Abstract
Early-onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD) is a myopathic disorder associated with mutations in MEGF10. By novel analysis of SNP array hybridization and exome sequence coverage, we diagnosed a 10-year old girl with EMARDD following identification of a novel homozygous deletion of exon 7 in MEGF10. In contrast to previously reported EMARDD patients, her weakness was more prominent proximally than distally, and involved her legs more than her arms. MRI of her pelvis and thighs showed muscle atrophy and fatty replacement. Ultrasound of several muscle groups revealed dense homogenous increases in echogenicity. Cloning and sequencing of the deletion breakpoint identified features suggesting the mutation arose by fork stalling and template switching. These findings constitute the first genomic deletion causing EMARDD, expand the clinical phenotype, and provide new insight into the pattern and histology of its muscular pathology.
Keywords: EMARDD, MEGF10, SNP array, exome sequencing, deletion analysis, myopathy
1. Introduction
Early onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD; MIM#614399) is caused by null mutations of MEGF10 [1]. Similar to spinal muscular atrophy with respiratory distress-1 (SMARD1), EMARDD is a congenital neuromuscular disease with a hallmark feature of infantile-onset diaphragmatic paralysis requiring mechanical ventilation [1,2]. Other shared features of these disorders include decreased fetal movements, finger contractures, and diffuse hypotonia with progressive weakness [3]. Since SMARD1 is a largely neuropathic condition [3], EMARDD is distinguished by its myopathic features [1].
MEGF10 encodes a transmembrane protein in the epidermal growth factor family [1]. In humans, this protein is expressed in spinal cord and brain, as well as cardiac, smooth and skeletal muscle [4]. The murine homologue has also been detected in Schwann cell precursors and muscle satellite cells, and has been shown to participate in satellite cell proliferation, differentiation and fusion into multi-nucleated myofibers [1]. Furthermore, knockdown of MEGF10 in zebrafish disrupted myofibrillar organization and striation [5].
We used a novel approach to SNP chip analysis along with exome sequencing to discover the first reported exonic deletion of MEGF10. Furthermore, phenotypic characterization of the affected individual expands the clinical and pathological phenotype of EMARDD.
2. Patients and Methods
The proband (UDP2437) presented at 10-years of age to the NIH Undiagnosed Diseases Program with neuromuscular decline. An older sister was similarly affected, while a brother was unaffected. Their asymptomatic parents were first cousins of Arabic origin (FIGURE 1A).
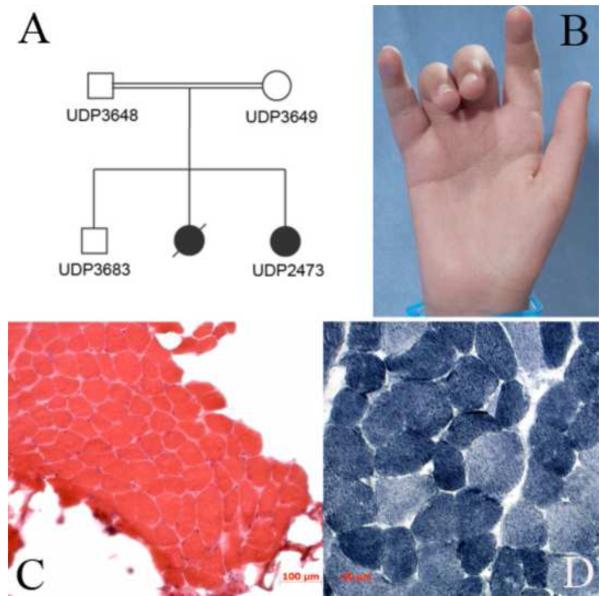
Fig. 1.
Pedigree, patient photographs and gastrocnemius muscle biopsy (a) Family pedigree (b) Finger contractures of the PIP joints of the 3rd and 4th fingers. (c) Haematoxylin and eosin staining of muscle biopsy revealed mild variability in fiber diameter with only occasional central nucleation. Large pockets of fatty tissue, but only minimal connective tissue buildup is seen between fibers (20× magnification) (d) NADH stain of muscle biopsy revealed no core formations. There was mild type I fiber type predominance (40× magnification).
