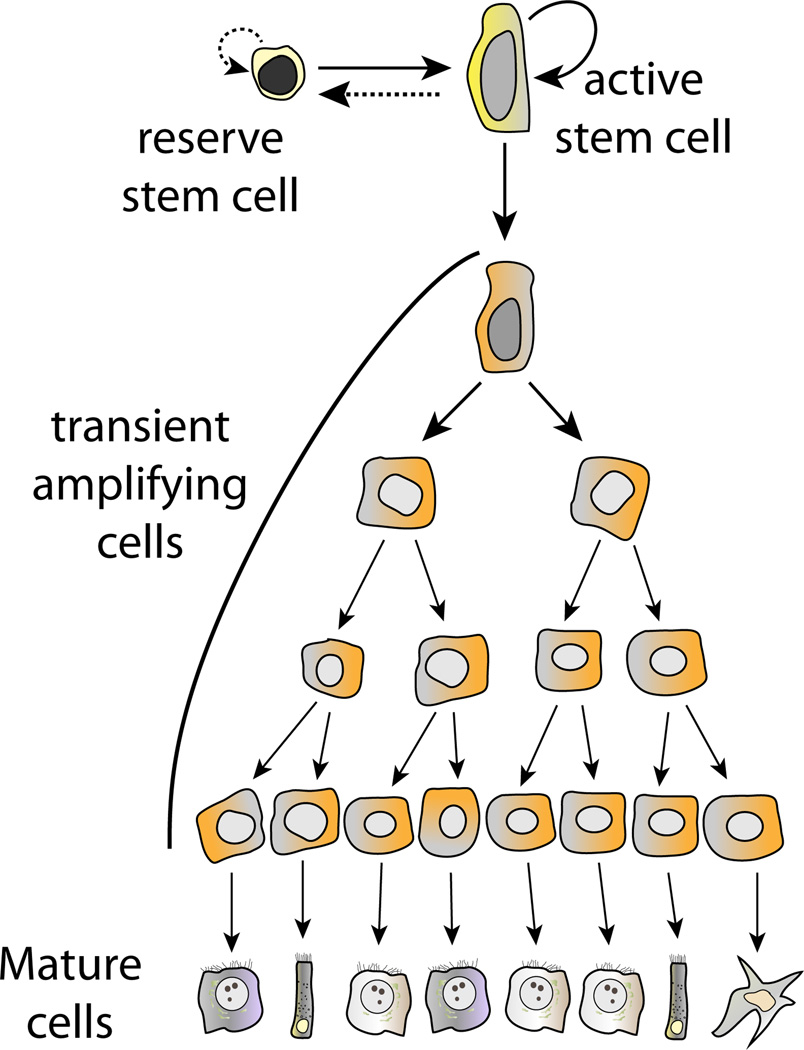
What then is the role of the label-retaining +4 intestinal stem cell (Potten et al., 2009)? Further investigation of this topic has revealed another important new concept in tissue stem cell biology: there can be more than one type of stem cell (Figure 2; Li and Clevers, 2010). Although it is clear that Lgr5+ stem cells are responsible for self-renewal during normal homeostasis, certain injuries, particularly irradiation, result in the activation of the label-retaining +4 stem cell (Takeda et al., 2011; Tian et al., 2011; Yan et al., 2012). Activated +4 cells are able to produce new Lgr5+ cells, both in vivo and in vitro. Hence, the +4 position intestinal stem cell is the prototype of a facultative tissue stem cell, called upon only in very specific circumstances (Figure 2). Although the +4 cells are normally quiescent, lineage tracing with Hopx as a specific marker for this population has shown that the relationship between the two stem cells is bidirectional (Takeda et al., 2011). Depending on the situation, Lgr5+ intestinal stem cells can produce Hopx+ +4 cells and vice versa (Figure 2).
Figure 2. Schematic of a System with Two Stem Cells.
