Graphical abstract
Keywords: Leishmaniasis, Polymerase chain reaction, Drug screening, Cell viability
Highlights
-
•
A duplex qRT-PCR was optimized for use in drug screening against Leishmania.
-
•
Drug activities were measured against the intracellular amastigote stage.
-
•
Cytotoxicity against the human host cells was simultaneously assessed.
-
•
Consistent and reproducible dose–response data were generated.
-
•
Suitability of this duplex PCR for anti-leishmanial drug screening was demonstrated.
Abstract
Currently available drugs for treatment of Leishmania infections are highly toxic and drug resistance to first line therapies has been observed. New, safer and more effective drugs are urgently needed to improve clinical resolution of the disease and reduce the risks associated with it. High-throughput screening of new compounds against cultured promastigotes is easy to perform, but the results are poorly predictive of in vivo efficacy. Intra-macrophage amastigote models provide a better proxy of the clinically relevant stage of disease and should be routinely implemented in the search for new anti-leishmanial agents, despite being labor intensive.
This study describes the use of a duplex quantitative Reverse-Transcriptase PCR (qRT-PCR) for assessment of drug activity against Leishmania intracellular amastigotes and their host cells. The assay simultaneously quantifies Leishmania 18S ribosomal RNA and the human β2-microglobulin (β-2M) mRNA, used for monitoring drug cytotoxicity and test performance. Accurate determination of parasite viability by the newly developed qRT-PCR was confirmed by parallel assessment of compound performance against standard microscopy. Highly reproducible anti-leishmanial activities were obtained with a set of structurally- and pharmacologically-diverse compounds, whose toxicity against host cells correlated with a low β-2M amplification. Sensitive and versatile, this duplex qRT-PCR offers a valuable tool for assessment of drug activities against Leishmania amastigotes and their host cells.
1. Introduction
Leishmaniasis is caused by protozoan parasites of the genus Leishmania. In humans, the parasite causes a wide spectrum of clinical syndromes, from self-healing skin ulcers to severe, life-threatening disease, following visceralization in the lymphoid organs. Approximately 12 million people are currently infected with the disease and an estimated 2 million new cases occur each year, of which 1.5 million of cutaneous leishmaniasis and 0.2–0.4 million of visceral leishmaniasis (Alvar et al., 2012). Chemotherapy remains the only effective way to treat all forms of disease, but current drugs are toxic, expensive and increasingly inefficacious due to parasite resistance, compelling the search for new anti-leishmanial agents (Grogl et al., 1992; Croft et al., 2006a; Monzote, 2009).
Development of new drugs relies on the screening of large numbers of compounds, for which robust and highly performing assays are required. Axenic, insect-stage promastigotes provide a simple model for evaluation of anti-leishmanial drug activity at high-throughput, but the resulting data have little value in animals, due to the metabolic and ecological differences that distinguish promastigotes from the mammalian amastigote stage (Croft et al., 2006b; Gupta and Nishi, 2011). Macrophages carrying Leishmania amastigotes resemble the clinically relevant phase of disease, and have therefore been recognized as most predictive of clinical efficacy (Gupta and Nishi, 2011; Seifert, 2011). Traditionally, drug activity against this stage is measured by microscopy counting of the percentage of infected macrophages and the number of amastigotes therein, a method which is labor-intensive and time-consuming (Sereno et al., 2007). In addition, establishment of parasite viability through staining procedures is difficult and often subjective, leading to potential inaccuracies in potency estimation (Gupta and Nishi, 2011). Recent introduction of reporter gene assays for engineered Leishmania spp. (Okuno et al., 2003; Ashuthos et al., 2005; Buckner and Wilson, 2005; Lang et al., 2005) has overcome this problem, but the use of parasite-recombinant markers that is associated with these techniques require stringent culturing conditions and prevent from assessing macrophage viability. Automated image-based assays provide a valuable alternative in this respect, enabling high-content examinations of compound activity against both Leishmania amastigotes and their host cells (Siqueira-Neto et al., 2012). Their implementation across non-dedicated facilities, however, remains limited, due to the sophisticated equipment required. PCR technology is nowadays widely available and has proven useful in monitoring anti-leishmanial drug efficacy at both in vitro and in vivo level (Dorlo et al., 2011; Sundarshan et al., 2011). Several real-time quantitative PCR (qPCR) assays have been developed for detection of viable Leishmania parasites and assessment of drug performance (Reimão et al., 2001; Ordóñez-Gutiérrez et al., 2009; Gomes et al., 2012), either through SYBR Green or fluorogenic probe technology. Importantly, these tests lacked an internal control that surveyed for test performance and monitored drug toxicity against the mammalian host cells.
