Abstract
Traditionally regarded as a genetic disease of the cardiac sarcomere, hypertrophic cardiomyopathy (HCM) is the most common inherited cardiovascular disease and a significant cause of sudden cardiac death. While the most common etiologies of this phenotypically diverse disease lie in a handful of genes encoding critical contractile myofilament proteins, approximately 50% of patients diagnosed with HCM worldwide do not host sarcomeric gene mutations. Recently, mutations in genes encoding calcium-sensitive and calcium-handling proteins have been implicated in the pathogenesis of HCM. Among these are mutations in TNNC1-encoded cardiac troponin C, PLN-encoded phospholamban, and JPH2-encoded junctophilin 2 which have each been associated with HCM in multiple studies. In addition, mutations in RYR2-encoded ryanodine receptor 2, CASQ2-encoded calsequestrin 2, CALR3-encoded calreticulin 3, and SRI-encoded sorcin have been associated with HCM, although more studies are required to validate initial findings. While a relatively uncommon cause of HCM, mutations in genes that encode calcium-handling proteins represent an emerging genetic subset of HCM. Furthermore, these naturally occurring disease-associated mutations have provided useful molecular tools for uncovering novel mechanisms of disease pathogenesis, increasing our understanding of basic cardiac physiology, and dissecting important structure-function relationships within these proteins.
Keywords: Calcium, genetics, hypertrophic cardiomyopathy, junctophilin, mutation, phospholamban, troponin
HYPERTROPHIC CARDIOMYOPATHY
Hypertrophic cardiomyopathy (HCM) is a common cardiovascular disease defined clinically as otherwise unexplained thickening of the ventricular walls and/or interventricular septum. First described by Teare as “asymmetrical hypertrophy of the heart” in eight young adults, five decades of intense clinical, translational, and basic science investigation have been dedicated to this disease [1]. HCM is the most common inherited cardiovascular disorder affecting approximately 1 in 500 persons. It is a significant cause of youthful sudden cardiac death (SCD) and the most common cause of SCD in athletes [2–4]. Grossly, HCM is characterized by asymmetrical left ventricular (LV) hypertrophy with possible involvement of the right ventricle. At the histological level, HCM is defined by myocyte hypertrophy and myofibrillar disarray with interstitial fibrosis.
HCM demonstrates phenotypic heterogeneity with a spectrum of clinical features. The degree of cardiac hypertrophy is variable between probands as is cardiac fibrosis, myocyte disarray, LV outflow tract obstruction, and sudden death susceptibility. Further, abnormal intramural coronary arteries, systolic anterior movement of the mitral valve, and arrhythmias can be variably present [5]. Presenting symptoms typically include chest pain, dyspnea, palpitations, and syncope; however, SCD may also occur in the absence of major symptoms [6]. Just as clinical presentation is variable, clinical progression is largely unpredictable, with some patients remaining asymptomatic over their lifetime while others present as neonates with profound cardiac hypertrophy [7]. Cardiac arrhythmias can occur such as atrial fibrillation and non-sustained ventricular tachycardia [8]. 5–15% of patients with HCM will experience further pathogenic remodeling ultimately leading to heart failure with LV wall thinning, chamber dilatation, and loss of cardiac contractility [9, 10]. In this way, HCM is not a static disease with consistent clinical findings, but rather one that may or may not include a number of clinical characteristics that can evolve over time.
The variable clinical phenotype of HCM reflects both the diverse genetic basis of the disease and the myriad of factors which can modify disease expression. Indeed, HCM has been appreciated as principally an autosomal dominant disease with variable expressivity and incomplete penetrance. Hundreds of mutations found in dozens of genes encoding various sarcomeric/myofilament, Z-disc, and calcium (Ca2+)-handling proteins have been associated with this disease (Table 1).
Table 1.
Summary of the Genetic Causes of HCM
| Gene | Locus | Protein | Frequency |
|---|---|---|---|
| Myofilament/Sarcomeric HCM | |||
| Giant Filament | |||
| TTN | 2q31 | Titin | Rare |
| Thick Filament | |||
| MYH7 | 14q11.2-q12 | β-Myosin heavy chain | 15–25% |
| MYH6 | 14q11.2-q12 | α-Myosin heavy chain | Rare |
| MYL2 | 12q23-q24.3 | Regulatory myosin light chain | Rare |
| MYL3 | 3p21.2-p21.3 | Essential myosin light chain | Rare |
| Intermediate Filament | |||
| MYBPC3 | 11p11.2 | Cardiac myosin-binding protein C | 15–25% |
| Thin Filament | |||
| TNNT2 | 1q32 | Cardiac troponin T | 3–5% |
| TNNI3 | 19p13.4 | Cardiac troponin I | 1–5% |
| TPM1 | 15q22.1 | α-Tropomyosin | 1–5% |
| ACTC | 15q14 | α-Cardiac actin | Rare |
| TNNC1 | 3p21.1 | Cardiac troponin C | Rare |
| Z-Disc HCM | |||
| ACTN2 | 1q42-q43 | α-Actinin 2 | Rare |
| ANKRD1 | 10q23.31 | Cardiac ankyrin repeat protein | Rare |
| CSRP3 | 11p15.1 | Muscle LIM protein | Rare |
| LBD3 | 10q22.2-q23.3 | LIM binding domain 3 | Rare |
| MYOZ2 | 4q26-q27 | Myozenin 2 | Rare |
| TCAP | 17q12-q21.1 | Telethonin | Rare |
| VCL | 10q22.1-q23 | Vinculin/metavinculin | Rare |
| Calcium-Handling HCM | |||
| CALR3 | 19p13.11 | Calreticulin 3 | Rare |
| CASQ2* | 1p13.3-p11 | Calsequestrin | Rare |
| JPH2 | 20q13.12 | Junctophilin 2 | Rare |
| PLN | 6q22.1 | Phospholamban | Rare |
| RYR2* | 1q43 | Ryanodine receptor 2 | Rare |
| SRI* | 7q21.1 | Sorcin | Rare |
Asterisk indicates additional studies may be needed before accepting as a rare HCM-susceptibility gene. Frequency estimates are derived from a population of all unrelated patients with clinically diagnosed HCM without further clinical refinement based on family history, degree of hypertrophy, age at diagnosis, or septal profile. Rare indicates <1% contribution.
HCM DUE TO CARDIAC MYOFILAMENT/SARCOMERE MUTATIONS
Myofilament/Sarcomeric-HCM
HCM is the first myocardial disorder in which the genetic basis of disease pathogenesis has been elucidated. The discovery of a genetic locus responsible for familial HCM was first identified over 20 years ago utilizing linkage analysis of a large, multi-generational family [11]. The following year, the first HCM-associated mutation localizing to the MYH7-encoded β-myosin heavy chain genetic locus was identified [12]. Since then, multiple studies have determined that the majority of HCM is due to mutations in genes encoding components of the cardiac sarcomere responsible for generating the molecular force of myocyte contraction. However, in a given cohort of unrelated patients with clinically diagnosed HCM, the frequency of mutations localizing to these genes, the so-called “sarcomere-positive” individuals, varies from approximately 25% to 65% depending on the cohort analyzed [13–16]. Worldwide, approximately 50% of patients diagnosed with HCM are sarcomeric mutation negative [17].