Both the active and reserve stem cells are capable of self-renewal. In most situations, including normal tissue maintenance, only the active stem cell contributes to renewal of the tissue, dividing continuously and giving rise to an amplifying compartment and mature cells. When the active stem cell is lost, a normally dormant (label-retaining) reserve stem cell is activated and replenishes the active stem cell compartment.
Multiple distinct stem cell populations are also found in the skin. Here the classic LRC—once thought to be the only stem cell—resides in the bulge of the hair follicle. Normally quiescent, these cells can spring into action after treatment with phorbol esters (Cotsarelis et al., 1990). Although bulge LRCs clearly can act as stem cells, an independent population, labeled by Lgr6, cycles continuously and renews all skin lineages for the life of the animal (Jaks et al., 2010; Snippert et al., 2010a).
Evidence for facultative stem cells also exists in several other tissues, including those with slow turnover in the adult such as the liver (Dorrell et al., 2011; Duncan et al., 2009) or muscle (Tedesco et al., 2010). In the liver, replication of mature hepatocytes is responsible for tissue maintenance and also many injury responses (Michalopoulos, 2010). However, normally dormant facultative adult stem cells can be activated by certain injuries (Furuyama et al., 2011; Malato et al., 2011), proliferate, and give rise to multiple differentiated lineages until the tissue is restored. By definition, facultative stem cells are quiescent and thus it appears that the label retention methodology is particularly well suited to identify this kind of stem cell. Because the utilization of more than one progenitor cell compartment during normal tissue maintenance and injury has now been shown in multiple examples, it will be important to carefully evaluate multiple kinds of clinically relevant injury paradigms in each tissue. Different physiological and pathological stressors might very well give significantly different answers regarding the identity and molecular regulation of the resident stem cell.
In Vitro Clonogenic Assays: Faithful or Misleading?
When lineage tracing tools and/or transplantation assays are not available, clonal self-renewal in vitro coupled with in vitro differentiation can be used as a surrogate to identify putative stem/progenitor cells. If these assays are combined with cell fractionation methods such as FACS, it becomes possible to identify the most clonogenic cells and interrogate their properties, including the signals required for self-renewal. After clonogenic growth, differentiation protocols can be tested to determine the signals that govern maturation. Much progress has been made in this area for many different tissue stem cell systems. New surface markers have been developed and colony-forming assays now exist for most tissues. Because genetic lineage tracing is not possible in humans and cell transplantation assays are not available for many tissues, clonogenic growth is also used as an assay for human tissue stem cells, even when their murine counterparts are tractable by the other methods. In many tissues, for example the liver and intestine, there is excellent congruence between the clonogenic assays and lineage tracing (Dorrell et al., 2011; Sato et al., 2009). However, there are also several examples where in vitro assays produce different answers than the more definitive in vivo genetic lineage tracing. Normal prostate gland epithelium contains two main differentiated cell types: luminal columnar epithelium and basal cells below the luminal layer (Wang et al., 2009). In the mouse prostate, Lin−/Sca-1+/CD49f+ cells are most clonogenic in sphere-forming assays and most capable of forming prostate tissue in renal grafts (Lawson et al., 2007), indicating that prostate stem cells reside in the basal layer. Using similar assays, human prostate stem cells are also thought to be basal in nature (Garraway et al., 2010). Interestingly, however, genetic lineage tracing experiments using the transcription factor Nkx3.1 as a marker indicate that a completely distinct, rare luminal epithelial cell is responsible for regenerating the prostate after androgen deprivation (Wang et al., 2009). Once again, these findings suggest that there may be more than one type of stem cell in this tissue. However, it is also possible that the clonogenic basal cells are not stem cells in vivo at all and that the in vitro assay does not faithfully reflect tissue biology. Similarly, in vitro experiments had long suggested that the ducts of the adult pancreas might be able to produce endocrine β cells by neogenesis. Several publications agree that ducts are the progenitors for the acinar exocrine pancreatic cells (Criscimanna et al., 2011; Furuyama et al., 2011; Kopp et al., 2011b; Xu et al., 2008), but there is discordance in regards to the origin of β cells. When definitive genetic lineage tracing tools were used to specifically identify the progeny of pancreatic ducts in vivo (Furuyama et al., 2011; Kopp et al., 2011b), it was clearly shown that they do not ever produce new β cells, even after extensive injury such as duct ligation or β cell ablation (Kopp et al., 2011a). These cautionary tales illustrate that in vitro clonogenic assays can be misleading and that cells identified in this fashion need to be validated as true tissue stem cells by more definitive methodology.