The present study validates the use for drug screening purposes of a duplex Reverse-Transcriptase qPCR (qRT-PCR) simultaneously assessing for toxicity against Leishmania intracellular amastigotes and their host cells. Amplification of Leishmania-specific 18S ribosomal RNA was combined with the human β2-microglobulin (β2-M) mRNA, producing a sensitive and highly accessible tool to accurately measure anti-proliferative effects against the relevant stage of leishmaniasis.
2. Materials and methods
2.1. Compounds
Amphotericin B (Calbiochem, Nottingham, UK), miltefosine, paromomycin, H-89 dihydrochloride hydrate and imipramine hydrochloride (all from Sigma–Aldrich Co., St. Louis, USA) were used for pharmacological validation of the qRT-PCR, alongside with the newly synthesized compounds (VUF11852, VUF11854, VUF11856, VUF11857 and VUF13577) kindly provided by Dr. K. Orrling (VU University, Amsterdam, The Netherlands) within the framework of a collaborative project (T4-302, Ti Pharma) for developing new phosphodiesterase inhibitors as potential anti-leishmanial drugs (Orrling et al., 2012). Drug stock solutions (10 mM) were constituted in water, ethanol or DMSO, according to the compound solubility. Further dilutions of the drugs were performed in culture medium, possibly supplemented with the corresponding solvent to yield equal medium composition over the dilution series. For infected macrophages used as untreated controls (without drugs), DMSO, ethanol or water were added to the culture medium at the same concentrations as for the drugs.
2.2. Leishmania promastigotes
Leishmania donovani 1S promastigotes (MHOM/SD/68/1S) were maintained axenically at 27 °C, by serial passages in RPMI 1640 medium (Gibco, Bleiswijk, The Netherlands) supplemented with 25 mM HEPES, 2 mM l-glutamine and 10% fetal calf serum (FCS, Sigma–Aldrich Co.).
2.3. Leishmania intracellular amastigote model
THP-1 cells (human acute monocyte leukemia cell line) were cultured in RPMI 1640 medium supplemented with 25 mM HEPES, 2 mM l-glutamine, 10% FCS, penicillin (50 IU/ml) and streptomycin (50 μg/ml), to a density of 500,000 cells/ml. Cells (100 μl) were transferred to a 96-well culture plate for later testing by PCR and to a Lab Tek 16-well chamber-slide (Nunc, Waltham, USA) for microscopy counting. Next, 100 μl of cell culture medium containing 20 ng/ml of phorbol myristate acetate (PMA, Sigma–Aldrich Co.) were added to each well in order to differentiate the THP-1 cells into adherent macrophage-like cells. After 2 days of incubation at 37 °C, 5% CO2, cells were washed 3 times with culture medium to remove non-adherent cells. Infection of macrophages was performed with 200 μl of Leishmania donovani 1S promastigotes at a density of 2.5 million parasites/ml. After 18 h of incubation at 37 °C, 5% CO2, cells were washed to remove excess of promastigotes and further incubated for 3 days with culture medium containing the appropriate concentration of test compound. Untreated (containing no drugs) and uninfected (containing no parasites and no drugs) controls were included in all drug experiments using the same culture medium. Two independent experiments testing a total of 10 different compounds were performed in triplicate for PCR and in duplicate for microscopy.
2.4. Analysis of intracellular amastigote content by microscopy
After 72 h of incubation at 37 °C, 5% CO2, the slides were removed from the culture chambers, methanol-fixed and Field-stained. Their examination was performed with a light microscope (Leica, Rijswijk, The Netherlands), using a 100× objective. Results were expressed by mean of the parasite index (percentage of infected macrophages × mean number of amastigotes per macrophage), after examining a minimum of 100 macrophages per sample. Infection was judged to be adequate if more than 70% of the macrophages in the untreated control were infected.