Sarcomeric-HCM genes are divided into sub-groups based on the type of protein encoded. These include proteins comprising the sarcomere’s thick myofilament (MYH7, MYL2-encoded regulatory myosin light chain, and MYL3-encoded essential myosin light chain [18,19]), intermediate myofilament (MYPBC3-encoded cardiac myosin binding protein C [20]), and thin myofilament (ACTC-encoded actin [21], TPM1-encoded alpha-tropomyosin [22], TNNT2-encoded cardiac troponin T [22], and TNNI3-encoded cardiac troponin I [23]). A handful of mutations in the “giant” myofilament, TTN-encoded cardiac titin have also been described [24, 25]. Despite early studies indicating that specific mutations held prognostic significance, or that mutations localizing to particular sarcomeric genes might predispose to a particular HCM phenotype, there is no major HCM genotype-phenotype correlation that has been universally validated [26].
While the frequency of mutations localizing to sarcomeric genes varies slightly depending on the cohort analyzed, mutations in MYBPC3 and MYH7 are the two most common genetic subtypes [13, 14, 16]. Wide-spread acceptance of sarcomeric genes, with the exception of the still-emerging role of TTN, has led to the development of multiple clinically/commercially available genetic tests. While this developmental benchmark represents the advances that have been made in HCM research, at least 50% of the HCM population, and the majority of patients with sigmoidal and apical septal morphologies remain, genotype-negative [27–29]. This has afforded the opportunity for identification of novel genes that may be responsible for HCM and has cultivated a wealth of research to identify these still enigmatic mechanisms of cardiac hypertrophy.
Cardiac Troponin C – The Most Recent Sarcomeric Protein Implicated in HCM
The cardiac troponin heterotrimeric myofilament complex is comprised of an elongated troponin T (TNNT2) subunit, an inhibitory troponin I (TNNI3) subunit, and a Ca2+-sensitive troponin C subunit (TNNC1, also denoted HcTnC) encoded by the gene TNNC1. While TNNT2 and TNNI3 were established as HCM-susceptibility genes over a decade ago, HCM-associated mutations localizing to TNNC1 have only recently been identified [30–33]. At the molecular level, TNNC1 is a sarcomeric Ca2+ sensor which, when bound to the cytosolic divalent cation at the single Ca2+-specific binding site, strengthens its interaction with TNNI3. This binding reduces the inhibitory function of TNNI3, releasing it from actin, and allows a shift in the troponin-tropomyosin complex which slides deeper into the actin groove exposing the myosin-binding sites. In this way, TNNC1 represents a critical molecular switch which initializes myofilament contraction [34].
The first case report identifying an HCM-associated mutation in TNNC1, L29Q, was subsequently shown to reduce TNNC1 Ca2+-sensitivity by blunting the functional effects of TNNI3 phosphorylation on TNNC1 [30, 31]. While the observed reduction Ca2+-sensitivity has proven controversial, it is possible that TNNC1-L29Q functionally disrupts the dynamic interaction of TNNI3 and TNNC1 [32, 33]. Subsequently, four additional mutations, TNNC1-A8V, C84Y, E134D, and D145E, were identified in a large cohort of 1025 unrelated cases diagnosed with HCM [35]. Concomitant comprehensive genetic analysis of 600 reference alleles did not identify any protein-altering genetic variation among healthy individuals. Cardiac fibers reconstituted with mutated TNNC1 demonstrated increased Ca2+-sensitivity to contractile force generation with A8V, C84Y and D145E mutations. This property of increased myofilament Ca2+-sensitivity fits well with the defective cardiac relaxation noted in both animal models and patients with HCM and is in contrast to dilated cardiomyopathy (DCM)-associated TNNC1 mutations which have reduced Ca2+-sensitivity and are associated with loss of contractility [36, 37]. Indeed, increased Ca2+-sensitivity may be a defining characteristic of HCM-associated TNNC1 mutations [38, 39]. Further characterization of TNNC1-A8V, C84Y, and D145E demonstrated mutation-induced alterations in protein secondary structure that modifies dynamic interactions with other cardiac thin filament proteins through potentially mutation-specific mechanisms [40]. In addition, TNNC1-D145E, and to a lesser degree E134D, may perturb Ca2+ association with the carboxy-terminal Ca2+-binding EF hands of TNNC1 [41]. Finally, the first frameshift mutation in TNNC1 was recently identified in a youthful proband who died of SCD [42]. This insertion results in alteration at residue Q122 leading to an out-of-frame scramble of following residues and premature protein truncation (Q122fs/30). While this mutation cosegregates with incidence of LV hypertrophy in the identified family, in-depth functional studies delineating the potential molecular and cellular impact of this mutation are yet to be completed.
Taken together, these studies demonstrate that mutations in TNNC1 are rare, occurring in ~0.4% of HCM patients. Despite this rarity, TNNC1 mutations demonstrate a frequency similar to other genes included in the current sarcomeric genetic test panel including TNNI3 (~1.5%), TPM1 (~0.77%), and ACTC (~0.26%) [27]. Based on the functional, apparently HCM-specific, impact of these mutations in multiple studies, and the absence of genetic variation in ostensibly healthy controls to date, TNNC1 has been incorporated into the HCM genetic test as an HCM-associated sarcomeric gene [43].
HCM DUE TO MUTATIONS IN NON-SARCOMERIC CALCIUM-HANDLING PROTEINS
Calcium Signaling in the Cardiocyte
A major role of Ca2+ within the cardiocyte is the initiation and coordination of myofilament contraction – a critical process of the cardiocyte and one that is accordingly tightly regulated (Reviewed [44, 45], Fig. 1). Opening of the voltage-gated L-type Ca2+ channel (LTCC) at the sarcolemma allows for an influx of extracellular Ca2+ across the cardiac dyad. This triggers Ca2+ release from the sarcoplasmic reticulum (SR) via RYR2-encoded ryanodine receptors type 2 (RyR2) in a process known as Ca2+-induced Ca2+-release (CICR). Cytosolic Ca2+ binds to TNNC1 of the myofilament and serves as the molecular initiator for mechanical contraction [46]. Systolic contraction is terminated by re-uptake of cytosolic Ca2+ into the SR via the SR Ca2+ ATPase (SERCA2) and removal of Ca2+ from the cell through the sodium-Ca2+ exchanger (NCX1). PLN-encoded phospholamban (PLN) negatively-regulates the Ca2+ uptake action of SERCA2. In this way, Ca2+ is the critical ion that couples membrane excitation with cardiocyte contraction, a process known as excitation-contraction coupling [47, 48].
Fig. (1). Cardiocyte calcium-induced calcium-release.
Ca2+ influx through sarcolemmal LTCCs triggers a large release of SR-stored Ca2+ by RyR2 which then triggers Ca2+-sensitive myofilament contraction. Contraction is terminated by Ca2+ uptake into the SR via SERCA2 or expulsion of Ca2+ into the extracellular space by NCX and PMCA. Blue circles represent Ca2+ while green circles represent Na+. Width of blue arrows is roughly proportional to the relative degree of Ca2+ flux.
Calcium-Handling Protein-Mediated HCM
Given the critical role of Ca2+ in excitation-contraction coupling and the expanding body of literature linking alterations in Ca2+-handling with hypertrophic remodeling, investigators have begun to explore whether mutations in genes encoding Ca2+-handling proteins might be associated with HCM. TNNC1, one of the first genes encoding a Ca2+ sensitive/handling protein to be firmly linked with HCM, was a logical target as it represents a Ca2+-sensitive element of the cardiac sarcomere (Fig. 2A). Further, it is part of the same thin filament macromolecular complex that several other canonical HCM myofilament proteins comprise. Since this first association, mutations in several other genes, including PLN, JPH2, RYR2, CASQ2, CALR3, and SRI, have been suggested to contribute to the pathogenesis of HCM to varying degrees (Tables 2 & 3). Given the complete absence of reported genetic variation in TNNC1 and PLN among healthy individuals to date, inclusion of these two genes in the clinically available genetic test may be warranted.