Tissues with No Need for Stem Cells?
One of the most interesting recent findings is that some adult tissues do not appear to have a stem cell at all, which may explain their vulnerability to cell loss. Most notable in this regard is the endocrine pancreas. This tissue also exemplifies an interesting variation of the genetic lineage tracing methodology. If a specific stem cell marker is not available, one can determine the existence or assess the importance of stem cells by using a marker that is highly specific for their mature, differentiated progeny (Figure 1B). In the case of the endocrine pancreas such a marker is insulin, which is expressed only in β cells. Adult β cells can be genetically labeled and their fate, and that of their derivatives, followed over time (Dor et al., 2004; Malato et al., 2011). This kind of lineage tracing experiment allows asking the question whether unlabeled cells contribute to the lineage over an extended period. Emergence of unlabeled β cells would indicate the presence of a stem cell or progenitor. Unexpectedly, such experiments based on labeling of insulin+ cells showed that β cells did not derive from insulin− stem cells or progenitors in postnatal life during normal tissue maintenance (Dor et al., 2004). Instead, newly born β cells were generated only by division of pre-existing β cells. This observation has challenged the previous notion that the adult pancreas harbors β cell stem cells or progenitors. Similar experiments were done for the liver utilizing transthyretin as a marker of mature hepatocytes (Malato et al., 2011). The experiments showed that liver homeostasis in adult mice is achieved by hepatocyte replication similar to the β cell situation (Dor et al., 2004). Unlike the pancreas, however, specific injuries revealed the emergence of unlabeled hepatocytes, which must be derived from facultative stem cells or progenitors. Lineage tracing with mature markers nicely complements genetic marking studies using stem cell-specific genes. The use of both approaches in the same tissue increases the confidence in models of tissue stem cell behavior if the results are congruent (Dorrell et al., 2011; Furuyama et al., 2011; Iverson et al., 2011; Malato et al., 2011; Shin et al., 2011). Thus, the liver has now been firmly added to the list of organs that harbors an adult tissue stem cell. Interestingly, similar certainty is still lacking for some very important tissues. For example, there currently is no convincing evidence that the podocytes of the renal glomerulus have an adult progenitor.
Niches
Once a stem cell is identified and localized, it becomes possible to study the extrinsic regulatory signals produced by the local microenvironment, the niche (reviewed in Lander et al., 2012). This particular aspect of tissue stem cell biology is especially important for cancer. Many of the factors that impact healthy adult tissue stem cells also play a role in tumor self-renewal (Burness and Sipkins, 2010; Cabarcas et al., 2011). Each tissue and each stem cell uses different niche signals, but the Wnt/β-catenin pathway has received the most attention overall and has been found to play a key role in the intestine, skin (Haegebarth and Clevers, 2009), and breast (Tanos and Brisken, 2008). The full elucidation of interactions between niches and stem cells will undoubtedly be one of the main topics of tissue stem cell research in the future. Therapeutic opportunities abound in this area because molecules to block or activate extrinsic signaling pathways could be used to affect the behavior of particular stem cells, both in terms of ablation (cancer) and expansion (tissue repair). An intriguing application of the knowledge garnered from the study of the intestinal stem cell niche is the bacterial delivery of β-catenin-lowering transkingdom RNAi to lower the risk of cancer in familial adenomatosis of the colon (FAP) (Xiang et al., 2009).