The proband was born at 35 weeks gestation via Caesarean section with gestation notable for decreased fetal movement. Her birth-weight was <5th centile and postnatal complications included diffuse hypotonia and contractures of the PIP joints of her 3rd and 4th fingers (FIGURE 1B). At four months of age, she developed severe dysphagia. At eight months, she underwent gastrostomy tube placement. After surgery, she required continuous ventilatory support due to eventration of the right hemi-diaphragm and ultimately, tracheostomy. Areflexia was evident at 3 years of age. She was able to stand with support, but never walked independently. She has bladder incontinence and progressive scoliosis requiring numerous surgical stabilizations.
On examination at 10 years, she was alert and engaging. She answered questions, performed commands, and had normal coordination and sensation. She had a high-arched palate, myopathic facies, and bilateral ptosis. Extraocular movements were full. She had diffusely decreased muscle bulk and tone associated with diffuse, but more proximal than distal weakness. Her legs were weaker than her arms. She could not sit independently, and had bilateral contractures of her elbows, hips, knees, ankles and the 3rd and 4th PIP joints of her hands (FIGURE 1B). Deep tendon reflexes were absent.
The patient’s older sister was a full-term child with delivery complicated by perinatal asphyxia. She had central hypotonia and axial hypertonia along with finger contractures similar to her sister. Tracheostomy and gastrostomy tube placement was performed at 6 months due to increasing weakness. An intercostal muscle biopsy at the time reportedly showed nonspecific abnormalities. She never stood independently and had progressive scoliosis. She died at 4 years of age (genetic and tissue samples were unavailable for the present study).
DNA isolation, exome sequencing and CNV analysis
Clinical research protocols were approved through the NHGRI Institutional Review Board, and family members gave informed written consent. Genomic DNA was extracted and used for copy number analysis and exome sequencing as previously described, with modifications [6,7,8]. PCR amplification, purification, and sequencing were performed per established protocols or as detailed in Supplementary Table 1.
3. Results
3.1 Clinical Studies
Gastrocnemius muscle biopsy at 11 years of age revealed large pockets of fatty tissue replacement in between fascicles, but minimal connective tissue buildup between individual fibers as well as mild variability in fiber diameter and occasional central nucleation, but no degenerating or regenerating fibers (FIGURE 1C-D). There was mild type I fiber type predominance that was validated by immunohistochemistry of slow and fast myosin heavy chains (data not shown). Serum creatine kinase, electrocardiogram, echocardiogram, and brain neuroimaging were normal.
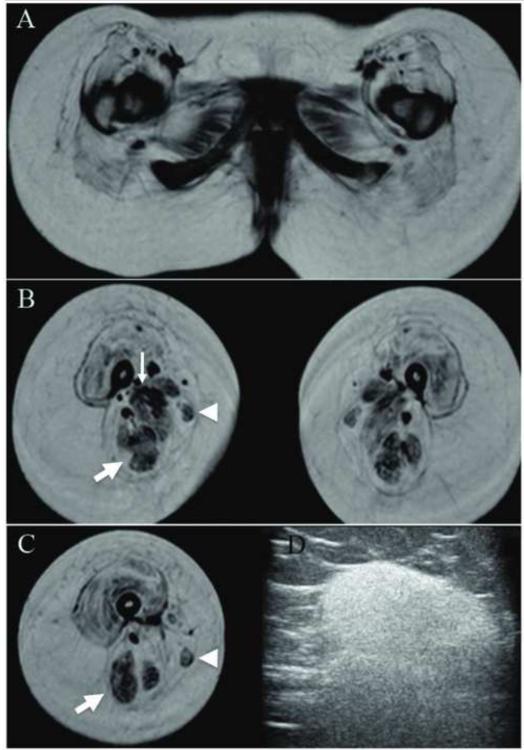
Electrophysiology and imaging studies revealed myopathic abnormalities. Nerve conduction velocity was normal, but peroneal motor responses had decreased amplitude. Electromyography showed few low amplitude and short duration motor units in the left vastus lateralis. Magnetic resonance imaging of the pelvis and thighs revealed bilateral symmetric muscular atrophy and nearly complete fatty replacement of the gluteal and quadriceps compartments; less atrophy and fatty replacement was present in the hamstring and gracilis muscles (FIGURE 2A-C). Muscle ultrasound revealed dense homogenous increase in echogenicity in nearly all muscles of the upper and lower extremity (FIGURE 2D).
Fig. 2.