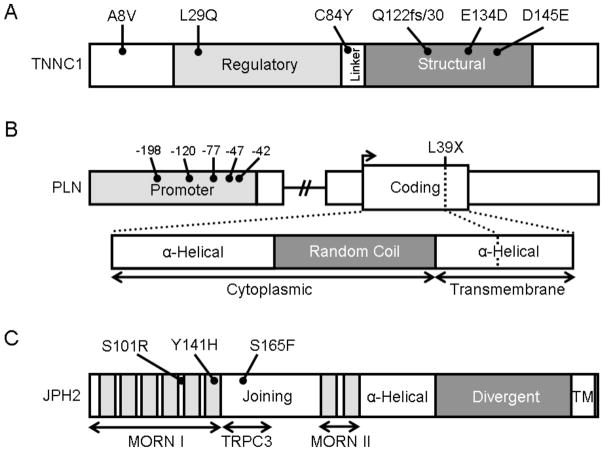
Fig. (2). Protein topology with all HCM-associated mutations for TNNC1, PLN, and JPH2.
A. Linear topology of the 6 exon-encoded, 161 amino acid TNNC1. Domains include the regulatory EF-hand (light gray), the linker, and the structural EF-hand domain (dark gray). B. Schematic of the proximal promoter (light gray), coding sequence, and protein topology of the 1 exon-encoded, 52 amino acid PLN. Domains of PLN include two α-helical domains separated by random coil (dark gray). The amino-terminal aspect of the protein is cytoplasmic while the carboxy-terminus is a hydrophobic, SR-associated domain. C. Linear topology of the 5 exon-encoded, 696 amino acid JPH2. Domains include the MORN I and II domains which each contain multiple MORN motifs (light gray) and are separated by the joining region domain. Contained within the joining region is the TRPC3 reciprocal binding domain identified in rodent models of skeletal muscle. A putative α-helical domain connects the MORN II domain with the divergent domain (dark gray) and a carboxy-terminal transmembrane domain (TM) associates with the SR.
Table 2.
Summary of Possible HCM-Associated Mutations Involving Calcium-Handling Proteins
| Mutation |
|---|
| Troponin C |
| A8V |
| L29Q |
| C84Y |
| Q122fs/30 |
| E134D |
| D145E |
| Junctophilin 2 |
| S101R |
| Y141H |
| S165F |
| (R436C) |
| (G505S) |
| Ryanodine Receptor 2 |
| (T1107M) |
| Calsequestrin 2 |
| (D63E) |
| Calreticulin 3 |
| (R73Q) |
| K82R |
| Sorcin |
| (F112L) |
| Phospholamban |
| (C>T −235) |
| A>C −198 |
| A>G −120 |
| (T>C −114) |
| A>G −77 |
| G>T −47* |
| C>G −42 |
| L39X* |
Parentheses indicate compound sarcomeric HCM-associated mutations identified in the proband, identification of the mutation in ostensibly healthy individuals, or unpublished.
, two mutation-positive probands identified.
Table 3.
Summary of Cohort Demographics
| Demographic/Feature | HCM | Myofilament | Calcium-Handling Proteins | |||
|---|---|---|---|---|---|---|
| Overall | TNNC1 | PLN | JPH2 | |||
| No. of probands | 388 | 186 | 17 | 6 | 8 | 3 |
| Proportion of cohort† | 100% | 47.8% | 1.64% | 0.40% | 0.47% | 0.77% |
| Proportion female | 44.7 | 44.6% | 35.3% | 16.7% | 50.0% | 33.3% |
| Age at diagnosis (yrs) | 41.2 ± 19 | 35.5 ± 16 | 41.3 ± 4 | 32.3 ± 9 | 53.5 ± 4 | 27.0 ± 2 |
| Maximum LVWT (mm) | 21.3 ± 6 | 22.8 ± 6 | 20.9 ± 1 | 19.5 ± 2 | 20.1 ± 2 | 26.0 ± 6 |
| Mean LVOTO (mmHg) | 44.5 ± 42 | 41.6 ± 41 | 34.0 ± 11 | 61.8 ± 21 | 21.9 ± 17 | 20.3 ± 11 |
| Family history of HCM | 35.2% | 48.9% | 47.1% | 50.0% | 50.0% | 33.3% |
| Family history of SCD | 21.2% | 27.7% | 11.8% | 33.3%‡ | 0% | 0% |
| Myectomy | 44.6% | 49.1% | 35.3% | 50.0% | 25.0% | 33.3% |
| ICD | 20.2% | 29.4% | 23.5% | 0% | 25.0% | 66.7% |
| Pacemaker | 24.9% | 24.6% | 11.8% | 0% | 12.5% | 33.3% |
No. of probands, the number of mutations-positive probands in a cohort of Mayo Clinic patients (HCM, Myofilament columns) and all mutation-positive probands not hosting compound sarcomeric mutations identified in the literature (Calcium-Handling Proteins columns).
, proportion of the Mayo Clinic HCM cohort; LVWT, maximum left ventricular wall thickness; LVOTO, left ventricular outflow tract obstruction; SCD, sudden cardiac death;
, SCD of two probands reported; ICD, implantable cardioverter defibrillator.
Phospholamban
Cardiocyte relaxation after contraction is an active process mediated by ATP-expending pumping of cytosolic Ca2+ into the SR lumen through the action of the ATPase SERCA2 or the exchange of Ca2+ out of the cell for Na+ through the function of NCX1. PLN is a highly regulated small protein binding partner of SERCA2 which, when bound, inhibits the Ca2+-reuptake action of SERCA2. This inhibitory action can be relieved by phosphorylation, and PLN serves as a substrate for the kinase-action of protein kinase A or calmodulin-dependent protein kinase 2 [49, 50]. Inhibition of PLN results in increased SERCA2-mediated Ca2+ removal from the cytosol and increased diastolic relaxation.
Rare mutations and common polymorphisms localizing to both the promoter and coding region of PLN have been associated with HCM (Fig. 2B). A handful of rare promoter variants have been identified in multiple independent cohorts of HCM patients including a C to T conversion at position −235 (C>T −235), A>C −198, A>G −120, T>C −114 [51], A>G −77 [52], G>T −47 (2 probands) [51], and C>G −42 [53]. Each of these rare variants was not found in ethnically-matched controls. Two of these probands, C>T −235 and T>C −144, hosted compound mutations in MYH7 and MYL2, respectively.
In addition to promoter variants, a mutation in the coding region of PLN has been associated with HCM. The nonsense mutation, PLN-L39X, was originally identified in a DCM proband and was found to cosegregate with incidence of both DCM and HCM in a large family [54]. Within this family, kindred hosting a homozygous L39X mutation demonstrated, or quickly progressed to, DCM/heart failure, while individuals hosting a heterozygous mutation were unaffected or demonstrated HCM. Subsequently, this mutation has been identified in two HCM probands from two independent cohorts of HCM index cases [51, 55]. In one study, the L39X mutation cosegregated in a small family demonstrating a solely HCM phenotype. To our knowledge, L39X represents the only HCM-associated PLN mutation which alters the protein sequence and may represent an HCM-associated mutation “hot spot” within the PLN primary sequence.