miRNAs: Regulators of Stem Cell Quiescence and Self-Renewal
Our newfound ability to isolate and highly purify tissue stem cells in some organs has also opened up the study of intrinsic signals. In addition to transcription factors, microRNAs (miRNAs) have emerged as key players in determining the fate of tissue stem cells and their progeny. Improved methods for profiling miRNAs in very small numbers of cells (Smith and Murray, 2012) have facilitated identification of major changes in specific miRNAs as stem cells are activated and embark on differentiation (Yi and Fuchs, 2011). A single microRNA, miR-489, maintains satellite cells (the facultative stem cells of skeletal muscle) in quiescence and its downregulation results in satellite cell activation (Cheung et al., 2012). Conversely, miR-1 and miR-206 are upregulated when satellite cells differentiate and suppress Pax7 message, a transcription factor that is important for maintaining satellite cell quiescence (Chen et al., 2010). In addition to their important roles in somatic stem cell activation and differentiation, miRNAs can also be important for self-renewal. Recent examples include miR-205, which is highly expressed in mammary stem cells and can drive self-renewal when overexpressed (Greene et al., 2010; Ibarra et al., 2007). In the skin, inducible overexpression of miR-125b in adult stem cells produces an increased stem cell pool (Zhang et al., 2011). miRNAs also regulate the regenerative abilities of mature cells, such as recently shown for miR-21 in hepatocytes (Ng et al., 2012).
Conclusions and Future Directions
The past 5 years have seen the transition from fairly nonspecific methods for the identification of tissue stem cells such as label retention to more definitive methodology, especially genetic lineage tracing. The ability to prospectively isolate tissue stem cells with high purity and to visualize them within their niches is producing incisive insights into the intrinsic and extrinsic (niche) regulation of their behavior. Recent progress in intestinal stem cell biology has been particularly impressive and it can be hoped that other important organ systems such as the lung, liver, and kidney will follow suit in the near future.
ACKNOWLEDGMENTS
This work was supported by NIH grant DK51592 (M.G.)
REFERENCES
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Burness ML, Sipkins DA. The stem cell niche in health and malignancy. Semin. Cancer Biol. 2010;20:107–115. doi: 10.1016/j.semcancer.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Cabarcas SM, Mathews LA, Farrar WL. The cancer stem cell niche—there goes the neighborhood? Int. J. Cancer. 2011;129:2315–2327. doi: 10.1002/ijc.26312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang DZ. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RB, Anderson E, Howell A, Potten CS. Regulation of human breast epithelial stem cells. Cell Prolif. 2003;36(Suppl 1):45–58. doi: 10.1046/j.1365-2184.36.s.1.5.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Criscimanna A, Speicher JA, Houshmand G, Shiota C, Prasadan K, Ji B, Logsdon CD, Gittes GK, Esni F. Duct cells contribute to regeneration of endocrine and acinar cells following pancreatic damage in adult mice. Gastroenterology. 2011;141:1451–1462. doi: 10.1053/j.gastro.2011.07.003. 1462, e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Dorrell C, Erker L, Schug J, Kopp JL, Canaday PS, Fox AJ, Smirnova O, Duncan AW, Finegold MJ, Sander M, et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 2011;25:1193–1203. doi: 10.1101/gad.2029411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvillié B, Attali M, Aiello V, Quemeneur E, Scharfmann R. Label-retaining cells in the rat pancreas: location and differentiation potential in vitro. Diabetes. 2003;52:2035–2042. doi: 10.2337/diabetes.52.8.2035. [DOI] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Garraway IP, Sun W, Tran CP, Perner S, Zhang B, Goldstein AS, Hahm SA, Haider M, Head CS, Reiter RE, et al. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate. 2010;70:491–501. doi: 10.1002/pros.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 2010;7:300–304. doi: 10.4161/rna.7.3.