Abstract
Cell replacement is an emerging therapy for type 1 diabetes. Pluripotent stem cells have received a lot of attention as a potential source of transplantable β-cells, but their ability to form teratomas poses significant risks. Here, we evaluated the potential of primary mouse gall bladder epithelial cells (GBCs) as targets for ex vivo genetic reprogramming to the β-cell fate. Conditions for robust expansion and genetic transduction of primary GBCs by adenoviral vectors were developed. Using a GFP reporter for insulin, conditions for reprogramming were then optimized. Global expression analysis by RNA-sequencing was used to quantitatively compare reprogrammed GBCs (rGBCs) to true β-cells, revealing both similarities and differences. Adenoviral-mediated expression of NEUROG3, Pdx1, and MafA in GBCs resulted in robust induction of pancreatic endocrine genes, including Ins1, Ins2, Neurod1, Nkx2-2 and Isl1. Furthermore, expression of GBC-specific genes was repressed, including Sox17 and Hes1. Reprogramming was also enhanced by addition of retinoic acid and inhibition of Notch signaling. Importantly, rGBCs were able to engraft long term in vivo and remained insulin-positive for 15 weeks. We conclude that GBCs are a viable source for autologous cell replacement in diabetes, but that complete reprogramming will require further manipulations.
Keywords: gall bladder, type 1 diabetes, beta cells, insulin, reprogramming
Section 1: INTRODUCTION
1.1. The initial success of the Edmonton protocol highlighted the potential of cell replacement therapy in type 1 diabetes (Shapiro et al., 2000). However, wider application of this approach is severely limited by the shortage of transplantable pancreatic β-cells (de Kort, de Koning, Rabelink, Bruijn, & Bajema, 2011). In addition, the transplantation of cadaveric islets requires life-long immune suppression (Ricordi & Strom, 2004). Ideally, therefore, a source of transplantable β-cells would be both autologous and abundant. Because of their in vitro growth capacity, pluripotent stem cells (PSCs) are an attractive potential source of transplantable β-cells. While significant progress has been made, the generation of true β-cells in vitro has remained elusive thus far (Alipio et al., 2010; D'Amour et al., 2006; Nostro et al., 2011). In addition, PSCs are capable of forming teratomas and represent an unknown risk in terms of tumorigenesis (Fujikawa et al., 2005; Kroon et al., 2008).
1.2. Direct genetic reprogramming of postnatal primary cells by forced expression of key developmental transcription factors has emerged as an alternative to in vitro differentiation of PSCs. This strategy has been used successfully to produce functional cells, including neurons, hepatocyte-like cells and cardiomyocytes from fibroblasts (Huang et al., 2011; Ieda et al., 2010; Vierbuchen et al., 2010). Similarly, in vivo reprogramming of hepatic cells by expressing different pancreatic transcription factors, including Neurog3 and Pdx1, was able to restore euglycemia in hyperglycemic mice (Ferber et al., 2000; Wang, Ehrhardt, Xu, & Kay, 2007; Vijay Yechoor et al., 2009). Additionally, in vivo reprogramming of exocrine acinar cells into insulin-positive cells by expression of Neurog3, Pdx1 and MafA was also able to reverse hyperglycemia in mice (Zhou, Brown, Kanarek, Rajagopal, & Melton, 2008). Furthermore, overexpression of Neurog3 and Pdx1 has been shown to enhance pancreatic differentiation of embryonic stem cells (Kubo et al., 2011). Other cell types have also been tested for amenability to reprogramming towards the β-cell fate, including adipose tissue-derived stem cells, placenta-derived multipotent stem cells, hepatocytes, intrahepatic biliary epithelial cells and gall bladder cells (Chandra et al., 2011; Chiou et al., 2010; Coad, Dutton, Tosh, & Slack, 2009; Motoyama et al., 2009; Nagaya, Katsuta, Kaneto, Bonner-Weir, & Weir, 2009; Shigeru et al., 2007).
1.3. The extrahepatic biliary tissue, including the gallbladder, is a particularly appealing source of cells for reprogramming to the pancreatic fate. The extrahepatobiliary system shares a common developmental origin with the ventral pancreas, from a cell termed the pancreatobiliary progenitor (Spence et al., 2009). Segregation of these distinct lineages is partly regulated by the Notch effector Hes1. Recently it was demonstrated that inhibition of Hes1 in cultured gall bladder cells (GBCs) was sufficient to induce some insulin expression (Coad, et al., 2009). While this work highlighted the potential of GBCs as a source of transplantable β-cells, the full spectrum of β-cell expressed genes or the in vivo functionality of these cells were not determined. Moreover, the cells showed only limited proliferative potential under the culture conditions used. For GBCs to be a viable substrate of future β-cell replacement therapies, they would have to be robustly expandable (V. Yechoor & Chan, 2010). Therefore, the true utility of GBCs as a source of transplantable β-cells remains unknown.
1.4. In this study, we investigated if mouse GBCs significantly expanded in vitro can still be reprogrammed towards the β-cell fate by using a combination of positive instructive signals as well as Notch inhibition. GBCs were transduced with adenoviruses expressing the transcription factors NEUROG3, Pdx1 and MafA and treated with retinoic acid and Notch inhibitors, resulting in their differentiation into islet-like cells. Reprogrammed cells had the ability to engraft, survive and remain insulin-positive up to 15 weeks post-transplantation. However, there were also differences between the reprogrammed GBCs and true β-cells. Our findings confirm that the gall bladder represents a promising source of autologous reprogrammable cells for the treatment of type 1 diabetes mellitus.
Section 2: MATERIALS & METHODS
2.1. Mouse gall bladder cell isolation and culture
Gall bladders from C57Bl6/6J-MIP-GFP male and female mice between the ages of 4–8 weeks were removed by a surgical incision and bile released by making a single cut in the wall. Gall bladders were rinsed twice in DPBS (Life Technologies, Grand Island, Ca) and then cut into several pieces. This material was then incubated at 37°C with 0.25% Trypsin/EDTA (Life Technologies, Grand Island, Ca) for 45 minutes to obtain a cell suspension. Cells were cultured using a modified protocol to that previously described (Manohar et al., 2011). Briefly, cells were plated on a 70–80% confluent irradiated LA7 rat epithelial feeder layer that had been previously irradiated at 60 Gy. Cells were cultured in DMEM/F12 (Life Technologies, Grand Island, Ca) supplemented with 0.5% FBS (Thermo Fisher Scientific, Cambridge, MA), 1% insulin-transferrin-selenium (Roche, Indianapolis, IN), 15mM HEPES (Thermo Fisher Scientific, Cambridge, MA) and antimicrobials (100U/ml penicillin, 100µg/ml streptomycin and 0.25µg/ml Amphotericin B; Cellgro, Manassas, VA) in a 37°C incubator with 5% CO2. Media was changed every two to three days. When GBCs were 70–90% confluent, they were passaged by incubation with 0.05% trypsin/EDTA (Life Technologies, Grand Island, Ca) at 37°C, followed by incubation with DNaseI at 37°C for 10 minutes to obtain a single cell suspension.
2.2. Fibroblast culture
For initiating fibroblast cultures, mouse tail-tips from euthanized C57Bl6/6J-MIP -GFP mice were washed with DPBS, cut into several pieces, and digested with 0.25% Trypsin/EDTA for 60 minutes at 37°C with regular mixing. Upon inactivation of trypsin by addition of serum, the tissue mix was spun at 1000 rpm for 5 mins, resuspended in DMEM supplemented with 15% FBS and antimicrobials, followed by plating in a 37°C incubator with 5% CO2. Media was changed every two to three days.
2.3. Adenovirus transduction of GBCs
Each E1-deleted adenovirus (serotype 5) consisted of the full-length cDNA (human NEUROG, rat Pdx1, mouse MafA; NEUROG3 and Pdx1 provided by Michael German, University of California at San Francisco; MafA provided by Roland Stein, Vanderbilt University Medical Center) driven by the CMV promoter. For additional amplification, each virus was expanded in HEK293 cells and purified using the FastTrap Purification Kit (Millipore, Billerica, MA) per the manufacturer’s protocol. Adenovirus titers were calculated based on spectrometry. Adenoviruses expressing NEUROG3 (MOI 1000), Pdx1 (MOI 500), and MafA (MOI 500) were incubated in 100 µg/ml DEAE-Dextran (Sigma-Aldrich, St. Louis, MO) for 30 minutes at room temperature with regular mixing prior to addition to the cell media for a final concentration of 10 µg/ml DEAE-Dextran. Media was changed 20–24 hours post transduction.