2.5. Analysis of intracellular amastigote content by qRT-PCR
After removal of culture medium, 0.75 ml of Trizol® (Invitrogen, Bleiswijk, The Netherlands) was added to all wells for cell lysis and the lysates transferred to 1.5 ml tubes. RNA was Trizol®-extracted and purified according to manufacturer’s instructions. Residual DNA was further removed from all extracts by DNase digestion with Turbo DNase (Ambion, Invitrogen), performed according to the manufacturer’s instructions. A duplex qRT-PCR was performed for quantification of both Leishmania and human RNAs. The assay targeted the 18S small sub unit (SSU) rRNA conserved in Leishmania spp. and the human β-2M mRNA. The Leishmania qRT-PCR consisted of the forward primer 5′-CCAAAGTGTGGAGATCGAAG-3′, the reverse primer 5′-GGCCGGTAAAGGCCGAATAG-3′ and the probe 5′-6FAM-ACCATTGTAGTCCACACTGC-3′-NFQ conjugated to a minor groove binder (MGB) to increase the binding affinity of the probe to the target (van der Meide et al., 2008). For β-2M, the sense primer 5′-GGCTATCCAGCGTACTCCAA-3′ and the antisense primer 5′-GATGAAACCCAGACACATAGCA-3′ located within exon 1 and 2 (Mahoney et al., 2004) were used, together with the fluorescent probe 5′-GATGAAACCCAGACACATAGCA-3′, labeled at the 5′-terminus with the reporter dye Texas red and at the 3′-terminus with the quenching dye. Tenfold lower quantities of β-2M primers and probe with respect to the Leishmania 18S ones were added to the reaction mixture, to avoid sensitivity of the 18S PCR being affected by competition for β-2M amplification. For the PCR reaction, 1.25 μl of isolated RNA sample was added to 11.25 μl of amplification mix (Bio-Rad iScriptTM One-Step RT-PCR Kit for Probes, Hercules, USA) containing 2× PCR reaction reagent (20 mM Tris–HCl (pH 8.4), 50 mM KCl, 0.8 mM dNTPs, 3 mM MgCl2 and 0.6 U/μl iTaq DNA polymerase), reverse transcriptase, 0.8 μM of each Leishmania primer and 0.2 μM of FAM-MGB probe, 80 nM of each β-2M primer and 20 nM of probe. Amplification and real-time measurement were performed with the CFX-1 Cycler (Bio-Rad) using the following conditions: 10 min at 50 °C, 5 min at 95 °C followed by 45 cycles, each consisting of 30 s at 95 °C and 45 s at 60 °C.
Toxicity thresholds for macrophage viability were set according to 90% cytotoxic concentrations (CC90) of tested drugs, corresponding to a 3 Ct value increase in β-2M amplification over the controls. Consistent reduction of β-2M signal over the triplicate samples and its dose-related dependency from drug concentration confirmed low RNA contents being caused by drug toxicity rather than poor test performance. Inter-replicate variation >3 Ct values for either Leishmania 18S or β-2M amplification was considered indicative of poor RNA quality or extraction performance, resulting in the exclusion of these samples.
2.6. Data analysis
The inhibition of parasite growth was determined by comparing the amount of viable parasites detected in the drug-treated samples with that of the untreated controls. Non-linear regression analysis (Graph-Pad Software Inc., San Diego, USA) was used for curve fitting and calculation of 50% inhibitory concentrations (IC50). The relationship between the two assays was studied by use of the individual log IC50 values. Correlation between data obtained with the two assays was assessed by the Pearson’s correlation test, while the Bland and Altman plot was used to study the level of agreement between the two methods and to show dispersion of data (Bland and Altman, 1986).
3. Results
3.1. Performance of the duplex qRT-PCR
The qRT-PCR targeting the Leishmania 18S rRNA has previously been validated for quantification of Leishmania parasites (van der Meide et al., 2005). The standard curve for amplification was linear over a 6-fold log10 range of Leishmania donovani 1S RNA.
In the present study, a dilution series of Trizol®-extracted RNA from Leishmania donovani 1S promastigotes was used as a standard for the duplex qRT-PCR in a background of 100 ng of THP-1 cell RNA extract, which corresponds to the average amount of human RNA found in each micro-titer well seeded with Leishmania amastigotes. The Leishmania 18S amplification reaction was confirmed linear with a correlation coefficient of 0.995 over the range of 1 ng to 1 fg RNA, corresponding to 10,000–0.01 parasites (Fig. 1A, blue curves, and Fig. 1B). To exclude that sensitivity of the Leishmania 18S-PCR could be negatively affected by competition for β-2M amplification, the standard samples were tested with and without β-2M primers in the PCR mix. Comparison of the two reaction mixes showed equally good amplification of Leishmania 18S for all standard samples, with no loss of sensitivity due to competition for β-2M amplification (data not shown). The β-2M mRNA signals derived from amplification of background THP-1 cell RNA in the standard samples were all comparable (Ct values ranging from 23.3 to 23.7), regardless of the amount of Leishmania RNA present (Fig. 1A, red curves). Only the standard samples with the highest parasite content (1 ng and 100 pg parasite RNA) showed slightly reduced amplifications of β-2M (Ct values of 25.6 and 24.1, respectively), due to internal competition (Fig. 1A, red curves 1–2).