Overall, among studies exploring index case cohorts of HCM, PLN promoter and coding-region mutations were identified in ~0.50% of HCM probands which is similar to the frequency of TNNC1, TPM1, and ACTC-associated HCM mutations [27, 35]. As with TNNC1, comprehensive genetic interrogation of 600 reference alleles did not identify any rare promoter variants or amino acid substitutions which might be considered “false positive” results for genetic testing. Given these findings, as well as the relatively small size of the PLN genetic locus, inclusion of PLN on the genetic test for HCM appears reasonable.
Junctophilin 2
Junctophilins (JPHs, also known as JPs) are a family of proteins found in all excitable cells from striated muscle to neurons [56, 57]. JPHs contain multiple amino-terminal membrane occupation and recognition nexus (MORN) motifs which localize the amino-terminus of protein to the plasma membrane. A carboxy-terminal membrane-spanning domain embeds the opposing end into the ER/SR [58, 59]. In this way, through bridging the subcellular space between the plasma membrane and the ER/SR, JPHs play a critical role in the maintenance of effective Ca2+-handling.
JPH2 is the major JPH family member in the heart and plays a key role in maintaining the critical ultrastructural geometry of the cardiac dyad needed for effective CICR [60]. JPH2 null mice are embryonic lethal and demonstrate alteration of the cardiac dyad distance as well as vacuolization of the SR. Cardiocytes demonstrate stochastic contraction and irregular, smaller Ca2+ transients [59]. Rodent models of HCM, DCM, and pressure-induced hypertrophy/heart failure are associated with loss of JPH2 expression [61–63]. Recently, this observation of reduced JPH2 expression has been extended to humans with HCM. Expression silencing of JPH2 in vitro is sufficient to cause cellular hypertrophy and reduced CICR [64]. In vivo conditional knock-down JPH2 mice rapidly develop heart failure with disrupted cellular architecture and reduced excitation-contraction coupling gain [65].
Three mutations localizing to the amino terminus of JPH2 have been associated with a largely Caucasian HCM cohort of 388 individuals – S101R, Y141H, and S165F [66] (Fig. 2C). These mutations, localizing to the first MORN motif domain and the linker domain, were identified in three unrelated individuals negative for sarcomeric or Z-disc mutations. Each of these mutations were found to reduce CICR amplitude and disrupt cellular ultrastructure utilizing in vitro myocyte models, and in the case of Y141H and S165F, induce cellular hypertrophy [66]. These findings are in close agreement with the increase in cell size, induction of pro-hypertrophic transcriptional markers, and reduced CICR seen with JPH2 expression knock-down [64]. Subsequent studies in skeletal muscle found that the S165 residue is part of the reciprocal binding domain for the transient receptor potential cation channel 3 (TRPC3), a non-selective cation channel in the plasma membrane which allows for influx of extracellular Ca2+ and Na+ to enter the cell [67–69]. Phylogenic and biochemical analyses identified the S165 residue as a protein kinase C phosphorylation site which is abolished by the S165F mutation resulting in reduced binding to TRPC3, decreased CICR, and hypertrophy in skeletal myocytes [57, 70].
Two additional genetic variants localizing to the divergent domain, JPH2-R436C and G505S, have been reported in two and four unrelated individuals, respectively, within an independent Japanese HCM cohort [71]. Importantly, the R436C variant was also identified in two ostensibly healthy Japanese control individuals in the study and may represent a polymorphism rather than a true pathogenic mutation [71]. While the G505S variant was not originally identified in the healthy control Japanese cohort, we have identified this variant in ~0.5% of ostensibly healthy Caucasian Americans and ~2.0% of African Americans (unpublished). As is the case with the R436C variant, given the ~0.2% frequency of HCM in the general population and the relative rarity of non-sarcomeric gene mutations, G505S is unlikely to be a pathogenic mutation in isolation.
Overall, these studies indicate that mutations in JPH2 are a rare cause of HCM, found in less than 1% of index cases. Due to the presence of both common and rare genetic variation at this genetic locus in reference alleles, inclusion of JPH2 on the HCM genetic test panel at this time is likely to make interpretation of a positive test challenging. Despite this practical consideration, these mutations have catalyzed molecular insight into an enigmatic protein with a clear physiologic and pathophysiologic role in both cardiac and skeletal muscle. Additional studies are needed to further clarify the role of JPH2 dysregulation in hypertrophic remodeling as well as to elucidate the potential role of perturbed JPH2 in other cardiovascular diseases.
Ryanodine Receptor 2
The major molecule responsible for CICR and Ca2+-mediated myofilament contraction is RyR2. Heritable mutations in RYR2 have been associated with catecholaminergic polymorphic ventricular tachycardia (CPVT) [72, 73], sudden infant death syndrome [74], and arrhythmogenic right ventricular cardiomyopathy/dysplasia [75]. While it has been hypothesized to be a rare cause of HCM by many investigators, there has not been a definitive study associating mutations in RYR2 with HCM. To our knowledge, the only HCM-associated mutation described to date is the RyR2-T1107M mutation. Originally identified in an individual negative for sarcomeric mutations, this mutation was absent in ostensibly healthy controls and co-segregated with incidence of HCM upon pedigree analysis [76]. Interestingly, this proband also demonstrated a clinical history of ventricular fibrillation and this variant has been previously described in a cohort of CPVT cases [77]. In addition to this study, a recent linkage study involving a large family with genotype-negative HCM identified a locus containing the gene for alpha actinin 2 (ACTN2) and RYR2 [78]. While mutations in ACTN2 were found to co-segregate in this study, it is possible that compound mutations in the over 60 RYR2 coding exons not genotyped in this study may still exist.
Calsequestrin 2 and Calreticulin 3
RyR2-mediated SR Ca2+-release is a highly regulated event and several modulators, both cytosolic and SR luminal, have been shown to alter RyR2 gating. While the carboxy-terminal domain of each RyR2 monomer comes together to form a central pore, RyR2 has a large cytosolic amino-terminus which modulates its Ca2+-gating action. The cytosolic domain has multiple phosphorylation sites as well as binding sites for various interacting proteins which can either increase or decrease channel opening probability [79–84]. Similarly, the luminal side of RyR2 is regulated by modulatory proteins which are critical for the proper functioning of RyR2.
CASQ2-encoded calsequestrin 2 (CASQ2) serves as a low-affinity, high-capacity Ca2+ buffer within the SR [85]. While its role in modulating cellular Ca2+ homeostasis is unclear, it is known that CASQ2 functions as a Ca2+ sensor for RyR2 as well as a modulator of free Ca2+ levels in the store. Over-expression of CASQ2 has been linked to increased SR luminal Ca2+ load and induction of cardiac hypertrophy in animal models [86, 87]. While only a handful of CASQ2 mutations have been reported, nearly all have been associated with the development of CPVT in cases negative for mutations in RYR2 [88, 89]. To our knowledge, only a single CASQ2 mutation, D63E, has loosely been associated with HCM [55]. The small family found to be positive for CASQ2-D63E also hosted two compound mutations in the MYBPC3 (MYPBC3-R326Q and Q1233X) which also co-segregated with incidence of disease. The presence of these compound mutations provide an alternative genetic explanation for the pathogenesis of HCM in this family and thus decrease the candidacy of CASQ2-D63E as a HCM-causing mutation.