2.4. GBC reprogramming
GBC cultures were grown to ~70% confluency. On this day, day 0, cells were transduced with the adenovirus/DEAE-Dextran mix. 20–24 hours later, the media was changed to include 2 µM retinoic acid (Sigma-Aldrich, St. Louis, MO) and 1% DMSO (Thermo Fisher Scientific, Cambridge, MA). Twenty four hours later, the media was changed to include 250 nM of the γ-secretase inhibitor dibenzazepine (DBZ; EMD Chemicals, Darmstadt, Germany) and 1% DMSO. All cells, attached and in suspension, were kept by spinning the media to pellet all cells prior to addition of new media.
2.5. Flow cytometry and FACS
For antibody labeling, GBCs were dissociated into single cells using a non-trypsin method (Cell dissociation buffer; Gibco, Grand Island, CA). Dissociated cells were resuspended in DMEM, supplemented with 2% FBS and 0.25 mg/ml DNaseI (Sigma-Aldrich, St. Louis, MO). Propidium iodide staining was used to label dead cells for exclusion. The forward scatter (FSC):pulse width gating excluded cell doublets from sorts, as previously described (Dorrell et al., 2008). Cells were analyzed with a FACScalibur or sorted by an inFluxV-GS (BD Biosciences, San Jose, CA for both) at 15 psi using a 100 µm nozzle. Data were analyzed using FlowJo (Treestar, Ashland, OR).
2.6. RNA isolation and qRT-PCR
For RNA isolation, cells were either directly FACS-sorted into Trizol Liquid Sample (Life Technologies, Grand Island, CA) or trypsinized and pelleted by centrifugation prior to cell lysis with Trizol. RNA was purified using RNeasy (Qiagen, Valencia, CA) per the manufacturer’s protocol. First strand cDNA synthesis was completed using MMLV reverse transcriptase and random oligonucleotide primers (Life Technologies, Grand Island, CA). Relative mRNA expression levels were determined by qRT-PCR using a BioRad iCycler with a single color MyiQ detection system. All reactions were performed with Platinum Taq DNA Polymerase (Life Technologies, Grand Island, CA) and SYBR Green using 45 cycles of 95°C for 15s, 68°C for 20s, and 72°C for 20s. The full list of mouse-specific primers is given in Table S4. Results were analyzed using gene expression relative to control Alas1 or Actb gene expression (ΔΔCT). qPCR data were expressed as mean fold change (2ΔΔct) ± 95% confidence.
2.7. Immunohistochemistry and immunofluorescent imaging
For cytospin imaging, either reprogrammed GFP+ cells or non-adenovirus transduced control cells were spun at 1000rpm for 5 mins onto Superfrost Plus slides (Thermo Fisher Scientific, Cambridge, MA). Cells were fixed in either 4% paraformaldehyde or 90% methanol at 4°C for 10 mins. Prior to labeling, cells were blocked in 5% BSA for 60 mins at 23°C. Primary labeling was performed overnight at 4°C in PBS supplemented with 2% BSA and 0.05% Triton-X using rabbit polyclonal antibodies against insulin (H-86; Santa Cruz Biotechnology, Santa Cruz, CA), C-peptide (BCBC collection #1042), Neurod1 (16508; Abcam, Cambridge, MA) and Somatostatin (A0566; DAKO, Carpinteria, CA). Secondary labeling was performed for 60 mins at 23°C in PBS supplemented with 2% BSA and 0.05% Triton-X with a 1:200 dilution of Alexa 555-conjugated goat anti-rabbit IgG (Cell Signaling, Danvers, MA). Nuclei were stained using Hoechst 33342 (Sigma-Aldrich, St. Louis, MO). For IHC analysis, formalin-fixed paraffin-embedded kidneys were sectioned and labeled with a primary antibody against insulin (H-86; Santa Cruz Biotechnology, Santa Cruz, CA) and detected using previously described methods (Overturf et al., 1996).
2.8. Enzyme-linked immunosorbent assay
Control and rGBCs were harvested at the indicated time points. Cells were washed twice in Krebs Ringer Buffer supplemented with 2.8 mM glucose (KRB-2.8) and incubated at 37°C for 90 minutes in KRB. Following two further washes in KRB-2.8, cells were incubated in either KRB-2.8 or Krebs Ringer Buffer supplemented with 16.7 mM glucose (KRB-16.7) for 60 minutes at 37°C. Cells were spun at 1200 rpm for 5 mins and the media collected and stored at −80°C. Insulin quantitative analyses were performed per the manufacturer’s instructions (Insulin Ultrasensitive ELISA, Alpco, Salem, NH).
2.9. Cell transplantation
GBCs were expanded to 70% confluency prior to reprogramming at the indicated passage number (Table S3). Cells were transduced with the three adenoviruses expressing NEUROG3, Pdx1 and MafA with or without retinoic acid as described in Table S1. One to three days later, cells were harvested and shipped on ice to the University of Massachusetts Medical School (Worcester, MA) for transplantation on the following day. Cell viability was determined by propidium iodide staining after shipping and prior to transplantation. 20×106 unsorted cells were transplanted under the renal capsule of diabetic NOD.Cg-Rag1tm1Mom IL2rgtm1WjlIns2Akita (NRG-Akita) and NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ Ins2Akita (NSG-Akita) mice, as previously described (Brehm et al., 2010). Non-fasting blood glucose levels were monitored following transplantation by blood glucose measurements with an ACCUCHEK active glucometer (Hoffman-LaRoche, Basel, Switzerland). The Institutional Animal Care and Use Committee approved all animal studies.
2.10. RNA-SEQ analysis
Libraries were single-read sequenced to 100bp on an Illumina hiSeq2000. Reads coming from ribosomal RNA and repeats were filtered out by aligning 50bp of all reads to mouse ribosomal sequence and the human contents of Repbase (release 14.10)(Jurka et al., 2005). The remaining reads where processed with RUM(Grant et al., 2011) using the default parameters to align reads to genome (mm9) and gene models and to produce transcript-level quantification in reads. Expression levels (in reads) were log2 transformed and then normalized using quantile normalization (R limma). After unclogging, data were converted to reads and averaged between replicates. Differential expression was determined using the Fisher exact test and Benjamini-Hochberg correction for multiple testing on genes with at least a two-fold difference between conditions. Gene lists from either the up- or down-regulated portion of each comparison were analyzed using Ingenuity Core Analysis (Ingenuity Systems, www.ingenuity.com). The RNA-Seq data was submitted to the European Bioinformatics Institute ArrayExpress (http://www.ebi.ac.uk/arrayexpress/).
2.11. Statistical Analysis
Statistical analyses were conducted with GraphPad Prism software v.4.0 or Microsoft Excel. Experimental differences were evaluated by student two-tailed t-test assuming equal variance. P values <0.05 were considered statistically significant.