Fig. 1.
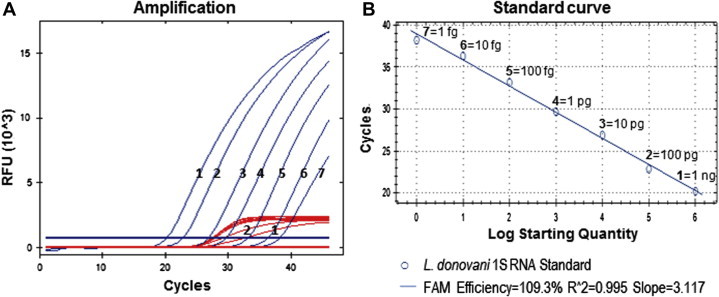
Duplex qRT-PCR targeting the Leishmania 18S rRNA (in blue) and the human β-2-microglobulin mRNA (in red): amplification curves (A) and linearity range of the Leishmania 18S amplification reaction (B) for a dilution series of Leishmania donovani 1S RNA ranging from 1 ng (1) to 1 fg (7) in a background of 100 ng of THP-1 cell RNA. Modest reduction of β-2M amplification (red curves 1–2) was observed in the presence of 1 ng and 100 pg of Leishmania RNA, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Pharmacological validation
To validate the use of the qRT-PCR for assessment of drug efficacy against Leishmania intracellular amastigotes, the PCR assay was compared with standard microscopy by a blind, side-by-side evaluation. A set of 10 compounds marked by different potencies and mechanisms of action was used to validate the performance of the duplex qRT-PCR. The compound panel included a series of protein kinase-, protein transporter- and phosphodiesterase-inhibitors (Orrling et al., 2012). Next to these compounds, three reference anti-leishmanial drugs were also assessed in the developed assay. A high correlation was found between the parasite load estimated by qRT-PCR and microscopy counting, regardless of which compound and concentration the parasites were exposed to (Fig. 2 and Table 1). The IC50 values yielded by the two assays during two independent experiments showed good agreement for most of the compounds tested (Table 1). Few discrepancies were observed (imipramine, VUF11852 and VUF11856), in which either the qRT-PCR or the microscopy data were marked by poor reproducibility with the replicate experiment. The Pearson’s correlation test confirmed a significant correlation between the log IC50 values obtained by the two assays (r = 0.95, n = 21, P <0.0001) (Fig. 3A). In the Bland and Altman plot (Fig. 3B), the mean difference in the log IC50 values of the 10 compounds yielded by the qRT-PCR and microscopy was −0.03 (limits of agreement −0.30 and 0.24). No tendency for a greater or smaller difference between the two methods was shown as the IC50 values increased.
Fig. 2.
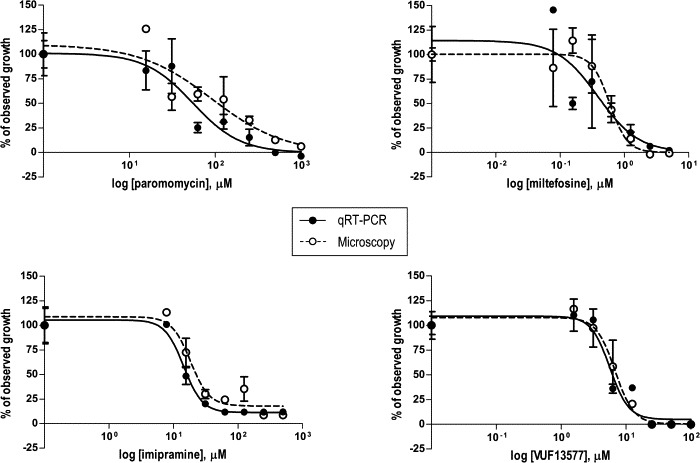
Drug dose–response curves of Leishmania donovani 1S intracellular amastigotes, as measured by the qRT-PCR assay (closed circles) and standard microscopy (open circles). Values are normalized by use of the upper and the lower best-fit values as 100% and 0% responses, respectively, and plotted as the means ± standard errors of the means of three replicates for the qRT-PCR assay and two replicates for microscopy. Representative data of one out of two experiments are shown.
Table 1.