CALR3-encoded calreticulin 3 (CALR3) is another SR luminal protein which serves as a Ca2+-buffering chaperone that regulates SR store Ca2+ levels as well as modulates function of various Ca2+ channels and pumps [90]. Two mutations in CALR3, R73Q and K82R, have been identified in two unrelated probands with HCM within a cohort [55]. As with the CASQ2-D63E mutation, CALR3-R73Q was identified in a proband who also hosted two mutations in MYBPC3 (D745G and P873H) which clouds any association between this particular CALR3 variant and the development of HCM. Among these three identified mutations, only CALR3-K82R was identified in a proband negative for the mutations in the canonical sarcomeric genes.
Sorcin
SRI-encoded sorcin is a member of the penta-EF hand protein family which undergoes conformational change in response to Ca2+ allowing it to translocate to membranes, such as the t-tubular sarcolemma and SR, and dynamically modulate CICR [91, 92]. Sorcin has been shown to interact with and modulate several proteins, including LTCC and RyR2, and plays a role in providing negative feedback for the termination of CICR [93, 94]. The SRI-F112L mutation was initially identified in two unrelated families with HCM and hypertension and was found to be absent in 200 control individuals [95]. However, subsequent large genomic/bioinformatic studies have identified this variant in ostensibly healthy individuals, and this variant has been generally accepted as a polymorphism [96, 97]. Further, mice over-expressing sorcin F112L did not develop LV hypertrophy or systemic hypertension, demonstrating only mild LV chamber dilatation with intact contractility [98]. Despite this, F112L disrupts the tertiary structure of the protein by X-ray crystal structure analysis resulting in reduced inhibitory function of the protein on CICR [98–100]. These results support the possibility that sorcin-F112L in isolation is not associated with the pathogenesis of HCM. It is possible that this mutation creates a genetic “second hit” in which HCM and hypertension develop due to alterations in CICR in concert with another HCM-associated mutation. Additional studies examining SRI in large cohorts of HCM index cases may yield true disease-associated mutations which perturb the regulatory function of sorcin in a pro-hypertrophic manner.
FUTURE DIRECTIONS
While real progress has been made in identifying the cause of individuals with HCM who lack sarcomeric gene mutations, alternative genetic subtypes still account for only a small subset of these patients. Additional candidate gene analyses may be useful in elucidating those individuals hosting mutations localizing to proteins that are already known to influence hypertrophic remodeling of the heart. In particular, two Ca2+-sensitive pro-hypertrophic cascades centered around calmodulin (CaM)-dependent protein kinase 2 (CaMKII) and the phosphatase calcineurin are rational targets [101–103]. The Ca2+/CaM-dependent CaMKII can phosphorylate and inhibit histone deacetylase 4 (HDAC4), a repressor of hypertrophic remodeling. This repression removes the tonic inhibition of HDAC4 on the pro-hypertrophic transcription factor MEF2 [104–106]. In a similar pathway, calcineurin, a Ca2+-/CaM-dependent protein phosphatase, induces cardiocyte hypertrophy through activation of the transcription factor NFAT3 and its cofactor GATA4 which colocalize to the nucleus to activate a similar pro-hypertrophic gene profile [107, 108].
In addition to the candidate gene approach, linkage analysis still has a role in determining familial HCM. While most large families with HCM have likely been subjected to linkage analysis already, there are still occasional robust reports of this technique used to identify novel genetic loci of disease association. Perhaps more sensitive techniques utilizing this same methodological principal, such as genome-wide association studies (GWAS) or whole exome sequencing, will identify novel causes of HCM pathogenesis. Each of these techniques allow for an unbiased examination of the entire genome or transcribed exome. These techniques are not limited to small genetic loci like “first generation” sequencing and are more informative for relatively less cost than “next generation” sequencing modalities. These methods also provide the opportunity to examine regions of the genome that are transcribed but not translated into protein (i.e. 5′ and 3′ untranslated gene regions or genes of non-coding RNAs) and, in the case of GWAS analysis, introns and large intergenic regions as well. While a large number of cardiovascular disease GWAS studies have been conducted to date, few disease-locus associations have been replicated and fewer still have become “actionable” biomarkers which play a clear role in clinical evaluation or management of a patient [109]. Perhaps the comprehensive analyses of transcribed genetic loci afforded by whole exome sequencing will aid in the identification of disease-associated genetic perturbations which have evaded current GWAS.
CONCLUSIONS
HCM is relatively common heritable cardiovascular disease defined by marked genotypic and phenotypic heterogeneity. While mutations in the canonical genes encoding elements of the cardiac sarcomere represent the most common genetic subtypes of HCM, mutations in genes encoding Ca2+-handling proteins have been identified in some patients that are negative for the clinically available HCM genetic test. While these mutations are rare, identification of the mutations has shed light on the physiology of the cardiocyte, and when perturbed, the development of Ca2+-mediated pathologic remodeling. Absence of genetic “background noise” and independent identification of mutations in various HCM cohorts argue favorably for incorporation of TNNC1 and PLN into clinical genetic testing. Additional investigation into these genes, as well as identification of novel Ca2+-handling protein-encoding genes associated with HCM, is warranted to uncover novel mechanisms of HCM pathogenesis.
Acknowledgments
FUNDING SOURCES
A.P.L. is supported by the American Heart Association Predoctoral Fellowship. M.J.A. is an Established Investigator of the American Heart Association and is supported by NIH grants R01-HD42569 and P01-HL94291, Fondation Leducq Award to the ‘Alliance for Calmodulin Kinase Signaling in Heart Disease,’ and Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program.
Footnotes
CONFLICT OF INTEREST
M.J.A. is a consultant for Biotronik, Boston Scientific, Medtronic, St. Jude Medical, Inc., and Transgenomic. Intellectual property derived from M.J.A.’s research program resulted in license agreements in 2004 between Mayo Clinic Health Solutions (formerly Mayo Medical Ventures) and PGxHealth (formerly Genaissance Pharmaceuticals, now Transgenomic).