Section 3: RESULTS
3.1. Expansion and transduction of mouse gall bladder cells
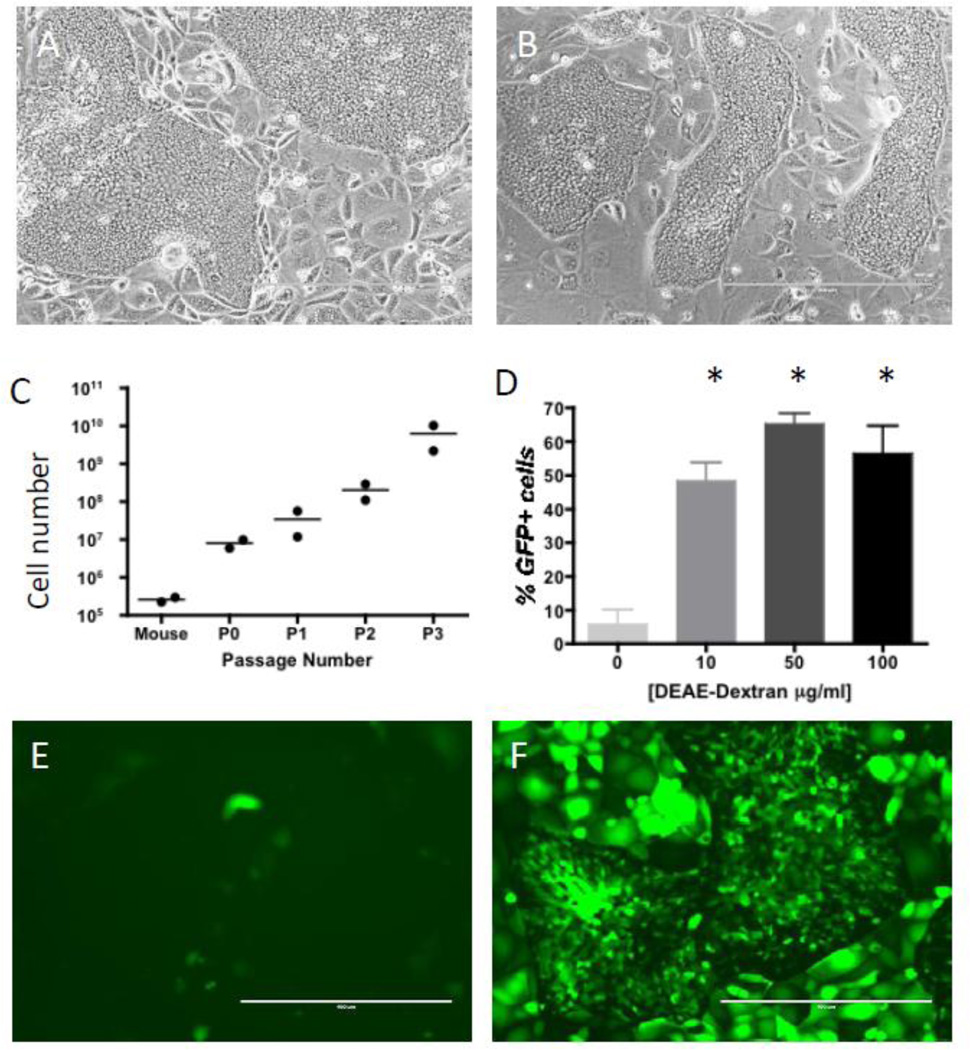
Cell replacement therapy for type 1 diabetes requires large numbers of functional cells. Therefore, the expandability of primary GBCs had to be demonstrated in order for the adult gall bladder to be a viable alternative to pluripotent stem cells (PSCs), which can be readily grown in vitro. A modified protocol first reported by Manohar et al. was used to isolate and expand mouse GBCs (Manohar, et al., 2011). GBCs were cultured on a feeder layer of irradiated LA7 rat epithelial cells (Dulbecco, Bologna, & Unger, 1979) that allowed their robust expansion. Upon initial harvest a typical mouse gall bladder yielded approximately 200–400,000 cells that showed unlimited ability to replicate and expand, normally generating hundreds of millions of cells by passage 3 (Figs. 1A–C). Expanded cells also maintained their epithelial characteristics and there was no evidence of epithelial-mesenchymal transition based on expression of known mesenchymal markers (Kalluri & Weinberg, 2009), including Acta2, Col1a1, Ddr2, S100a4 and Vim (Table S1). Next, the suitability of adenoviral vectors for the introduction of transgenes in expanded GBCs was assessed. Less than 10% of the cells were positive for the marker transgene GFP when using standard transduction protocols (Figs. 1D,E). DEAE-dextran has previously been shown to enhance the transduction of other epithelial cells by adenoviral vectors (Gregory et al., 2003; Kaplan et al., 1998). Concentrations greater than 10 µg/ml allowed transduction of more than 50% of the GBCs (Figs. 1D,F). Although higher concentrations of DEAE-Dextran gave even greater transduction efficiencies, they were also associated with increased cell mortality (data not shown). Therefore, for all the reprogramming experiments, a final DEAE-Dextran concentration of 10 µg/ml was used for adenovirus transduction of GBCs.
Figure 1. Expansion and adenovirus transduction of GBCs.
(A,B) Phase contrast images of passage 0 (A) and passage 3 (B) typical GBC colonies. (C) GBCs were robustly expanded, generating hundreds of millions of cells by passage 3 (P3). The data shown is for two independent gall bladders. (D) Without any DEAE-Dextran, only ~ 5% of cells were transduced by adenovirus expressing GFP at 500 MOI. However, with increasing concentrations of DEAE-Dextran, there was a significant increase in the number of GBCs transduced (* P<0.05; N=3). (E,F) Fluorescent images showing (E) few GBCs GFP+ without DEAE-Dextran addition, whereas at 10 µg/ml (F), ~70% of the GBCs were GFP+. Scale bar for all images is 400 µm.
3.2. NEUROG3, Pdx1 and MafA are required for optimal expression of both Ins1 and Ins2
In order to determine the optimal combination of reprogramming transcription factors, a mouse reporter strain (MIP-GFP (Hara et al., 2003)) in which the mouse Ins1 promoter drives expression of GFP was utilized. Multiple pancreatic developmental transcription factors were tested singly and in combinations, including Pdx1 (Jonsson, Carlsson, Edlund, & Edlund, 1994), NEUROG3 (Apelqvist et al., 1999), MafA (Nishimura et al., 2006), and Nkx6-1 (Sander et al., 2000). In control GBCs, no GFP expression was detected, indicating absence of transcriptional activity from the Ins1 promoter (Fig. 2). Previous work has shown that forced expression of Neurog3 or Pdx1 is sufficient to differentiate liver cells towards a pancreatic fate in vivo (Ferber, et al., 2000; Wang, et al., 2007). Although expression of these two factors together induced low levels of GFP expression, adenoviral-mediated expression of MafA, together with NEUROG3 and Pdx1, was required for optimal GFP expression in GBCs in vitro (Fig. 2A). Additional transduction of GBCs with Nkx 6-1 as a fourth factor did not yield increased reprogramming (data not shown). A second reporter strain was also generated by breeding a mouse expressing Cre recombinase under control of the rat Ins2 promoter with a dual reporter Tomato-RFP/GFP mouse in which expression of GFP is only detected in cells in which a Cre-mediated recombination event has occurred (Muzumdar, Tasic, Miyamichi, Li, & Luo, 2007; Postic et al., 1999). Control cultured GBCs were always GFP negative, whereas GBCs reprogrammed with NEUROG3, Pdx1 and MafA (NPM) activated the Ins2 promoter (Fig. S1). These results indicated that both insulin promoters were active in reprogrammed cells. For all subsequent experiments, GBCs expanded from MIP-GFP mice (Ins1 promoter) were used.
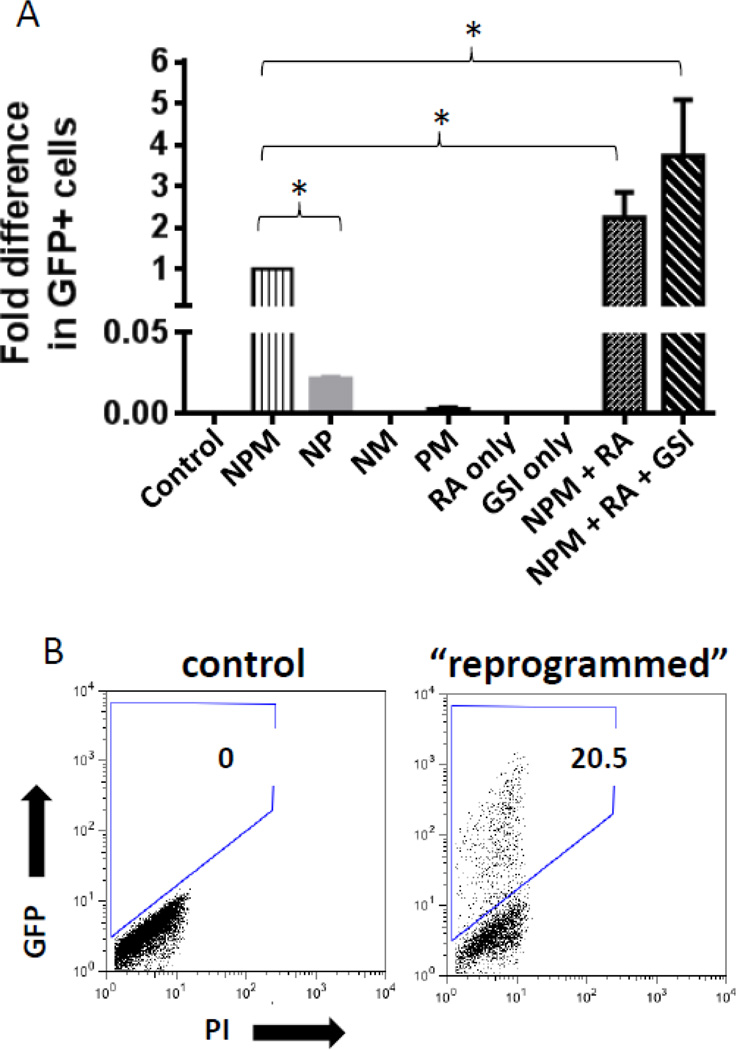
Figure 2. Optimization of GBC reprogramming.