Comparison of IC50 values (μM) of test drugs against Leishmania donovani 1S intracellular amastigotes, as measured by the qRT-PCR and microscopy in two independent experiments. Data are shown with three significant digits.
| Test compounds | qRT-PCR (μM) |
Microscopy (μM) |
||
|---|---|---|---|---|
| Exp. 1 | Exp. 2 | Exp. 1 | Exp. 2 | |
| Amphotericin B | 0.0590 | 0.233 | 0.0200 | 0.180 |
| Miltefosine | 1.19 | 0.639 | 0.820 | 0.590 |
| Paromomycin | 81.8 | 53.4 | 41.9 | 112 |
| H-89 | 10.8 | 6.90 | 14.6 | 9.92 |
| Imipramine | 25.6 | 5.50 | 17.8 | 19.1 |
| VUF 11852 | 21.0 | 27.6 | 8.93 | 31.6 |
| VUF 11854 | 15.0 | 13.3 | 13.5 | 16.7 |
| VUF 11856 | 4.82 | 27.8 | 20.6 | 40.9 |
| VUF 11857 | 4.39 | 5.88 | 6.21 | 7.95 |
| VUF 13577 | 6.70 | 9.01 | 7.34 | 7.05 |
Fig. 3.
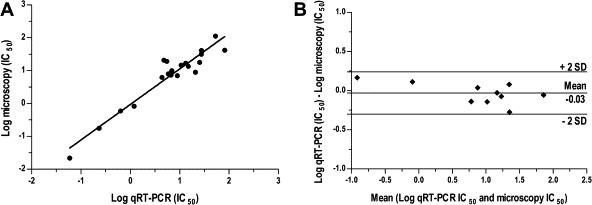
(A) Correlation between the logarithm of the individual IC50 values (in μM) measured by the qRT-PCR and the microscopy assays for 10 different compounds (n = 21; r = 0.95; P <0.0001). (B) Bland and Altman graph of the mean log IC50 values, as determined by qRT-PCR and microscopy for 10 different compounds, plotted against their log IC50 difference. The estimated mean difference (−0.03) and its confidence interval (± 2 standard deviation) are shown as horizontal lines.
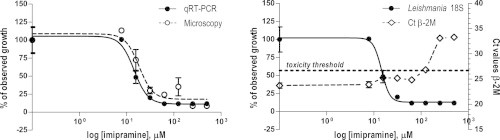
The mean Ct value for β-2M amplification ranged from 24.5 in the first experiment to 22.7 in the second experiment for untreated Leishmania-harboring macrophages and uninfected cells. When serial dilutions of the test compounds were incubated with the Leishmania-infected cells, a dose-related increase in β-2M Ct values could be observed (Fig. 4), suggesting reduced viability of macrophages as a result of drug exposure. The cytotoxic effect displayed by these compounds and indicated by an at least 10-fold lower β-2M amplification (corresponding to ⩾3 Ct value increase over the controls) corroborated microscopy observation, whereby poor or no cell morphology was confirmed (data not shown). Based on these microscopy observations, a 10-fold lower β-2M amplification was considered indicative of overall cell toxicity.
Fig. 4.
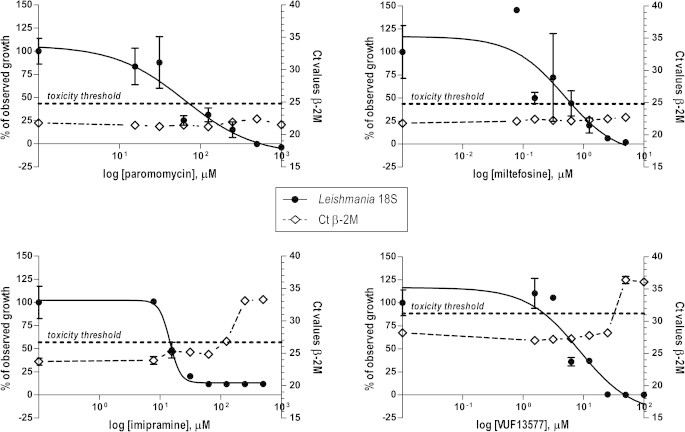
Drug dose–response curves of Leishmania donovani 1S intracellular amastigotes, as measured by the qRT-PCR assay (closed circles, left y axis) and the corresponding Ct values for β-2M amplification (open rhombus, right y axis). Values indicating Leishmania growth (closed circles) are normalized by use of the upper and the lower best-fit values as 100 and 0% responses, respectively. Data are plotted as the means ± standard errors of the means of three replicates for the qRT-PCR assay and two replicates for microscopy. Representative data of one out of two experiments are shown.