References
- 1.Teare D. Asymmetrical hypertrophy of the heart in young adults. Br Heart J. 1958;20:1–8. doi: 10.1136/hrt.20.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–9. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 3.Maron B, Roberts W, McAllister H, Rosing D, Epstein S. Sudden death in young athletes. Circulation. 1980;62:218–29. doi: 10.1161/01.cir.62.2.218. [DOI] [PubMed] [Google Scholar]
- 4.Maron B, Epstein S, Roberts W. Causes of sudden death in competitive athletes. J Am Coll Cardiol. 1986;7:204–14. doi: 10.1016/s0735-1097(86)80283-2. [DOI] [PubMed] [Google Scholar]
- 5.Bos JM, Ommen SR, Ackerman MJ. Genetics of hypertrophic cardiomyopathy: one, two, or more diseases? Curr Opin Cardiol. 2007;22:193–9. doi: 10.1097/HCO.0b013e3280e1cc7f. [DOI] [PubMed] [Google Scholar]
- 6.Maron BJ. Sudden death in young athletes. N Engl J Med. 2003;349:1064–75. doi: 10.1056/NEJMra022783. [DOI] [PubMed] [Google Scholar]
- 7.Maron BJ. Hypertrophic cardiomyopathy: A systematic review. JAMA. 2002;287:1308–20. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 8.Olivotto I, Cecchi F, Casey SA, Dolara A, Traverse JH, Maron BJ. Impact of atrial fibrillation on the clinical course of hypertrophic cardiomyopathy. Circulation. 2001;104:2517–24. doi: 10.1161/hc4601.097997. [DOI] [PubMed] [Google Scholar]
- 9.McKenna WJ, Borggrefe M, England D, Deanfield J, Oakley CM, Goodwin JF. The natural history of left ventricular hypertrophy in hypertrophic cardiomyopathy: an electrocardiographic study. Circulation. 1982;66:1233–40. doi: 10.1161/01.cir.66.6.1233. [DOI] [PubMed] [Google Scholar]
- 10.Spirito P, Maron BJ, Bonow RO, Epstein SE. Occurrence and significance of progressive left ventricular wall thinning and relative cavity dilatation in hypertrophic cardiomyopathy. Am J Cardiol. 1987;60:123–9. doi: 10.1016/0002-9149(87)90998-2. [DOI] [PubMed] [Google Scholar]
- 11.Jarcho JA, McKenna W, Pare JA, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–8. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 12.Geisterfer-Lowrance AA, Kass S, Tanigawa G, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 13.Morner S, Richard P, Kazzam E, et al. Identification of the genotypes causing hypertrophic cardiomyopathy in northern Sweden. J Mol Cell Cardiol. 2003;35:841–9. doi: 10.1016/s0022-2828(03)00146-9. [DOI] [PubMed] [Google Scholar]
- 14.Erdmann J, Daehmlow S, Wischke S, et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin Genet. 2003;64:339–49. doi: 10.1034/j.1399-0004.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 15.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80:739–44. doi: 10.1016/S0025-6196(11)61527-9. [DOI] [PubMed] [Google Scholar]
- 16.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–32. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 17.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80:463–9. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 18.Poetter K, Jiang H, Hassanzadeh S, et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–9. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 19.Olson TM, Karst ML, Whitby FG, Driscoll DJ. Myosin light chain mutation causes autosomal recessive cardiomyopathy with mid-cavitary hypertrophy and restrictive physiology. Circulation. 2002;105:2337–40. doi: 10.1161/01.cir.0000018444.47798.94. [DOI] [PubMed] [Google Scholar]
- 20.Watkins H, Conner D, Thierfelder L, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 21.Mogensen J, Klausen IC, Pedersen AK, et al. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103:R39–R43. doi: 10.1172/JCI6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thierfelder L, Watkins H, MacRae C, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 23.Kimura A, Harada H, Park JE, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–82. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 24.Satoh M, Takahashi M, Sakamoto T, Hiroe M, Marumo F, Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: Identification of a novel disease gene. Biochem Biophys Res Comm. 1999;262:411–7. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 25.Arimura T, Bos JM, Sato A, et al. Cardiac ankyrin repeat protein gene (ANKRD1) mutations in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54:334–342. doi: 10.1016/j.jacc.2008.12.082. [DOI] [PubMed] [Google Scholar]
- 26.Landstrom AP, Ackerman MJ. Mutation type is not clinically useful in predicting prognosis in hypertrophic cardiomyopathy. Circulation. 2010;122:2441–50. doi: 10.1161/CIRCULATIONAHA.110.954446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108:445–51. doi: 10.1161/01.CIR.0000080896.52003.DF. [DOI] [PubMed] [Google Scholar]
- 28.Van Driest SL, Vasile VC, Ommen SR, et al. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–10. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 29.Binder J, Ommen SR, Gersh BJ, et al. Echocardiography-guided genetic testing in hypertrophic cardiomyopathy: septal morphological features predict the presence of myofilament mutations. Mayo Clin Proc. 2006;81:459–67. doi: 10.4065/81.4.459. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Geßner R. First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat. 2001;17:524. doi: 10.1002/humu.1143. [DOI] [PubMed] [Google Scholar]
- 31.Schmidtmann A, Lindow C, Villard S, et al. Cardiac troponin C-L29Q, related to hypertrophic cardiomyopathy, hinders the transduction of the protein kinase A dependent phosphorylation signal from cardiac troponin I to C. FEBS Journal. 2005;272:6087–97. doi: 10.1111/j.1742-4658.2005.05001.x. [DOI] [PubMed] [Google Scholar]
- 32.Baryshnikova OK, Li MX, Sykes BD. Modulation of cardiac troponin C function by the cardiac-specific N-terminus of troponin I: Influence of PKA phosphorylation and involvement in cardiomyopathies. J Mol Biol. 2008;375:735–51. doi: 10.1016/j.jmb.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 33.Liang B, Chung F, Qu Y, et al. Familial hypertrophic cardiomyopathy-related cardiac troponin C mutation L29Q affects Ca2+ binding and myofilament contractility. Physiol Genomics. 2008;33:257–66. doi: 10.1152/physiolgenomics.00154.2007. [DOI] [PubMed] [Google Scholar]
- 34.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 35.Landstrom AP, Parvatiyar MS, Pinto JR, et al. Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations in TNNC1-encoded troponin C. J Mol Cell Cardiol. 2008;45:281–8. doi: 10.1016/j.yjmcc.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim CC, Yang H, Yang M, et al. A novel mutant cardiac troponin C disrupts molecular motions critical for calcium binding affinity and cardiomyocyte contractility. Biophys J. 2008;94:3577–89. doi: 10.1529/biophysj.107.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyer EC, Jacques AM, Hoskins AC, et al. Functional analysis of a unique troponin C mutation, GLY159ASP, that causes familial dilated cardiomyopathy, studied in explanted heart muscle. Circ Heart Fail. 2009;2:456–64. doi: 10.1161/CIRCHEARTFAILURE.108.818237. [DOI] [PubMed] [Google Scholar]
- 38.Gomes AV, Potter JD. Molecular and cellular aspects of troponin cardiomyopathies. Ann NY Acad Sci. 2004;1015:214–24. doi: 10.1196/annals.1302.018. [DOI] [PubMed] [Google Scholar]