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 2009;174:715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson SV, Comstock KM, Kundert JA, Schmidt EE. Contributions of new hepatocyte lineages to liver growth, maintenance, and regeneration in mice. Hepatology. 2011;54:655–663. doi: 10.1002/hep.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaks V, Kasper M, Toftgård R. The hair follicle-a stem cell zoo. Exp. Cell Res. 2010;316:1422–1428. doi: 10.1016/j.yexcr.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Hao E, Thorel F, Herrera PL, Sander M. Progenitor cell domains in the developing and adult pancreas. Cell Cycle. 2011a;10:1921–1927. doi: 10.4161/cc.10.12.16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011b;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Lander AD, Kimble J, Clevers H, Fuchs E, Montarras D, Buckingham M, Calof AL, Trumpp A, Oskarsson T. What does the concept of the stem cell niche really mean today? BMC Biol. 2012;10:19. doi: 10.1186/1741-7007-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IC, Bickenbach JR. Label-retaining keratinocytes and Langerhans cells in mouse epithelia. Cell Tissue Res. 1985;242:551–556. doi: 10.1007/BF00225420. [DOI] [PubMed] [Google Scholar]
- Malato Y, Naqvi S, Schürmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Invest. 2011;121:4850–4860. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am. J. Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Potten CS. Slowly cycling (label-retaining) epidermal cells behave like clonogenic stem cells in vitro. Cell Prolif. 1994;27:279–289. doi: 10.1111/j.1365-2184.1994.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J. Clin. Invest. 2012;122:1097–1108. doi: 10.1172/JCI46039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Hendry JH. Differential regeneration of intestinal proliferative cells and cryptogenic cells after irradiation. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1975;27:413–424. doi: 10.1080/09553007514550411. [DOI] [PubMed] [Google Scholar]
- Potten CS, Gandara R, Mahida YR, Loeffler M, Wright NA. The stem cells of small intestinal crypts: where are they? Cell Prolif. 2009;42:731–750. doi: 10.1111/j.1365-2184.2009.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, AsselinLabat ML, Wu L, Lindeman GJ, Visvader JE. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shin S, Walton G, Aoki R, Brondell K, Schug J, Fox A, Smirnova O, Dorrell C, Erker L, Chu AS, et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Murray DW. An overview of microRNA methods: expression profiling and target identification. Methods Mol. Biol. 2012;823:119–138. doi: 10.1007/978-1-60327-216-2_9. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010a;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010b;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos T, Brisken C. What signals operate in the mammary niche? Breast Dis. 2008;29:69–82. doi: 10.3233/bd-2008-29108. [DOI] [PubMed] [Google Scholar]
- Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J. Clin. Invest. 2010;120:11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terskikh VV, Vasiliev AV, Vorotelyak EA. Label retaining cells and cutaneous stem cells. Stem Cell Rev. 2011 doi: 10.1007/s12015-011-9299-6. in press. Published online July 9, 2011. 10.1007/s12015-011-9299-6. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooy D, Weiss S. Why stem cells? Science. 2000;287:1439–1441. doi: 10.1126/science.287.5457.1439. [DOI] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- Xiang S, Keates AC, Fruehauf J, Yang Y, Guo H, Nguyen T, Li CJ. In vitro and in vivo gene silencing by TransKingdom RNAi (tkRNAi) Methods Mol. Biol. 2009;487:147–160. doi: 10.1007/978-1-60327-547-7_7. [DOI] [PubMed] [Google Scholar]
- Xu X, D’Hoker J, Stangé G, Bonné S, De Leu N, Xiao X, Van de Casteele M, Mellitzer G, Ling Z, Pipeleers D, et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Fuchs E. MicroRNAs and their roles in mammalian stem cells. J. Cell Sci. 2011;124:1775–1783. doi: 10.1242/jcs.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Stokes N, Polak L, Fuchs E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell. 2011;8:294–308. doi: 10.1016/j.stem.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]