Imaging of muscles. (a) T1-weighted magnetic resonance imaging of the pelvis and thighs. Marked bilateral symmetric muscular atrophy and nearly complete fatty replacement is present in the gluteal and quadriceps compartments. Less atrophy and fat infiltration was present in the adductor (thin arrow) and hamstring compartments (thick arrow). The gracilis (arrowhead) also appeared relatively preserved compared to the severely involved sartorius muscle. (b) Ultrasound (12Hz linear transducer) of the right biceps muscle revealed diffuse homogeneous increase in echogenicity.
3.2 Extreme novel analysis of SNP arrays identifies biallelic exonic deletions
To identify SNP results that could directly affect genes, we selected the 27,486 SNP probes that resided within 50 bp of an exon from among 773,233 Illumina Human Omni-Express12_v1_H array SNPs. We then determined which of these 27,486 SNPs behaved well (i.e., exhibited successful hybridization) when examined in 663 identically extracted and processed genomic samples. This identified >2000 SNP hybridization datapoints, which we studied in 134 individuals to detect unique loss of hybridization; which was found for 31 SNP probes in 24 patient DNA samples (Supplementary Table 2). PCR amplification and Sanger sequencing of each locus identified biallelic deletions encompassing the SNP locus for 5, an adjacent SNP that would interfere with hybridization to the SNP probe for 17, and no identifiable cause for loss of hybridization for 9 (Supplementary Table 2).
The biallelic deletions in these patients involved the 3′ UTR of HBA2, exon 2 of KLK15 and exon 7 of MEGF10. Aside from the HBA2 deletions being associated with α-thalassemia trait, MEGF10 was the only gene previously associated with disease and was therefore diagnostically helpful.
3.3 Identification and characterization of MEGF10 deletion
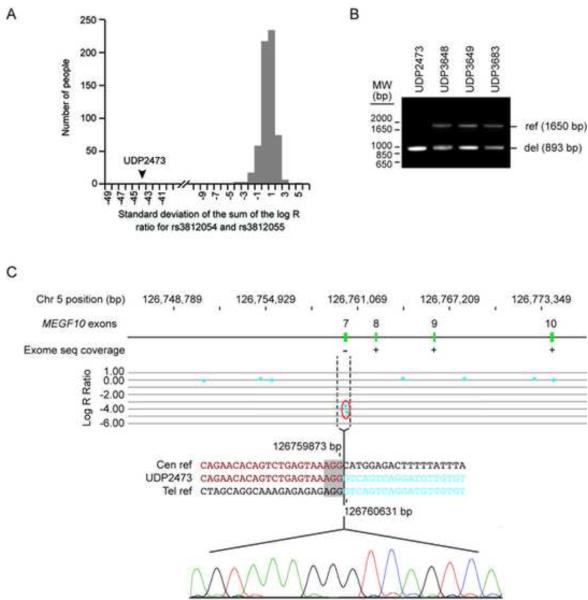
Two SNPs, rs3812054 and rs3812055, reside in exon 7 of MEGF10. As quantified by the standard deviation of the sum of the log R ratio for these two probes, neither probe hybridized to DNA derived from individual UDP2437, whereas each probe showed consistent hybridization to DNA from 663 other samples (FIGURE 3A-C). Supporting these data, exome sequencing of UDP2437 showed no coverage of MEGF10 exon 7 (FIGURE 3C), whereas the data for all UDP individuals tested (N=180) showed uniformly excellent coverage of this exon [9].
Fig. 3.
Identification of a MEGF10 exon 7 deletion arising by FosTeS x 1. (a) Plot of the count of individuals run on the OmniExpress12v-1 SNP chip versus the standard deviation of the sum of the log R ratio for two loci rs3812054 and rs3812055. 663 individuals from the UDP were measured at both SNPs and are represented in the plot. All samples were within 5 standard deviations of the mean except UDP2473 whose sum is −44 standard deviations from the mean. (b) PCR amplification of genomic DNA obtained using primers flanking the breakpoint. The 1650 bp band indicates an undeleted (ref) allele, while the 893 bp band indicates a deleted (del) allele. (c) Diagram of exons 7-10 (green) of the MEGF10 gene placed to scale on the hg18 build of chromosome 5 and characterization of the deletion breakpoints (dashed vertical lines). The presence or absence of exome sequencing data is indicated below each MEGF10 exon by a “+” or “−”, respectively. The middle panel shows a plot of the log R ratio for high resolution SNP array analysis; each point represents a SNP probe. The normalized data for each probe are represented along a horizontal line that indicates its position on chromosome 5. Loss of copy number is indicated by deviation below mean logR ratio of −1. The lower panel shows the results of breakpoint sequencing; the sequence across the breakpoint in UDP2473 (middle and chromatogram) is compared to the centromeric (upper) and telomeric (lower) reference sequences. The positions of the breakpoints on chromosome 5 are indicated above and below the centromeric and telomeric reference sequences, respectively. The microhomology (grey box) at the junction suggests a FoSTeS x 1 mechanism of deletion.