(A) Comparison of the percentage of GFP (=insulin1) expressing GBCs after reprogramming. GFP+ percentage after reprogramming with the three transcription factors - NEUROG3, Pdx1 and MafA (NPM) was arbitrarily set to 1. The combination of all three factors yielded significantly higher reprogramming rates compared to any combination of two factors (* P<0.05; N=3). RA increased the percentage of GFP+ GBCs in cultures by approximately 2.2 fold (* P<0.05; N=3). Addition of DBZ to the reprogramming mix on day 2 also increased the number of GFP+ GBCs. Addition of RA or GSI alone had no effect on reprogramming. (B) GBCs from MIP-GFP mice were GFP-negative by flow cytometry, indicating no activity of the Ins1 promoter. Three days after reprogramming, 20.5% of total GBCs in the plate became GFP+ using the optimized protocol depicted in Fig. S2.
3.3. Retinoic acid and Notch inhibition enhance reprogramming of GBCs
Small molecules have previously been successively used for reprogramming PSCs towards a pancreatic lineage (Kroon, et al., 2008; Nostro, et al., 2011). The effect of retinoic acid (RA) on reprogramming was first tested. By including RA in the reprogramming media at a concentration of 2 µM, there was a significant 2.2 fold increase in the percentage of GFP+ GBCs reprogrammed with NEUROG3, Pdx1 and MafA (NPM), compared to GBCs reprogrammed with NPM alone (Fig. 2A). The effect of inhibiting Notch signaling by using the gamma secretase inhibitor dibenzazepine (DBZ) was examined next (van Es et al., 2005). Inhibition of Notch signaling at day 2 of reprogramming caused a further significant increase in GFP+ reprogrammed cells (Fig. 2A). RA or DBZ in the absence of NPM did not cause any GFP+ cells to appear. The optimized reprogramming timeline is shown in Fig. S2 and a representative FACS plot of GFP expression after reprogramming is shown in Fig. 2B. As the cells were reprogramed towards the pancreatic fate, the morphology of the cells changed and the GBCs lost their distinctive tight colony structure, eventually loosing adherence and floating in suspension in the culture. We next analyzed the expression of GFP over a period of 12 days after reprogramming in two different gall bladder samples (Fig. S3). The first GFP+ cells appeared within 48 hours after reprogramming was begun, with the maximum number of GFP+ cells detected 24 hours later. After this time, the percentage of GFP+ cells decreased, coinciding with increased cell death under the culture conditions used (data not shown).
3.4. GBCs are rapidly reprogrammed towards the β-cell-fate
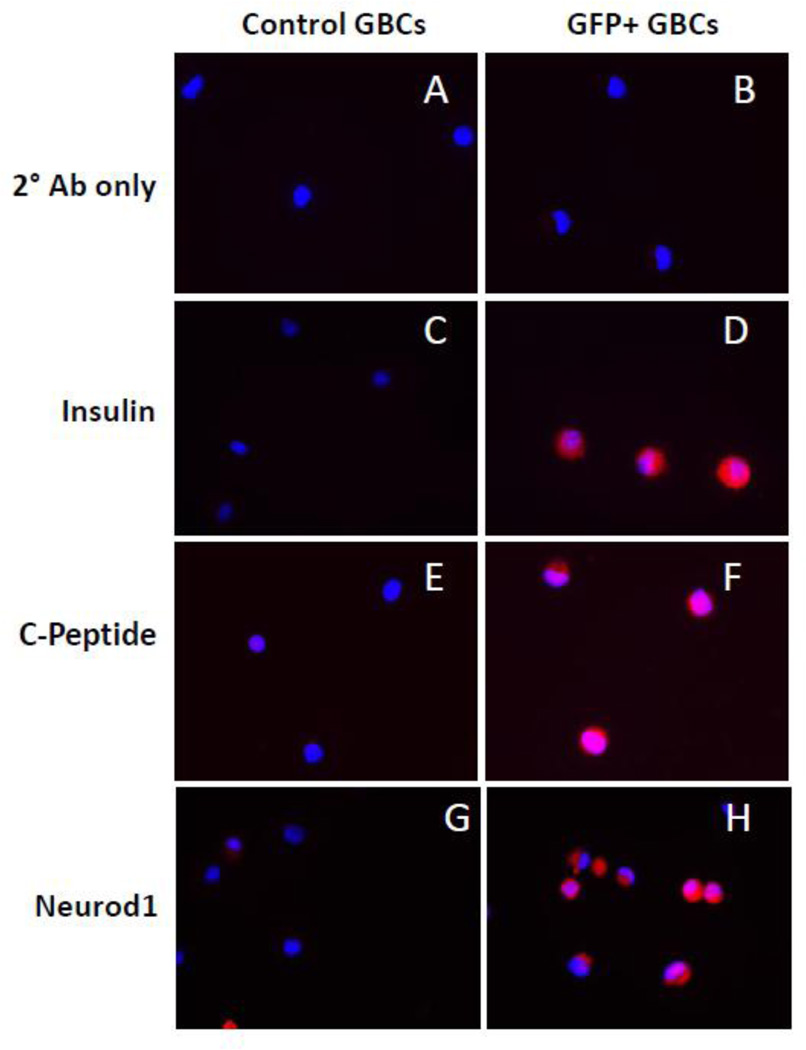
Four days after reprogramming, reprogrammed GFP+ GBCs were analyzed by flow cytometry and FACS-sorted for RNA and protein analyses. Adenovirus GFP-only transduced GBCs were used as the control. Using RNA-Seq, true reprogramming of these GBCs was evident by induction of genes involved in several aspects of β-cell function (Table 1) including proinsulin production (Ins1, Ins2), insulin processing (Pcsk1, Pcsk2, Cpe), transcription factors (Nkx2-2, Neurod1, Isl1), glucose metabolism (Gck), ATP-sensitive K+ channels (Kcnj11), calcium channels (Cacna1a) and insulin secretion (Chga, Scg3). RNA-Seq data was validated for several genes by qRT-PCR (Fig. S4A,B). In accordance with the gene expression data, rGBCs also expressed the proteins for insulin, c-peptide, and Neurod1 (Fig. 3). In fact, all GFP+ FACS-sorted cells were positive for these three proteins, indicating that the MIP-GFP reporter system was an excellent read-out for GBC-derived islet-like cells (Fig. 3). These results clearly demonstrated the activation of many genes specific for pancreatic β-cells. However, complete reprogramming involves not only the activation of desired genes, but also the silencing of genes specific for the cell type of origin. Within three days post reprogramming, GFP+ GBCs showed significant decreases in expression of several genes that are normally expressed in this cell type (Table S2). In addition, the pancreatobiliary transcription factors Sox17 and Hes1, required for normal gall bladder development and homeostasis, were also reduced. The RNA-Seq data for these genes was also validated by PCR (Fig. S4B).
Table 1.
Expression levels of β-cell-related genes in rGBCs relative to control GBCs. FC, fold change.
| Gene | Transcript | Description | FC | p value |
|---|---|---|---|---|
| Ins1 | NM 008386 | insulin I | 4837.3 | 4.9E-04 |
| Ins2 | NM_001185083 | Insulin II proprotein convertase subtilisin/kexin | 7082.3 | 5.5E-06 |
| Pcsk1 | NM_013628 | type 1 | 16.6 | 1.9E-01 |
| Pcsk2 | proprotein convertase subtilisin/kexin | |||
| NM 008792 | type 2 | 687.4 | 8.6E-02 | |
| Cpe | NM 013494 | carboxypeptidase E | 11.0 | 5.2E-03 |
| Nkx2-2 | NM 010919 | NK2 transcription factor related, locus 2 | 11625.5 | 9.9E-04 |
| Neurod1 | NM_010894 | neurogenic differentiation 1 | 40063.5 | 2.3E-04 |
| Isl1 | ISL1 transcription factor, | |||
| NM 021459 | LIM/homeodomain | 2749.6 | 1.6E-03 | |
| Pdx1 | NM 008814 | pancreatic and duodenal homeobox 1 | 8.5 | 7.1E-03 |
| Gck | NM_010292 | Glucokinase | 28.3 | 4.1E-02 |
| Kcnj11 | NM_010602 | potassium inwardly rectifying channel, subfamily J, member 11 | 10333.2 | 3.1E-04 |
| Cacna1a | NM_007578 | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | 24.7 | 1.5E-04 |
| Slc2a2 | NM_031197 | solute carrier family 2 (facilitated glucose transporter), member 2 | 60.5 | 5.1E-03 |
| Chga | NM 007693 | chromogranin A | 2157.3 | 1.0E-03 |
| Scg3 | NM_001164790 | secretogranin III | 52681.2 | 1.7E-05 |
Figure 3. Characterization of protein expression changes in reprogrammed GBCs.
Control or GFP+ GBCs were stained with no antibody (A,B) or with antibodies against Insulin (C,D), C-peptide (E,F), or Neurod1 (G,H). Representative images are shown for each cell type.