4. Discussion
In vitro amastigote-based assays for screening of novel anti-leishmanial compounds have been widely recognized as highly predictive of in vivo drug efficacy, although experimental constrains still limit their broad implementation (Gupta and Nishi, 2011). New technologies for improving the quality of these assays have been developed, including several molecular tools for evaluation of anti-leishmanial drug activities in both in vitro and in vivo models. The present work describes a further advancement of these genetic assays, in which viability of Leishmania intracellular forms, the amastigotes, and their host cells is simultaneously assessed by a duplex qRT-PCR, targeting the Leishmania-specific 18S rRNA and the human β-2M mRNA, which serves as internal control for test performance and drug cytotoxicity. Pharmacological validation of the assay, performed with a 10-compound set blindly assessed against standard microscopy, resulted in a good correlation between anti-leishmanial dose–response curves obtained by the two assays and the corresponding IC50 values, even when considering the highly subjective and inaccurate nature of microscopy scoring (Gupta and Nishi, 2011). Duplicate experiments showed fair intra-assay agreement for both PCR and microscopy, although for few compounds, a 4- to 5-fold difference in IC50 values was observed, possibly caused by differences in the infection load. Intra-assay variation of parasite burdens is not uncommon when using this amastigote-macrophage model, as parasites need to actively enter the host cells and proliferate therein, upon conversion into amastigotes. This may have important implications for the IC50 outcome, as confirmed by the critical role played by the number of promastigotes used to infect the cells, the particular Leishmania strain they belong to and the cell line bearing the infection (Seifert et al., 2010).
Compared to alternative drug screening assays, microscopy counting offers the advantage of enabling host cell morphology to be visualized, so that both anti-leishmanial activity and drug cytotoxicity can be simultaneously assessed. In this respect, this duplex qRT-PCR represents a valuable alternative, as it allows for simultaneous detection of parasite and cell viability. Substantial drug cytotoxicity, microscopically confirmed by the poor morphology of human macrophages, was detected by the qRT-PCR, which yielded an at least ten-fold reduced β-2M amplification rate as opposed to the untreated controls.
Extensive debate on the use of DNA over RNA for drug resistance studies (Reimão et al., 2001; van der Meide et al., 2008; Romero et al., 2010) has resulted in the general belief that DNA remains stable over a long period of time, pending on environmental conditions, and it is therefore unsuited for testing viability of micro-organisms, including Leishmania spp. Our preliminary experiments conducted with leishmanicidal doses of amphotericin B indicated that 18S DNA levels of Leishmania donovani 1S-infected macrophages differed less than a factor 10 from the untreated controls (data not shown), whereas a 1000-fold difference in 18S rRNA content was observed when the same samples were assayed by qRT-PCR and microscopy. Though in agreement with the general opinion, these findings contradict what was previously observed by others, who used real-time PCR to amplify DNA extracts and assess anti-leishmanial drug activity. Gomes et al. (2012) targeted the Leishmania-18S DNA in a Leishmania infantum amastigote-mouse model, obtaining amphotericin B IC50 values (0.02 μM) that corroborate our and previously published results. Whether these data resulted from a comparable reduction in parasite amount, however, is unknown since the report does not mention absolute parasite numbers. Ordóñez-Gutiérrez et al. (2009) were able to demonstrate by PCR only a 3-fold decrease in Leishmania amastigote DNA content, as opposed to the 10-fold difference observed by microscopy during exposure to cystatin. Finally, Prina et al. (2007) also used a DNA-based protocol for testing parasite viability and thereby demonstrate that Leishmania DNA is rapidly degraded in vitro following parasite death. These observations, however, were based on the use of l-Leucine methyl esther only, an experimental drug inducing rapid death of Leishmania mexicana and Leishmania amazonensis spp. Commercially available anti-leishmanial drugs, such as those tested in this study, are instead known to act progressively over a longer period of time (Monzote, 2009), resulting in potentially different effects on parasite/host cell viability and corresponding metabolisms. Furthermore, the use in the three above-mentioned studies of mouse cells as a macrophage source rather than the human THP-1 cell line employed here, may partially account for the discrepancies observed, given that profound differences between rodent and human macrophages have been described (Murray and Wynn, 2011).