- 39.Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD. Mutations in Troponin that cause HCM, DCM AND RCM: What can we learn about thin filament function? J Mol Cell Cardiol. 2010;48:882–92. doi: 10.1016/j.yjmcc.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 40.Pinto JR, Parvatiyar MS, Jones MA, Liang J, Ackerman MJ, Potter JD. A functional and structural study of troponin C mutations related to hypertrophic cardiomyopathy. J Biol Chem. 2009;284:19090–100. doi: 10.1074/jbc.M109.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swindle N, Tikunova SB. Hypertrophic cardiomyopathy-linked mutation D145E drastically alters calcium binding by the C-domain of cardiac troponin C. Biochemistry. 2010;49:4813–20. doi: 10.1021/bi100400h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung W, Kitner C, Maron B. Novel frameshift mutation in troponin C ( TNNC1) associated with hypertrophic cardiomyopathy and sudden death. Cardiol Young. 2011;2193:345–8. doi: 10.1017/S1047951110001927. [DOI] [PubMed] [Google Scholar]
- 43.Bos JM, Towbin JA, Ackerman MJ. Diagnostic, prognostic, and therapeutic implications of genetic testing for hypertrophic cardiomyopathy. J Am Coll Cardiol. 2009;54:201–11. doi: 10.1016/j.jacc.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 44.Bers D. Excitation-Contraction Coupling and Cardiac Contractile Force. Boston: Kluwer Academic Publisher; 2001. [Google Scholar]
- 45.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 46.Fabiato A. Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am J Physiol Cell Physiol. 1983;245:C1–14. doi: 10.1152/ajpcell.1983.245.1.C1. [DOI] [PubMed] [Google Scholar]
- 47.Wang S-Q, Song L-S, Lakatta EG, Cheng H. Ca2+ signalling between single L-type Ca2+ channels and ryanodine receptors in heart cells. Nature. 2001;410:592–96. doi: 10.1038/35069083. [DOI] [PubMed] [Google Scholar]
- 48.Stern MD, Song L-S, Cheng H, et al. Local control models of cardiac excitation-contraction coupling. J Gen Physiol. 1999;113:469–89. doi: 10.1085/jgp.113.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simmerman HK, Collins JH, Theibert JL, Wegener AD, Jones LR. Sequence analysis of phospholamban. Identification of phosphorylation sites and two major structural domains. J Biol Chem. 1986;261:1333341. [PubMed] [Google Scholar]
- 50.Inui M, Chamberlain BK, Saito A, Fleischer S. The nature of the modulation of Ca2+ transport as studied by reconstitution of cardiac sarcoplasmic reticulum. J Biol Chem. 1986;261:1794–800. [PubMed] [Google Scholar]
- 51.Landstrom A, Adekola B, Bos J, Ommen S, Ackerman M. PLN-encoded phospholamban mutation in a large cohort of hypertrophic cardiomyopathy cases: Summary of the literature and implications for genetic testing. Am Heart J. 2011;161:165–171. doi: 10.1016/j.ahj.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minamisawa S, Sato Y, Tatsuguchi Y, et al. Mutation of the phospholamban promoter associated with hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2003;304:1–4. doi: 10.1016/s0006-291x(03)00526-6. [DOI] [PubMed] [Google Scholar]
- 53.Medin M, Hermida-Prieto M, Monserrat L, et al. Mutational screening of phospholamban gene in hypertrophic and idiopathic dilated cardiomyopathy and functional study of the PLN - 42 C>G mutation. Eur J Heart Fail. 2007;9:37–43. doi: 10.1016/j.ejheart.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Haghighi K, Kolokathis F, Pater L, et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest. 2003;111:869–876. doi: 10.1172/JCI17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiu C, Tebo M, Ingles J, et al. Genetic screening of calcium regulation genes in familial hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2007;43:337–43. doi: 10.1016/j.yjmcc.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Weisleder N, Takeshima H, Ma J. Immuno-proteomic approach to excitation-contraction coupling in skeletal and cardiac muscle: Molecular insights revealed by the mitsugumins. Cell Calcium. 2008;43:1–8. doi: 10.1016/j.ceca.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garbino A, van Oort RJ, Dixit SS, Landstrom AP, Ackerman MJ, Wehrens XHT. Molecular evolution of the junctophilin gene family. Physiol Genomics. 2009;37:175–86. doi: 10.1152/physiolgenomics.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishi M, Mizushima A, Nakagawara K, Takeshima H. Characterization of human junctophilin subtype genes. Biochem Biophys Res Commun. 2000;273:920–7. doi: 10.1006/bbrc.2000.3011. [DOI] [PubMed] [Google Scholar]
- 59.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 60.Ziman AP, Gómez-Viquez NL, Bloch RJ, Lederer WJ. Excitation-contraction coupling changes during postnatal cardiac development. J Mol Cell Cardiol. 2010;48:379–86. doi: 10.1016/j.yjmcc.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minamisawa S, Oshikawa J, Takeshima H, et al. Junctophilin type 2 is associated with caveolin-3 and is down-regulated in the hypertrophic and dilated cardiomyopathies. Biochem Biophys Res Commun. 2004;325:852–856. doi: 10.1016/j.bbrc.2004.10.107. [DOI] [PubMed] [Google Scholar]
- 62.Xu M, Zhou P, Xu S-M, et al. Intermolecular failure of L-type Ca2 channel and ryanodine receptor signaling in hypertrophy. PLoS Biology. 2007;5:e21. doi: 10.1371/journal.pbio.0050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei S, Guo A, Chen B, et al. T-tubule remodeling during transition from hypertrophy to heart failure. Circ Res. 2010;107:520–531. doi: 10.1161/CIRCRESAHA.109.212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landstrom A, Kellen C, Dixit S, et al. Junctophilin-2 expression silencing causes cardiocyte hypertrophy and abnormal intracellular calcium-handling. Circ Heart Fail. 2011;4(2):214–23. doi: 10.1161/CIRCHEARTFAILURE.110.958694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Oort RJ, Garbino A, Wang W, et al. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice/clinical perspective. Circulation. 2011;123:979–988. doi: 10.1161/CIRCULATIONAHA.110.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Landstrom AP, Weisleder N, Batalden KB, et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol. 2007;42:1026–1035. doi: 10.1016/j.yjmcc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo JS, Kim DH, Allen PD, Lee EH. TRPC3-interacting triadic proteins in skeletal muscle. Biochem J. 2008;411:399–405. doi: 10.1042/bj20071504. [DOI] [PubMed] [Google Scholar]
- 68.Woo J, Hwang J, Ko J, Kim D, Ma J, Lee E. Glutamate at position 227 of junctophilin-2 is involved in binding to TRPC3. Mol Cell Biochem. 2009;328(1–2):25–32. doi: 10.1007/s11010-009-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee EH, Cherednichenko G, Pessah IN, Allen PD. Functional coupling between TRPC3 and RyR1 regulates the expressions of key triadic proteins. J Biol Chem. 2006;281:10042–10048. doi: 10.1074/jbc.M600981200. [DOI] [PubMed] [Google Scholar]
- 70.Woo JS, Hwang JH, Ko JK, et al. S165F mutation of junctophilin 2 affects Ca2+ signalling in skeletal muscle. Biochem J. 2010;427:125–134. doi: 10.1042/BJ20091225. [DOI] [PubMed] [Google Scholar]
- 71.Matsushita Y, Furukawa T, Kasanuki H, et al. Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J Hum Genet. 2007;52:543–548. doi: 10.1007/s10038-007-0149-y. [DOI] [PubMed] [Google Scholar]
- 72.Laitinen PJ, Brown KM, Piippo K, et al. Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation. 2001;103(4):485–90. doi: 10.1161/01.cir.103.4.485. [DOI] [PubMed] [Google Scholar]
- 73.Priori SG, Napolitano C, Tiso N, et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- 74.Tester DJ, Dura M, Carturan E, et al. A mechanism for sudden infant death syndrome (SIDS): Stress-induced leak via ryanodine receptors. Heart Rhythm. 2007;4:733–739. doi: 10.1016/j.hrthm.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tiso N, Stephan DA, Nava A, et al. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2) Hum Mol Genet. 2001;10:189–94. doi: 10.1093/hmg/10.3.189. [DOI] [PubMed] [Google Scholar]