3.5. The pancreatic endocrine phenotype of rGBCs is polyhormonal and non-glucose-responsive
Pancreatic endocrine cells produced by in vitro differentiation of PSCs are mostly polyhormonal (D'Amour, et al., 2006; Nostro, et al., 2011). In order to determine whether genetic reprogramming of GBCs resulted in the emergence of pancreatic cell types other than β-cells, qPCR analysis of mRNA expression was done initially on non-FACS sorted cells. In addition to insulin, other endocrine hormones were also produced, including Sst, Ppy, and Ghrl gene transcripts (Fig. S5A). Expression levels of the exocrine genes Cela2a (chymotrypsin-like elastase family, member 2A), Cpa2 (pancreatic carboxypeptidase A2) and Prss1 (trypsin 1) were also assessed (Fig. S5B). While Cela2a mRNA was not detected in reprogrammed cells, and there was no significant increase in Prss1 expression between control and transduced cells, there was a significant increase in expression of Cpa2. However, the overall expression of Cpa2 in rGBCs was less than 0.001% of that detected in normal pancreas. In order to determine if rGBCs were polyhormonal, insulin-positive GFP+ GBCs were immunostained for somatostatin. Indicative of a polyhormonal phenotype, insulin-positive cells were also somatostatin positive (Fig. S5C). RNA-Seq data from FACS-sorted MIP-GFP+ cells confirmed that insulin-positive cells also expressed Sst at levels greater than 20,000 higher than control GBCs (p<0.01).
When PSCs are differentiated toward the pancreatic endocrine fate in vitro they are often reported to be capable of tonic insulin secretion, but do not display increased secretion in response to glucose (Kubo, et al., 2011; Nostro, et al., 2011). In order to determine whether reprogrammed GBCs are superior to PSC derivatives in vitro, insulin secretion was measured after exposure to glucose. Similar to cells produced by in vitro differentiation of PSCs, rGBCs were able to secrete insulin, as detected using an insulin-specific ELISA (Fig. S6). However, the amount of insulin detected was not significantly different after stimulation with higher glucose concentrations. Hence, like their PSC derived counterparts rGBCs also did not display GSIS, at least at early time points after reprogramming.
3.6. RNA-SEQ analysis revealed only partial reprogramming into mature β-cells
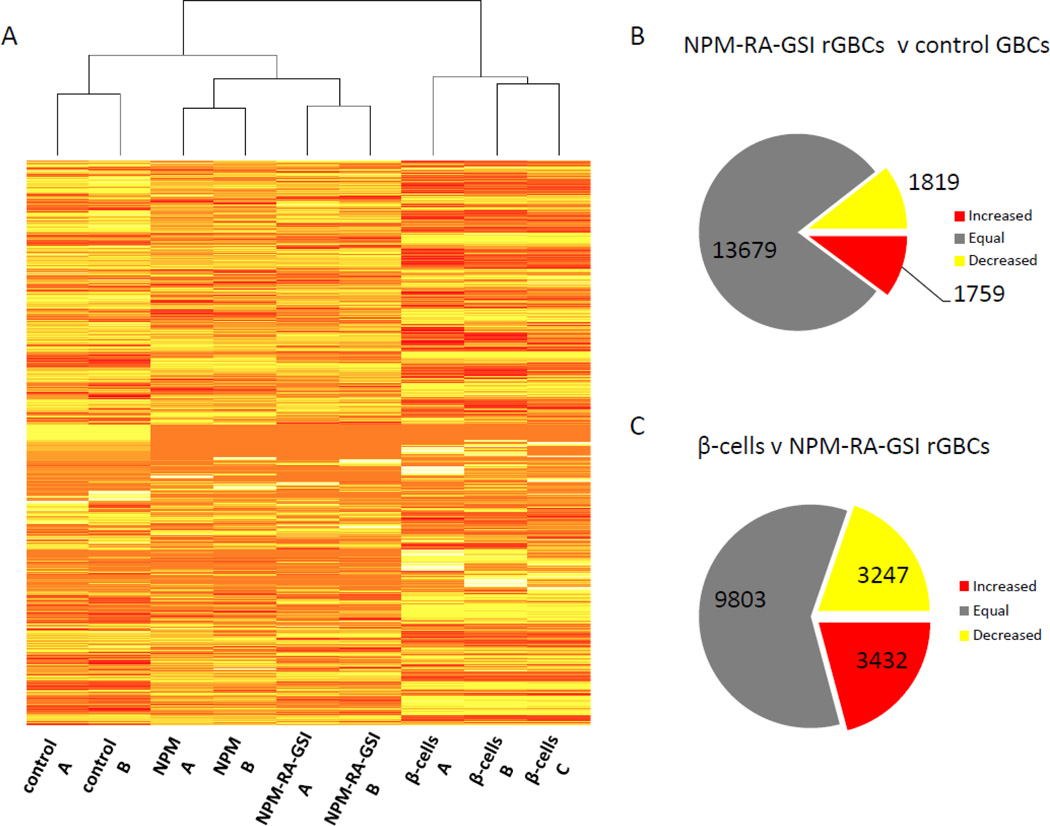
Both in vitro differentiated PSCs and the genetically reprogrammed GBCs described here display polyhormonal gene expression and lack of glucose responsiveness. However, the molecular mechanism(s) underlying this imperfect β-cell phenotype are currently unknown. In order to better understand the key differences between rGBCs and true β-cells and to uncover new targets for future reprogramming strategies, we assessed the complete mRNA expression profile of rGBCs. RNA-Sequencing technology and bioinformatics were utilized to compare the transcriptomes of adenovirus GFP-only transduced control GBCs, NPM rGBCs (without RA and DBZ), NPM-RA-GSI GBCs (with RA and DBZ) and β-cells. The heatmap depicted in Fig. 4A illustrates that while several groups of genes were expressed in similar fashion between mature β-cells and rGBCs, others were not. Differential expression analysis between control GBCs, NPM-RA-GSI GBCs and β-cells yielded a large number of differentially expressed genes as shown in Fig. 4B,C. Approximately 1,800 genes were up or down regulated with reprogramming after four days. However, a further 3,200 to 3,400 genes remained differentially expressed between β-cells and rGBCs. A functional analysis of the 1,759 genes that were up-regulated in rGBCs indicates that a statistically significant portion were associated with ‘neuronal functions’, e.g. ‘neurotransmission’ (p= 1.97e-6) and ‘morphology of neurons’ (p= 1.7e-1). Since the development of the endocrine pancreas and nervous systems shares many common factors (Atouf, Czernichow, & Scharfmann, 1997), this finding was consistent with the induction of pancreatic endocrine development. However the transcriptional signature of a more mature β-cell phenotype was also observable in that the functions ‘quantity of carbohydrate’ (p= 1.8e-2; 73 genes) and ‘quantity of hormone’ (p=3.8e-3; 69 genes) were present in rGBCs. The most significant canonical pathway was ‘MODY signaling’ (p=3.0e-7; 11 genes). Targets of the transcription factors Neurod1, Neurog3, Isl1, Pdx1, Nkx2-2, Pax6, Minx1, and Mafa (p=8.0e-4) were enriched in this set as well. On the other hand, the most significant functions among the down-regulated genes were related to proliferation, e.g., ‘carcinoma’ (p=1.7e-12; 452 genes), ‘proliferation cells’ (p= 6.8e-17; 363 genes), and ‘cell death’ (p=8.3e-17; 431 genes). ‘Immune response’ function was also down-regulated (p=1.1e-8; 227 genes). Targets of the estrogen receptor (ER) (p=1.0e-8) and transcription factor Hnf4a (p=2.0e-8) were enriched in this set as well. Despite these signatures suggestive of pancreatic endocrine differentiation, direct comparison of β-cells to rGBCs indicated that many genes related to ‘glucose metabolism disorder’ (p=8.2e-4; 204 genes) and ‘diabetes mellitus’ (p=9.7e-3; 159 genes) were differentially expressed. These differences are indicative of incomplete reprogramming.
Figure 4. RNA-Seq analysis of rGBCs.
(A) Adenovirus GFP-only infected GBCs (controls A&B) were compared to GBCs reprogrammed with either NPM only (NPM) or with NPM, RA and DBZ (NPM-RA-GSI). Actual pancreatic β-cells (β-cells A&B&C) were used as the optimal target population. The cladogram above the heat-map indicated rGBCs formed an intermediate population of cells with characteristics of both GBCs and β-cells. The number of genes differentially expressed between rGBCs and control GBCs (B) and between β-cells and rGBCs (C) is depicted, showing, for example, that after reprogramming 1,759 genes showed increased expression, 1,819 genes showed decreased expression, and 13,679 genes were unchanged in rGBCs compared to control GBCs.