The present study was conducted on a selection of compounds, for which comparable anti-leishmanial activities were obtained by qRT-PCR and microscopy. Further evaluation of the assay is now required to establish its performance with different Leishmania models and testing compounds. Efforts to automatize the technique are also equally crucial to ensure this qRT-PCR may be suitable for high-throughput drug screening. Accurate RNA isolation is key to this assay, but several automated RNA extraction systems capable of yielding high-quality RNA are nowadays available on the market. This opens important avenues for the further development of this assay.
In conclusions, the present results demonstrate the value of this qRT-PCR as a sensitive and reliable tool for assessing drug activities against Leishmania intracellular amastigotes and their host cells, simplifying the use of the clinically relevant stage of leishmaniasis for drug discovery programs.
Acknowledgments
The authors would like to thank Dr. Kristina Orrling (VU University, Amsterdam, The Netherlands) for kindly providing the VUF compound series of phosphodiesterase inhibitors and Dorien Faber (La Isla Foundation, León, Nicaragua) for conducting the early PCR assay development. This work was supported by the Top Institute Pharma (The Netherlands) and conducted within the framework of the Project T4-302 “Phosphodiesterase inhibitors for Neglected Tropical Diseases”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that no competing interests exist.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Erika van den Bogaart, Email: E.v.d.Bogaart@kit.nl.
Gerard J. Schoone, Email: G.Schoone@kit.nl.
Emily R. Adams, Email: E.Adams@liverpool.ac.uk.
Henk D.F.H. Schallig, Email: H.Schallig@kit.nl.
References
- Alvar J., Vélez I.D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., den Boer M., the WHO Leishmaniasis Control Team Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5):e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashuthos, Gupta S., Ramesh, Sundar S., Goyal N. Use of Leishmania donovani field isolates expressing the luciferase reporter gene in in vitro drug screening. Antimicrob. Agents Chemother. 2005;49:3776–3783. doi: 10.1128/AAC.49.9.3776-3783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- Buckner F.S., Wilson A.J. Colorimetric assay for screening compounds against Leishmania amastigotes grown in macrophages. Am. J. Trop. Med. Hyg. 2005;72(5):600–605. [PubMed] [Google Scholar]
- Croft S.L., Sundar S., Fairlamb A.H. Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 2006;19(1):111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft S.L., Seifert K., Yardley V. Current scenario of drug development for leishmaniasis. Indian J. Med. Res. 2006;123(3):399–410. [PubMed] [Google Scholar]
- Dorlo T.P.C., Van Thiel P.P., Schoone G.J., Stienstrsa Y., van Vugt M., Beijnen J.H., de Vries P.J. Dynamics of parasite clearance in cutaneous leishmaniasis patients treated with miltefosine. PLoS Negl. Trop. Dis. 2011;5(12):e1436. doi: 10.1371/journal.pntd.0001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L.I., Gonzaga F.M., de Morais-Teixeira E., de Souza-Lima B.S., Freire V.V., Rabello A. Validation of quantitative real-time PCR for the in vitro assessment of antileishmanial drug activity. Exp. Parasitol. 2012;131(2):175–179. doi: 10.1016/j.exppara.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Grogl M., Thomason T.N., Franke E.D. Drug resistance in leishmaniasis: its implication in systemic chemotherapy of cutaneous and mucocutaneous disease. Am. J. Trop. Med. Hyg. 1992;47:117–126. doi: 10.4269/ajtmh.1992.47.117. [DOI] [PubMed] [Google Scholar]
- Gupta S., Nishi Visceral leishmaniasis: experimental models for drug discovery. Indian J. Med. Res. 2011;133:27–39. [PMC free article] [PubMed] [Google Scholar]
- Lang T., Goyard S., Lebastard M., Milon G. Bioluminescent Leishmania expressing luciferase for rapid and high throughput screening of drugs on amastigote-harbouring macrophages and for quantitative real-time monitoring of parasitism features in living mice. Cell Microbiol. 2005;7:383–392. doi: 10.1111/j.1462-5822.2004.00468.x. [DOI] [PubMed] [Google Scholar]
- Mahoney D., Carey K., Fu M.H., Snow R., Parise G., Tarnopolsky M. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol. Genomics. 2004;18(2):226–231. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- Monzote L. Current treatment of leishmaniasis: a review. Open Antimicrob. Agents J. 2009;1:9–19. [Google Scholar]