- 76.Fujino N, Ino H, Hayashi K, et al. Abstract 915: A novel missense mutation in cardiac ryanodine receptor gene as a possible cause of hypertrophic cardiomyopathy: Evidence from familial analysis. Circulation. 2006;114:II-165. [Google Scholar]
- 77.Medeiros-Domingo A, Bhuiyan ZA, Tester DJ, et al. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either ctecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: A comprehensive open reading frame mutational analysis. J Am Coll Cardiol. 2009;54:2065–74. doi: 10.1016/j.jacc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiu C, Bagnall RD, Ingles J, et al. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: A genome-wide analysis. J Am Coll Cardiol. 2010;55:1127–35. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 79.Marx SO, Reiken S, Hisamatsu Y, et al. Phosphorylation-dependent regulation of ryanodine receptors. J Cell Biol. 2001;153:699–708. doi: 10.1083/jcb.153.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valdivia HH, Kaplan JH, Ellis-Davies GC, Lederer WJ. Rapid adaptation of cardiac ryanodine receptors: modulation by Mg2+ and phosphorylation. Science. 1995;267:1997–2000. doi: 10.1126/science.7701323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marx SO, Reiken S, Hisamatsu Y, et al. PKA phosphorylation dissociates FKBP12. 6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–76. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- 82.Bers DM, Eisner DA, Valdivia HH. Sarcoplasmic reticulum Ca2+ and heart failure: Roles of diastolic leak and Ca2+ transport. Circ Res. 2003;93:487–90. doi: 10.1161/01.RES.0000091871.54907.6B. [DOI] [PubMed] [Google Scholar]
- 83.Kaftan E, Marks AR, Ehrlich BE. Effects of rapamycin on ryanodine receptor/Ca2+-release channels from cardiac muscle. Circ Res. 1996;78:990–7. doi: 10.1161/01.res.78.6.990. [DOI] [PubMed] [Google Scholar]
- 84.Wehrens XHT, Lehnart SE, Reiken SR, Marks AR. Ca2+/calmodulin-dependent protein kinase II phosphorylation regulates the cardiac ryanodine receptor. Circ Res. 2004;94:e61–70. doi: 10.1161/01.RES.0000125626.33738.E2. [DOI] [PubMed] [Google Scholar]
- 85.Franzini-Armstrong C, Kenney LJ, Varriano-Marston E. The structure of calsequestrin in triads of vertebrate skeletal muscle: a deep-etch study. J Cell Biol. 1987;105:49–56. doi: 10.1083/jcb.105.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones LR, Suzuki YJ, Wang W, et al. Regulation of Ca2+ signaling in transgenic mouse cardiac myocytes overexpressing calsequestrin. J Clin Invest. 1998;101:1385–93. doi: 10.1172/JCI1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sato Y, Ferguson DG, Sako H, et al. Cardiac-specific overexpression of mouse cardiac calsequestrin is associated with depressed cardiovascular function and hypertrophy in transgenic mice. J Biol Chem. 1998;273:28470–7. doi: 10.1074/jbc.273.43.28470. [DOI] [PubMed] [Google Scholar]
- 88.Postma AV, Denjoy I, Hoorntje TM, et al. Absence of calsequestrin 2 causes severe forms of catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2002;91:e21–6. doi: 10.1161/01.res.0000038886.18992.6b. [DOI] [PubMed] [Google Scholar]
- 89.di Barletta MR, Viatchenko-Karpinski S, Nori A, et al. Clinical phenotype and functional characterization of CASQ2 mutations associated with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2006;114:1012–9. doi: 10.1161/CIRCULATIONAHA.106.623793. [DOI] [PubMed] [Google Scholar]
- 90.Nakamura K, Zuppini A, Arnaudeau S, et al. Functional specialization of calreticulin domains. J Cell Biol. 2001;154:961–72. doi: 10.1083/jcb.200102073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meyers MB, Zamparelli C, Verzili D, Dicker AP, Blanck TJJ, Chiancone E. Calcium-dependent translocation of sorcin to membranes: functional relevance in contractile tissue. FEBS Lett. 1995;357:230–4. doi: 10.1016/0014-5793(94)01338-2. [DOI] [PubMed] [Google Scholar]
- 92.Farrell E, Antaramian A, Rueda A, Gomez A, Valdivia H. Sorcin inhibits calcium release and modulates excitation-contraction coupling in the heart. J Biol Chem. 2003;278:34660–6. doi: 10.1074/jbc.M305931200. [DOI] [PubMed] [Google Scholar]
- 93.Meyers M, Puri T, Chien A, et al. Sorcin associates with the pore-forming subunit of voltage-dependent L-type Ca2+ channels. J Biol Chem. 1998;30:18930–5. doi: 10.1074/jbc.273.30.18930. [DOI] [PubMed] [Google Scholar]
- 94.Lokuta A, Meyers M, Sander P, Fishman G, Valdivia H. Modulation of cardiac ryanodine receptors by sorcin. J Biol Chem. 1997;272:25333–8. doi: 10.1074/jbc.272.40.25333. [DOI] [PubMed] [Google Scholar]
- 95.Mohiddin S, Antaramian A, Farrell E, et al. Abstract - A naturally-occurring sorcin missense mutation (F112L) is associated with hypertrophic cardiomyopathy, hypertension, and impaired modulation of cardiac ryanodine receptor. Circulation. 2002;106:II-319. [Google Scholar]
- 96.Buetow KH, Edmonson MN, Cassidy AB. Reliable identification of large numbers of candidate SNPs from public EST data. Nat Genet. 1999;21:323–5. doi: 10.1038/6851. [DOI] [PubMed] [Google Scholar]
- 97.Brookes AJ, Lehvaslaiho H, Siegfried M, et al. HGBASE: a database of SNPs and other variations in and around human genes. Nucl Acids Res. 2000;28:356–60. doi: 10.1093/nar/28.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Collis LP, Meyers MB, Zhang J, et al. Expression of a sorcin missense mutation in the heart modulates excitation-contraction coupling. FASEB J. 2007;21:475–87. doi: 10.1096/fj.06-6292com. [DOI] [PubMed] [Google Scholar]
- 99.Valdivia H, Farrell E, Antaramian A, et al. Sorcin and ryanodine receptors in heart failure. J Muscle Res Cell Motil. 2004;25:605–7. [PubMed] [Google Scholar]
- 100.Franceschini S, Ilari A, Verzili D, et al. Molecular basis for the impaired function of the natural F112L sorcin mutant: X-ray crystal structure, calcium affinity, and interaction with annexin VII and the ryanodine receptor. FASEB J. 2008;22:295–306. doi: 10.1096/fj.07-8988com. [DOI] [PubMed] [Google Scholar]
- 101.Maier LS, Bers DM. Calcium, calmodulin, and calcium-calmodulin kinase II: heartbeat to heartbeat and beyond. J Mol Cell Cardiol. 2002;34:919–39. doi: 10.1006/jmcc.2002.2038. [DOI] [PubMed] [Google Scholar]
- 102.Zhang T, Brown JH. Role of Ca2+/calmodulin-dependent protein kinase II in cardiac hypertrophy and heart failure. Cardiovasc Res. 2004;63:476–86. doi: 10.1016/j.cardiores.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 103.Houser S, Molkentin J. Does contractile Ca2+ control calcineurin-NFAT signaling and pathological hypertrophy in cardiac myocytes? Sci Signal. 2008;1:pe31. doi: 10.1126/scisignal.125pe31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Passier R, Zeng H, Frey N, et al. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J Clin Invest. 2000;105:1395–406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Little GH, Bai Y, Williams T, Poizat C. Nuclear calcium/calmodulin-dependent protein kinase II{delta} preferentially transmits signals to histone deacetylase 4 in cardiac cells. J Biol Chem. 2007;282:7219–31. doi: 10.1074/jbc.M604281200. [DOI] [PubMed] [Google Scholar]
- 106.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–88. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Molkentin JD, Lu J-R, Antos CL, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bourajjaj M, Armand A-S, da Costa Martins PA, et al. NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J Biol Chem. 2008;283:22295–303. doi: 10.1074/jbc.M801296200. [DOI] [PubMed] [Google Scholar]
- 109.Landstrom AP, Ackerman MJ. GWAS or Gee Whiz, PSAS or Pshaw: Elucidating the biologic and clinical significance of genetic variation in cardiovascular disease. Heart Rhythm. 2009;6:1751–3. doi: 10.1016/j.hrthm.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]