3.7. GBCs are more readily reprogrammed towards the β-cell fate than fibroblasts
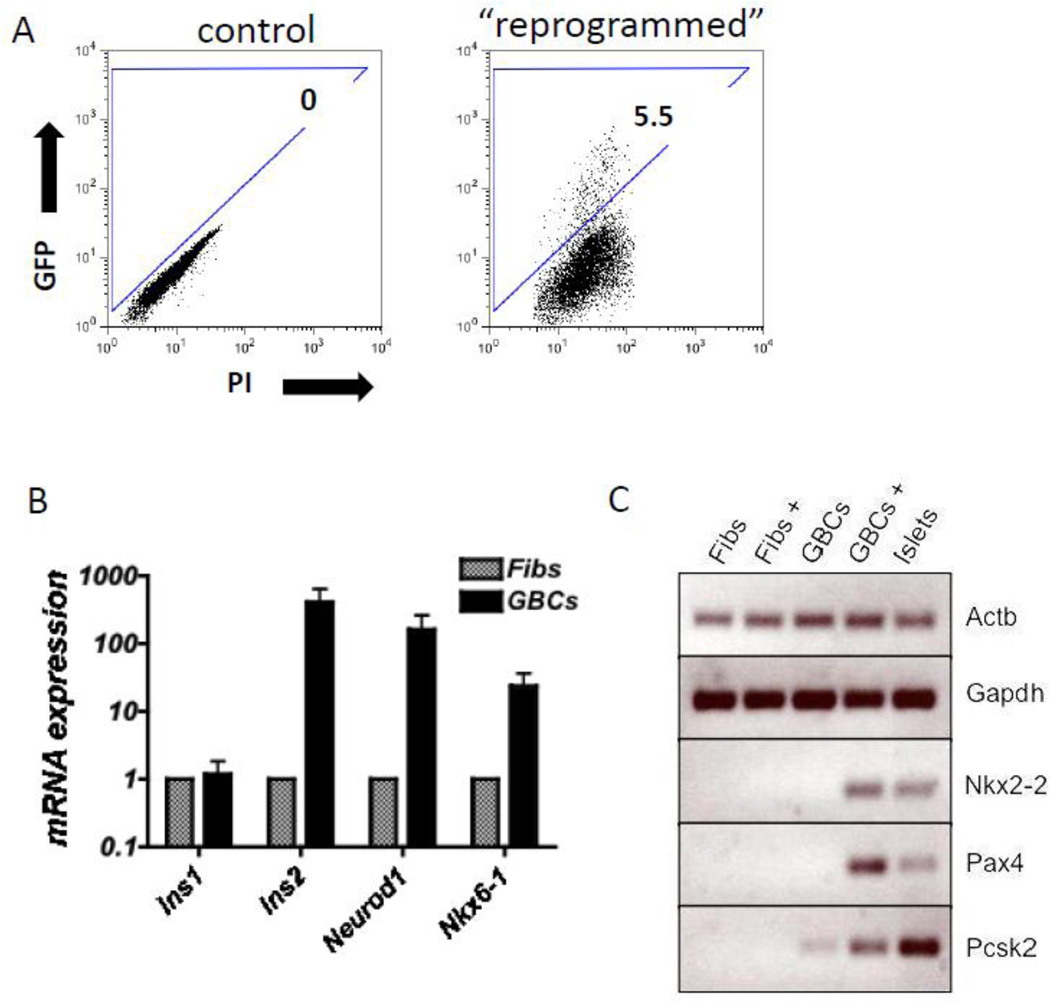
Skin fibroblasts are a cell type which is even more readily available than GBCs. Others previously demonstrated that fibroblasts can be differentiated into some functional cell types such as neurons, hepatocyte-like cells, and cardiomyocytes (Huang, et al., 2011; Ieda, et al., 2010; Vierbuchen, et al., 2010). To determine whether skin cells may be a suitable substrate for generating β-cells by reprogramming, tail tip adult mouse fibroblasts from MIP-GFP mice were expanded and exposed to the same reprogramming regimen as GBCs. Three days later, these cells were FACS-sorted based on GFP expression and gene expression analyzed by RT-PCR and qPCR (Fig. 5A–C). Interestingly, expression of NPM also was able to induce promoter activity at the Ins1 locus, as determined by GFP expression and detection of Ins1 mRNA after three days (Fig. 5A,B). However, other important β-cell transcripts such as Ins2, Neurod1 and Nkx6-1 were significantly less induced in GFP+ fibroblasts than in rGBCs (Fig. 5B). Moreover, other key β-cell transcripts expressed in rGBCs, including Nkx2-2, Pax4 and Pcsk2, were not detected at all in GFP+ fibroblasts using RT-PCR (Fig. 5C). Overall, these results indicated that GBCs were clearly more responsive to pancreatic endocrine reprogramming than fibroblasts.
Figure 5. Attempted reprogramming of fibroblasts to the β -cell fate.
(A) By flow cytometric analysis, control MIP-GFP fibroblasts did not contain any GFP+ cells. Adenovirus-NPM-transduced fibroblasts contained 5.5% GFP+ cells three days after reprogramming. (B) FACS-sorted GFP+ fibroblasts expressed Ins1 mRNA by qPCR. The other β-cell-markers Ins2, Neurod1, and Nkx6-1 were expressed at significantly lower levels in GFP+ fibroblasts compared to GFP+ rGBCs (normalized to Alas1) (P<0.01; N=3 replicates for each sample; error bars represent the standard deviation). (C) Using RT-PCR analysis, fibroblasts transduced with NPM (Fibs+) did not express Nkx2-2, Pax4 or Pcsk2 in contrast to GBCs transduced with NPM (GBC+). Actb and Gapdh were used as the housekeeping loading controls.
3.8. Transplanted reprogrammed cells can engraft, survive and produce insulin in diabetic mice
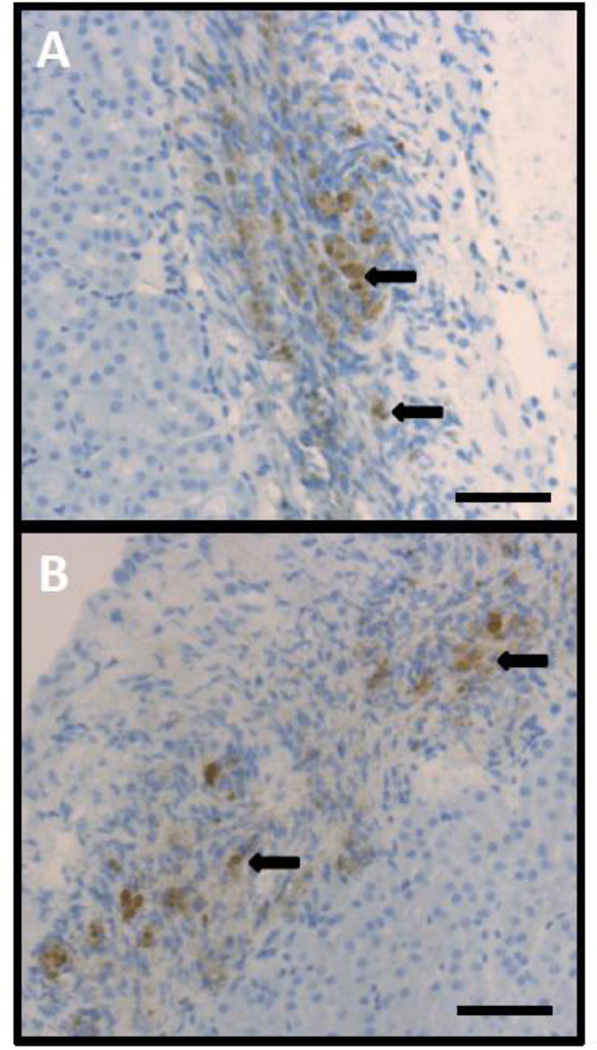
Finally, as it has been demonstrated that in vitro maturation for several months is needed to generate functional β-cells from PSCs (D'Amour, et al., 2006), it was tested whether transplantation of in vitro differentiated rGBCs could reverse hyperglycemia in diabetic mice. GBCs reprogrammed with NPM and incubated with or without RA were harvested from culture after two to three days and transplanted under the renal capsule of diabetic NOD.Cg-Rag1tm1Mom IL2rgtm1WjlIns2Akita (NRG-Akita) and NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ Ins2Akita (NSG-Akita) mice (Table S3). Blood glucose levels were monitored weekly. Of the 19 transplanted mice, only one mouse showed a temporary reversal of hyperglycemia that was not sustained (experiment PW0; Fig. S7). Between 8–15 weeks after transplantation, all mice were euthanized and the kidneys analyzed by immunohistochemistry by staining with an anti-insulin antibody. Interestingly, although the transplanted cells were unable to permanently reverse the hyperglycemia in any of the recipients, 8/19 of the mice had insulin-positive cells in the graft region of the kidney (Fig. 6A,B) compared with the absence of insulin positive cells detected in any of the control transplanted GBCs (data not shown). To determine if transplanted cells were still polyhormonal, grafts were stained with an antibody against somatostatin (Fig. S8). Some somatostatin-positive cells were also detected in these grafts, but their number was fewer than for insulin-positive cells in the same mice.