- Murray P., Wynn T. Obstacles and opportunities for understanding macrophage polarization. J. Leukocyte Biol. 2011;89(4):557–563. doi: 10.1189/jlb.0710409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno T., Goto Y., Matsumoto Y., Otsuka H., Matsumoto Y. Applications of recombinant Leishmania amazonensis expressing EGFP and the β-galactosidase gene for drug screening and histopathological analysis. Exp. Anim. 2003;52(2):109–118. doi: 10.1538/expanim.52.109. [DOI] [PubMed] [Google Scholar]
- Ordóñez-Gutiérrez L., Martínez M., Rubio-Somoza I., Díaz I., Mendez S., Alunda J.M. Leishmania infantum: antiproliferative effect of recombinant plant cystatins on promastigotes and intracellular amastigotes estimated by direct counting and real-time PCR. Exp. Parasitol. 2009;123(4):341–346. doi: 10.1016/j.exppara.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Orrling K.M., Jansen C., Vu X.L., Balmer V., Bregy P., Shanmugham A., England P., Bailey D., Cos P., Maes L., Adams E., van den Bogaart E., Chatelain E., Ioset J.R., van de Stolpe A., Zorg S., Veerman J., Seebeck T., Sterk G.J., de Esch I.J., Leurs R. Catechol pyrazolinones as trypanocidals: fragment-based design, synthesis, and pharmacological evaluation of nanomolar inhibitors of trypanosomal phosphodiesterase B1. J. Med. Chem. 2012;55(20):8745–8756. doi: 10.1021/jm301059b. [DOI] [PubMed] [Google Scholar]
- Prina E., Roux E., Mattei D., Milon G. Leishmania DNA is rapidly degraded following parasite death: an analysis by microscopy and real-time PCR. Microbes Infect. 2007;9:1307–1315. doi: 10.1016/j.micinf.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Reimão J.Q., Colombo F.A., Pereira-Chioccola V.L., Tempone A.G. In vitro and experimental therapeutic studies of the calcium channel blocker bepridil: detection of viable Leishmania (L.) chagasi by real-time PCR. Exp. Parasitol. 2001;128(2):111–115. doi: 10.1016/j.exppara.2011.02.021. [DOI] [PubMed] [Google Scholar]
- Romero I., Téllez J., Suárez Y., Cardona M., Figueroa R., Zelazny N. Viability and burden of Leishmania in extralesional sites during human dermal leishmaniasis. PLoS Negl. Trop. Dis. 2010;4(9):e819. doi: 10.1371/journal.pntd.0000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K. Structures, targets and recent approaches in anti-leishmanial drug discovery and development. Open Med. Chem. J. 2011;5:31–39. doi: 10.2174/1874104501105010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K., Escobar P., Croft S.L. In vitro activity of anti-leishmanial drugs against Leishmania donovani is host cell dependent. J. Antimicrob. Chemother. 2010;65(3):508–511. doi: 10.1093/jac/dkp500. [DOI] [PubMed] [Google Scholar]
- Sereno D., Cordeiro da Silva A., Mathieu-Daude F., Ouaissi A. Advances and perspectives in Leishmania cell based drug-screening procedures. Parasitol. Int. 2007;56(1):3–7. doi: 10.1016/j.parint.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Siqueira-Neto J.L., Moon S., Jang J., Yang G., Lee C., Moon H.K., Genovesio A., Cechetto J., Freitas-Junior L.H. An image-based high content screening assay for compounds targeting intracellular Leishmania donovani amastigotes in human macrophages. PLoS Negl. Trop. Dis. 2012;6(6):e1671. doi: 10.1371/journal.pntd.0001671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundarshan M., Weirather J.L., Wilson M.E., Sundar S. Study of parasite kinetics with antileishmanial drugs using real-time quantitative PCR in Indian visceral leishmaniasis. J. Antimicrob. Chemother. 2011;66(8):1751–1755. doi: 10.1093/jac/dkr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meide W., Schoone G., Faber W., Zeegelaar J., De Vries H., Özbel Y., Lai A., Fat R., Leíla I., Coelho L., Kassi M., Schallig H. Quantitative nucleic acid sequence-based assay as a new molecular tool for detection and quantification of Leishmania parasites in skin biopsy samples. J. Clin. Microbiol. 2005;43(11):5560–5566. doi: 10.1128/JCM.43.11.5560-5566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meide W., Guerra J., Schoone G., Farenhorst M., Coelho L., Faber W., Peekel I., Schallig H. Comparison between quantitative nucleic acid sequence-based amplification, real-time reverse transcriptase PCR, and real-time PCR for quantification of Leishmania parasites. J. Clin. Microbiol. 2008;46(1):73–78. doi: 10.1128/JCM.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