Using PCR amplification (primers MEGF10del_7F and MEGF10del_8R) and Sanger sequencing of the amplification products, we defined a homozygous 757 bp deletion encompassing exon 7 of MEGF10 in UDP2437 (Chr5 (GRCh37):g.126732224_126732470del; FIGURES 3B-C). cDNA sequencing from patient myoblasts (capable only with the use of cyclohexamide to prevent nonsense mediated decay) indicated these changes result in a transcript missing exon 7 and a frameshift mutation at amino acid 139 followed by the substitution of the next 114 amino acids and ending with a premature stop codon at amino acid 253 (pCys139Vals*115; data not shown). The deletion breakpoints shared 3 bp of homology and did not reside in a repetitive element. Unaffected family members were heterozygous for the deletion.
4. Discussion
In the case described above, restricting analysis of SNP hybridization data to probes that routinely hybridize with high efficiency enabled detection of biallelic deletions in the proband that were validated by exome sequencing and PCR amplification. In the future, the development of SNP chips containing probes within each exon (e.g., HumanOmni-ExpressExome-8v1) will facilitate these analyses. Nonetheless, even in the absence of probes in all exons, we identified a novel homozygous deletion of MEGF10 exon 7 as the molecular cause of EMARDD in this consanguineous family.
The breakpoint boundaries of this deletion showed 3 bp of homology and resided outside of larger repetitive elements such as SINEs and LINEs. Non-allelic homologous recombination usually occurs within repetitive elements such as Alu repeats and is generally associated with longer homology tracts at the breakpoint [10,11]. Non-homologous end joining is characterized by small insertions and the absence of microhomology at the breakpoint junction [12]. Although originally described in the context of complex rearrangements involving duplication, triplication and deletion [13], fork stalling and template switching (FoSTeS) has recently been considered responsible for many simple deletions within the human genome [14]. Deletions arising from FoSTeS, characterized by microhomology at the breakpoint junction, frequently occur outside of large repetitive elements [10]. Based on these observations, we hypothesize that FoSTeS was the cause of this ancestral MEGF10 deletion.
Each parent of this consanguineous relationship had this ancestral deletion in a heterozygous state (the proband’s brother was also heterozygous for the this deletion), while the proband (and likely her deceased sister) was homozygous for the ancestral deletion resulting in autosomal recessive EMARDD. MEGF10 mutations have been previously associated with a range of neuromuscular phenotypes, including EMARDD [1,5]. Similar to previously reported EMARDD patients with null mutations of MEGF10, the proband had decreased fetal movements, early hypotonia, finger contractures, scoliosis, dysphagia, areflexia, progressive weakness, and infantile respiratory distress requiring mechanical ventilation [1]. The patient was also noted to have myopathic facies, a high-arched palate, and ptosis. Electrophysiology was also consistent with EMARDD.
In contrast to previously reported EMARDD patients, the proposita’s weakness had a more proximal than distal quality, with a predominance of lower extremity over upper extremity weakness [1]. Her phenotype also varied from the less severe neuromuscular phenotype seen in patients with missense mutations in MEGF10 [5]. Those patients had minicores on muscle biopsy, did not require persistent respiratory ventilation, and presented with profound neck weakness, but milder axial and proximal appendicular weakness.
Muscle histology of the proband was also more consistent with other EMARDD patients, with only non-specific myopathic changes without minicores [1,5]. There was also occasional central nucleation. Imaging of her pelvis and thighs showed considerable atrophy and fatty replacement of all muscles, although gluteal and quadriceps compartments were more affected than the hamstring, adductor, and gracilis muscles. This may be a new finding since imaging and muscle assessments in advanced EMARDD cases have not yet been reported; more patients will be required to determine if this pattern is a consistent feature of this phenotype.