Figure 6. Immunohistochemistry of transplanted rGBCs.
(A,B) Insulin-positive cells (examples marked with black arrows) were detected in 8/19 of the grafts and never in the adjacent kidney. Two representative images are shown here. Scale bars are 100 µm.
Section 4: DISCUSSION
4.1. The results presented here establish a combined genetic and small molecule approach to differentiate mouse GBCs towards the pancreatic β-cell lineage (summarized in Fig. S2). The expression of key transcription factors has been shown to be a powerful method to directly reprogram one cell type to another, without the need to go through a pluripotent intermediate (Huang, et al., 2011; Ieda, et al., 2010; Vierbuchen, et al., 2010; Xie, Ye, Feng, & Graf, 2004). It is interesting that the three transcription factors capable of reprogramming GBCs into islet-like cells are the same as those required for in vivo reprogramming of the exocrine pancreas into endocrine pancreas (Zhou, et al., 2008), as well as in vitro reprogramming of pancreatic exocrine cells into β-like cells (Akinci, Banga, Greder, Dutton, & Slack, 2011). Therefore, together these results implicate Neurog3, Pdx1 and MafA as the key genetic reprogramming factors needed for the de novo generation of mouse β-cells. In addition to expressing these three transcription factors, the frequency of GFP+ rGBCs was augmented by the timed addition of RA and DBZ, a Notch signaling effector. Both these pathways are essential for normal pancreatic development (Apelqvist, et al., 1999; Martin et al., 2005; Stafford & Prince, 2002). Manipulation of these pathways has also been used to differentiate pluripotent stem cells towards a differentiated β-cell fate, indicating manipulation of both RA-responsive genes and Notch signaling to also be critical for de novo β-cell differentiation (Kroon, et al., 2008; Nostro, et al., 2011).
4.2. Moreover, Notch signaling, particularly involving Hes1, has been implicated as a key regulator of cell fate choice during differentiation of the pancreatobiliary progenitor (Spence, et al., 2009). In accordance with a shift towards pancreatic differentiation of rGBCs, there was a significant decrease in expression of Hes1, supporting a previous study that has shown down-regulation of Hes1 to be a critical step in differentiation of insulin-positive cells from extrahepatobiliary tissues (Coad, et al., 2009). The results presented here further strengthen the case for using GBCs in β-cell reprogramming and expand the previous observations. Firstly, our rGBCs consistently express both Ins1 and Ins2, which was not evident in the results of Coad and colleagues (Coad, et al., 2009). Moreover, induction of additional key β-cell transcription factors not described in the Coad study, including Neurod1, Nkx2-2 and Isl1, was observed here. Additionally, the ability to expand GBCs significantly, while retaining their reprogrammability, was not part of the earlier work.
4.3. One of the interesting features of the results presented here is the rapid fate conversion of GBCs into insulin-positive, islet-like cells. Within only 96 hours of genetic manipulation by transduction with NPM, de-repression of the insulin loci occurred as evidenced by expression of GFP and detection of both Ins1 and Ins2 mRNA. Other gene expression changes also indicated that these cells were differentiating towards the β-cell lineage, including increased expression of the transcription factors Neurod1, Nkx2-2, Pax4, and Isl1. In addition, rGBCs increased expression of the proprotein convertases Pcsk1 and Pcsk2 which are needed to process the immature proinsulin into insulin and c-peptide. In line with these gene expression changes, processed insulin could be detected by immunocytochemistry and by ELISA in rGBCs. The rapid changes in the gene expression profiles are similar to those observed in other direct reprogramming efforts. For example, Ieda et al. detected alphaMHC-GFP+ cardiomyocytes after only three days of reprogramming from fibroblasts (Ieda, et al., 2010). Together with these other studies, the results confirm the power of using key developmental transcription factors for direct reprogramming of various cell types (Huang, et al., 2011; Ieda, et al., 2010; Vierbuchen, et al., 2010; Xie, et al., 2004).
4.4. It is evident that the cells obtained with the current protocol were not mature, fully functioning β-cells after 96 hours. Firstly, the expression of both Ins1 and Ins2 mRNA was at a reduced level compared to that seen in pancreatic islets. Secondly, rGBCs were not responsive to glucose stimulation, a common problem for artificially generated β-cells (Kubo, et al., 2011; Nostro, et al., 2011). Thirdly, the RNA-Seq data showed many genes that were either under-expressed or over-expressed compared to mature adult β-cells. Fourthly, rGBCs were polyhormonal, another common characteristic of in vitro reprogrammed cells (D'Amour, et al., 2006; Nostro, et al., 2011). Finally, rGBCs were unable to reverse the hyperglycemic state of diabetic mice after transplantation. However, it is possible that the engrafted cells were simply too few in number to have a substantial effect on blood glucose levels. In fact, insulin-positive cells could be detected in the grafts of transplanted mice. Alternatively, some form of in vivo maturation may be required for generation of functioning β-cells, as has been shown to be needed for the transplantation of ESC-derived β-cells into diabetic mice (Kroon, et al., 2008). Further studies are required to investigate the in vivo maturation of transplanted rGBCs.
4.5. A common phenotype of most in vitro-derived β-like cells is the absence of GSIS (Kubo, et al., 2011; Nostro, et al., 2011). Similar to these studies, we could not reprogram GBCs to acquire GSIS, an essential feature of typical pancreatic β-cells. In the future, therefore, it will be imperative to determine the missing factor(s) required to instigate glucose sensing, insulin packaging and insulin secretion. The comparison of rGBCs to β-cells from type 2 diabetic patients may be appropriate. Type 2 diabetic patients exhibit defective insulin secretion, exemplified by the loss of GSIS. We analyzed the expression of known genes that when mutated can result in loss of insulin secretion. One factor that was under-expressed in rGBCs relative to β-cells was transmembrane protein 27, encoded by Tmem27 Also known as Collectrin, this factor controls insulin exocytosis and has been implicated as a key component of glucose-stimulated insulin secretion (Fukui et al., 2005). Decreased Tmem27 gene expression has also been detected in islets from type 2 diabetic patients (Altirriba et al., 2010). Other genes that were not expressed at comparable levels to actual β-cells were Slc30a8 and Nnat that are involved in vesicle maturation and insulin secretion respectively (Artner et al., 2010). Future reprogramming studies should be aimed at the induction of these genes that are essential for mature β-cell function.
4.6. In conclusion, the data presented here support a novel strategy to generate pancreatic β-like cells. The gall bladder is a readily accessible and non-essential tissue that contains cells amenable to large-scale expansion and reprogramming to a pancreatic fate. In addition, our data indicate that this endodermal derivative is more amenable to reprogramming than skin fibroblasts. However, the reprogramming is currently only partial and the rGBCs did not become fully functional, mature β-cells in vitro. Although there were many up-regulated marker genes detected in rGBCs, global RNA-Seq analysis was needed to properly characterize the cells relative to true functional β-cells, something that is commonly missing in other reprogramming studies. Nonetheless, the results outlined here will be useful for further experiments aimed at generating a direct differentiation-based cell therapy for type 1 diabetes in human patients.
Supplementary Material
Highlights.
Mouse gall bladder cells (GBCs) can be robustly expanded ex vivo.
GBCs can be reprogrammed towards the β-cell fate by expression of NEUROG3, Pdx1 and MafA.
Reprogramming is enhanced by addition of small molecules.
Reprogrammed GBCs can engraft and express insulin in vivo for at least 15 weeks.
ACKNOWLEDGEMENTS
We thank Pamela Canaday (Flow Cytometry Resource at OHSU) for cell sorting. We thank Jessie Coleman for technical support with the mouse colony. We also thank Milton Finegold and Angela Major (NIDDK-sponsored Digestive Disease Core Laboratory of the Texas Medical Center (DK56338)) for histology support and to Elisabetta Manduchi (UPenn) for helping to transfer data. We are grateful to Craig Dorrell (OHSU) for thoughtful discussions. This work was supported by grants from NIH/NIDDK grants U01 DK072477 (MG), NIH grants AI46629 (DLG, LDS), DK89572 (DLG, LDS), an institutional Diabetes Endocrinology Research Center (DERC) grant DK32520 (DLG, MAB, LDS), grants from the Helmsley Foundation (MG, DLG, LDS) and the Juvenile Diabetes Research Fund (MG).