In conclusion, this work utilized novel analysis of SNP arrays and exome sequencing to identify the first genomic rearrangement of MEGF10 causing EMARDD. Furthermore, this work confirms the previous report associating MEGF10 and EMARDD, while also expanding the clinical, histological, and imaging features of this very rare disease. Although generalization of the clinical findings associated with MEGF10 mutations requires further study in additional patients, the molecular data indicate that testing of EMARRD patients should include deletion analysis when MEGF10 sequencing does not detect other mutations.
Supplementary Material
| UDP4424 | rs12007545 | no detected deletion or adjacent SNV |
| UDP4941 | rs4073250 | adjacent homozygous SNP (rs34526867) |
| UDP0005 | rs11789583 | no detected deletion or adjacent SNV |
| UDP5022 | rs2234564 | no detected deletion or adjacent SNV |
| UDP5454 | rs2571108 | no detected deletion or adjacent SNV |
| UDP5454 | rs8093 | no detected deletion or adjacent SNV |
| UDP0810 | rs25707 | no detected deletion or adjacent SNV |
| UDP3587 | rs8049284 | adjacent homozygous SNV (chr16:79276382 C>A) |
ACKNOWLEDGMENTS
We are grateful to Shannon McNeil, Ronald Austin, Jose Salas, and Cheryl Hipple for excellent administrative assistance; Joy Bryant, Barrington Burnett, Roxanne Fischer, Chevalia Robinson, Gretchen Golas, and Camilo Toro for clinical and technical assistance and critical analysis. We would especially thank the family of our patient for the loving care for their children and cooperation with our work.
During this work TMP, DA, TM, KFF, CFB, and CT were supported by the NIH Undiagnosed Diseases Program; TM, GG, MS, NFH, PFC, PC, JKT, JCM, and WAG were supported by the Intramural Research Program of the National Human Genome Research Institute of the National Institutes of Health. CGB, SD, YH, IB, and KM were supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke of the National Institutes of Health. TMP was also supported by the Diana and Steve Marienhoff Fashion Industries Guild Endowed Fellowship in Pediatric Neuromuscular Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST The authors declare no conflict of interest.
REFERENCES
- 1.Logan CV, Lucke B, Pottinger C, et al. Mutations in MEGF10, a regulator of satellite cell myogenesis, cause early onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD) Nat Gen. 2011;43:1189–92. doi: 10.1038/ng.995. [DOI] [PubMed] [Google Scholar]
- 2.Hartley L, Kinali M, Knight R, et al. A congenital myopathy with diaphragmatic weakness not linked to the SMARD1 locus. Neuromuscul. Disord. 2007;17:174–9. doi: 10.1016/j.nmd.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Grohmann K, Varon R, Stolz P, et al. Infantile spinal muscular atrophy with respiratory distress type 1 (SMARD1) Ann Neurol. 2003;54:719–24. doi: 10.1002/ana.10755. [DOI] [PubMed] [Google Scholar]
- 4.Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O. Prediction of the Coding Sequences of Unidentified Human Genes. XX. The Complete Sequences of 100 New cDNA Clones from Brain Which Code for Large Proteins in vitro. DNA Res. 2001;8:85–95. doi: 10.1093/dnares/8.2.85. [DOI] [PubMed] [Google Scholar]
- 5.Boyden SE, Mahoney LJ, Kawahara G, et al. Mutations in the satellite cell gene MEGF10 cause a recessive congenital myopathy with minicores. Neurogenetics. 2012;13:115–24. doi: 10.1007/s10048-012-0315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierson TM, Simeonov DR, Sincan M, et al. Exome sequencing and SNP analysis detect novel compound heterozygosity in fatty acid hydroxylaseassociated neurodegeneration. Eur J Hum Gen. 2012;20:476–79. doi: 10.1038/ejhg.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teer JK, Bonnycastle LL, Chines PS, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–31. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston JJ, Teer JK, Cherukuri PF, et al. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet. 2010;86:743–8. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherukuri PF, Sincan M, Accardi JP, et al. Systematic identification and definition of consistently well-characterized protein-coding exons using next generation sequencing technology. Journal of Exomes and Genomes. Submitted. [Google Scholar]
- 10.Liu P, Carvalho CM, Hastings P, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–20. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am J Hum Genet. 2011;89:580–8. doi: 10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 14.Nagamani SC, Zhang F, Shchelochkov OA, et al. Microdeletions including YWHAE in the Miller-Dieker syndrome region on chromosome 17p13.3 result in facial dysmorphisms, growth restriction, and cognitive impairment. J Med Genet. 2009;46:825–33. doi: 10.1136/jmg.2009.067637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.