ABBREVIATIONS
- PSCs
pluripotent stem cells
- GBC
gall bladder cell
- RA
retinoic acid
- DBZ
dibenzazepine
- GSIS
glucose-stimulated insulin secretion
- qPCR
quantitative PCR
- RT-PCR
reverse transcription PCR
- FACS
fluorescence-activated cell sorting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akinci E, Banga A, Greder LV, Dutton JR, Slack JM. Reprogramming of pancreatic exocrine cells towards a beta cell character using Pdx1, Ngn3 and MafA. Biochem J. 2011 doi: 10.1042/BJ20111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipio Z, Liao W, Roemer EJ, Waner M, Fink LM, Ward DC, et al. Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic beta-like cells. Proc Natl Acad Sci U S A. 2010;107(30):13426–13431. doi: 10.1073/pnas.1007884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altirriba J, Gasa R, Casas S, Ramirez-Bajo MJ, Ros S, Gutierrez-Dalmau A, et al. The role of transmembrane protein 27 (TMEM27) in islet physiology and its potential use as a beta cell mass biomarker. Diabetologia. 2010;53(7):1406–1414. doi: 10.1007/s00125-010-1728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400(6747):877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Artner I, Hang Y, Mazur M, Yamamoto T, Guo M, Lindner J, et al. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes. 2010;59(10):2530–2539. doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atouf F, Czernichow P, Scharfmann R. Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/repressor element silencing transcription factor, a neuron-restrictive silencer. [Research Support, Non-U.S. Gov't] The Journal of biological chemistry. 1997;272(3):1929–1934. doi: 10.1074/jbc.272.3.1929. [DOI] [PubMed] [Google Scholar]
- Brehm MA, Bortell R, Diiorio P, Leif J, Laning J, Cuthbert A, et al. Human immune system development and rejection of human islet allografts in spontaneously diabetic NOD-Rag1null IL2rgammanull Ins2Akita mice. Diabetes. 2010;59(9):2265–2270. doi: 10.2337/db10-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Swetha G, Muthyala S, Jaiswal AK, Bellare JR, Nair PD, et al. Islet-like cell aggregates generated from human adipose tissue derived stem cells ameliorate experimental diabetes in mice. PLoS One. 2011;6(6):e20615. doi: 10.1371/journal.pone.0020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou SH, Chen SJ, Chang YL, Chen YC, Li HY, Chen DT, et al. MafA promotes the reprogramming of placenta-derived multipotent stem cells into pancreatic islets-like and insulin-positive cells. J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coad RA, Dutton JR, Tosh D, Slack JM. Inhibition of Hes1 activity in gall bladder epithelial cells promotes insulin expression and glucose responsiveness. Biochem Cell Biol. 2009;87(6):975–987. doi: 10.1139/o09-063. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24(11):1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- de Kort H, de Koning EJ, Rabelink TJ, Bruijn JA, Bajema IM. Islet transplantation in type 1 diabetes. BMJ. 2011;342:d217. doi: 10.1136/bmj.d217. [DOI] [PubMed] [Google Scholar]
- Dorrell C, Erker L, Lanxon-Cookson KM, Abraham SL, Victoroff T, Ro S, et al. Surface markers for the murine oval cell response. Hepatology. 2008;48(4):1282–1291. doi: 10.1002/hep.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulbecco R, Bologna M, Unger M. Differentiation of a rat mammary cell line in vitro. Proc Natl Acad Sci U S A. 1979;76(3):1256–1260. doi: 10.1073/pnas.76.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, et al. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med. 2000;6(5):568–572. doi: 10.1038/75050. [DOI] [PubMed] [Google Scholar]
- Fujikawa T, Oh SH, Pi L, Hatch HM, Shupe T, Petersen BE. Teratoma formation leads to failure of treatment for type I diabetes using embryonic stem cell-derived insulin-producing cells. Am J Pathol. 2005;166(6):1781–1791. doi: 10.1016/S0002-9440(10)62488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K, Yang Q, Cao Y, Takahashi N, Hatakeyama H, Wang H, et al. The HNF-1 target collectrin controls insulin exocytosis by SNARE complex formation. Cell Metab. 2005;2(6):373–384. doi: 10.1016/j.cmet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't] 2011;27(18):2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory LG, Harbottle RP, Lawrence L, Knapton HJ, Themis M, Coutelle C. Enhancement of adenovirus-mediated gene transfer to the airways by DEAE dextran and sodium caprate in vivo. Mol Ther. 2003;7(1):19–26. doi: 10.1016/s1525-0016(02)00021-7. [DOI] [PubMed] [Google Scholar]
- Hara M, Wang X, Kawamura T, Bindokas VP, Dizon RF, Alcoser SY, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. Am J Physiol Endocrinol Metab. 2003;284(1):E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. [Research Support, N.I.H., Extramural Research Support, U.S. Gov't, P.H.S. Review] Cytogenetic and genome research. 2005;110(1–4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Review] The Journal of clinical investigation. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JM, Pennington SE, St George JA, Woodworth LA, Fasbender A, Marshall J, et al. Potentiation of gene transfer to the mouse lung by complexes of adenovirus vector and polycations improves therapeutic potential. Hum Gene Ther. 1998;9(10):1469–1479. doi: 10.1089/hum.1998.9.10-1469. [DOI] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotech. 2008;26(4):443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Kubo A, Stull R, Takeuchi M, Bonham K, Gouon-Evans V, Sho M, et al. Pdx1 and Ngn3 Overexpression Enhances Pancreatic Differentiation of Mouse ES Cell-Derived Endoderm Population. PLoS One. 2011;6(9):e24058. doi: 10.1371/journal.pone.0024058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar R, Komori J, Guzik L, Stolz DB, Chandran UR, LaFramboise WA, et al. Identification and expansion of a unique stem cell population from adult mouse gallbladder. Hepatology. 2011;54(5):1830–1841. doi: 10.1002/hep.24568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, et al. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284(2):399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Motoyama H, Ogawa S, Kubo A, Miwa S, Nakayama J, Tagawa Y-i, et al. In vitro reprogramming of adult hepatocytes into insulin-producing cells without viral vectors. Biochemical and Biophysical Research Communications. 2009;385(1):123–128. doi: 10.1016/j.bbrc.2009.04.146. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nagaya M, Katsuta H, Kaneto H, Bonner-Weir S, Weir GC. Adult mouse intrahepatic biliary epithelial cells induced in vitro to become insulin-producing cells. J Endocrinol. 2009;201(1):37–47. doi: 10.1677/JOE-08-0482. [DOI] [PubMed] [Google Scholar]
- Nishimura W, Kondo T, Salameh T, El Khattabi I, Dodge R, Bonner-Weir S, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293(2):526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nostro MC, Sarangi F, Ogawa S, Holtzinger A, Corneo B, Li X, et al. Stage-specific signaling through TGFbeta family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138(5):861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12(3):266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274(1):305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- Ricordi C, Strom TB. Clinical islet transplantation: advances and immunological challenges. Nat Rev Immunol. 2004;4(4):259–268. doi: 10.1038/nri1332. [DOI] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127(24):5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Shapiro AM, Lakey JR, Ryan EA, Korbutt GS, Toth E, Warnock GL, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343(4):230–238. doi: 10.1056/NEJM200007273430401. [DOI] [PubMed] [Google Scholar]
- Shigeru Y, Tomoyuki A, Phillip PC, Hideaki K, Arun S, Susan B-W, et al. NeuroD and reaggregation induce beta-cell specific gene expression in cultured hepatocytes. Diabetes/Metabolism Research and Reviews. 2007;23(3):239–249. doi: 10.1002/dmrr.678. [DOI] [PubMed] [Google Scholar]
- Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, et al. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17(1):62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12(14):1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AY, Ehrhardt A, Xu H, Kay MA. Adenovirus transduction is required for the correction of diabetes using Pdx-1 or Neurogenin-3 in the liver. Mol Ther. 2007;15(2):255–263. doi: 10.1038/sj.mt.6300032. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yechoor V, Chan L. Minireview: beta-cell replacement therapy for diabetes in the 21st century: manipulation of cell fate by directed differentiation. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov't Review] Molecular endocrinology. 2010;24(8):1501–1511. doi: 10.1210/me.2009-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yechoor V, Liu V, Espiritu C, Paul A, Oka K, Kojima H, et al. Neurogenin3 Is Sufficient for Transdetermination of Hepatic Progenitor Cells into Neo-Islets In Vivo but Not Transdifferentiation of Hepatocytes. Developmental Cell. 2009;16(3):358–373. doi: 10.1016/j.devcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to [bgr]-cells. Nature. 2008;455(7213):627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.