Graphical abstract
Keywords: Leishmania braziliensis, Antimony, Resistance
Highlights
-
•
Functional cloning identifies a Leishmania-specific 58 kD antimony resistance marker, ARM58.
-
•
ARM58 overexpression confers antimony resistance.
-
•
A neighbouring, structurally related gene lacks antimony resistance function.
-
•
ARM58 comprises 4 related domains of unknown function, classified as DUF1935.
-
•
Cosmid-based functional cloning is feasible in L. braziliensis in spite of RNAi.
Abstract
Protozoa of the Leishmania genus cause a variety of disease forms that rank at the top of the list of neglected tropical diseases. Anti-leishmanial drugs based on pentavalent antimony have been the mainstay of therapy for over 60 years and resistance against them is increasingly encountered in the field. The biochemical basis for this is poorly understood and likely diverse. No stringent correlation between genetic markers and antimony resistance has so far been shown, prompting us to use a functional cloning approach to identify markers of resistance. Using gene libraries derived from drug-resistant and drug-sensitive Leishmania braziliensis clinical isolates in a functional cloning strategy, we repeatedly selected one gene locus located on chromosome 20 whose amplification confers increased antimony (III) resistance in vitro to an otherwise sensitive L. braziliensis clone. The gene responsible for the effect encodes a previously hypothetical protein that we dubbed LbrARM58. It comprises four repeats of a domain of unknown function, DUF1935, one of them harbouring a potential trans-membrane domain. The gene is so far unique to the Leishmania genus, while a structurally related gene without antimony resistance functionality is also found in Trypanosoma spp. Overexpression of LbrARM58 also confers antimony resistance to promastigotes and intracellular amastigotes of the related species Leishmania infantum, indicating a conserved function in Old World and New World Leishmania species. Our results also show that in spite of their RNAi system, L. braziliensis promastigotes can serve as acceptor cells for episomally propagated cosmid libraries, at least for the initial stages of functional cloning efforts.
1. Introduction
Leishmaniasis is a parasitic disease caused by protozoa of the genus Leishmania and found on four continents with an estimated annual incidence of between 900,000 and 1.8 million cases (Alvar et al., 2012). Leishmaniasis is a poverty related disease with many endemic countries lacking the infrastructure and funds for effective treatment and control measures. The clinical manifestations range from the generalised visceral leishmaniasis (VL) or Kala-Azar, to cutaneous leishmaniasis (CL) and mucocutaneous infections (MCL) and are mostly determined by the infecting Leishmania species.
Therapy against leishmaniasis still relies heavily on two formulations of pentavalent antimony (SbV), meglumine antimoniate (Glucantime®) and sodium stibogluconate (Pentostam®). Developed in the first half of the twentieth century, these compounds are still the mainstay of therapy in most endemic regions. Resistance against SbV has been on the rise since the 1970s. For Indian Kala-azar, the high rate of antimonial treatment failure (>60%) has all but eliminated those cost-effective drugs from the arsenal of clinicians (Sundar, 2001; Croft et al., 2006).
Pentavalent antimony, SbV, is assumed to be a pro-drug, requiring reduction to SbIII either by the parasite or by the host cell. SbV has very little toxicity for promastigotes of Leishmania donovani, apparently due to their inability to reduce SbV, whilst amastigotes are susceptible to varying degrees depending on their reducing capacity (Roberts et al., 1995; Shaked-Mishan et al., 2001). In L. tarentolae, a non-human pathogenic species, increased levels of trypanothion were found to aid in the detoxification of SbIII (Haimeur et al., 1999), as did increased levels of P-glycoproteins that act as extrusion pumps for SbIII (Grondin et al., 1997; Ouellette et al., 1998). Conversely, the reduction of aquaglyceroporin 1 levels is linked to reduced uptake of SbIII and increased resistance in L. major (Gourbal et al., 2004).
The diversity of pathways linked to antimony resistance is further compounded by a recent finding that some L. donovani isolates from treatment failure cases can induce expression of host cell multi-drug resistance genes (Mookerjee Basu et al., 2008; Mukherjee et al., 2013), thus relying on host cell functions for resistance to therapy.
Compared with Northern India, treatment failure of antimony-based drugs is less prevalent in South America. One of the chief disease agents in Brazil, L. (Viannia) braziliensis, is known to be sensitive to SbV treatment in vitro and responsive in the clinical practice (Croft et al., 2006; Azeredo-Coutinho et al., 2007). Nevertheless, primary antimony therapy has a failure rate of ∼20–25%, depending on the endemic region (Soto et al., 2005; Arevalo et al., 2007; Llanos-Cuentas et al., 2008), and sensitivity varies between patient isolates, even before the start of the treatment (Yardley et al., 2006; Azeredo-Coutinho et al., 2007). Infection with L. braziliensis was associated with the highest antimonial treatment failure rate (30.4%) among Peruvian patients with cutaneous leishmaniasis (Arevalo et al., 2007). The correlation of in vitro drug susceptibility with therapeutic success is still debated as the evaluation of antimony susceptibility by in vitro macrophage-amastigote assays does not necessarily reflect the in vivo situation.
In L. braziliensis, natural antimony resistance does not appear to be linked to phylogenetic markers, but emerges independently in different lineages of this species (Adaui et al., 2011b). Additional analyses show that expression of known and suspected marker genes for SbIII/SbV resistance display isolate-specific fluctuations, possibly representing a great clonal variety (Adaui et al., 2011a). Differential gene expression analysis of 13 genes in 21 L. braziliensis isolates showed a significant correlation between drug resistance and elevated expression of ornithine carboxylase and trypanothion reductase, but not stringent correlations (Adaui et al., 2011c). Therefore, as in L. donovani, antimony resistance in L. braziliensis is a multi-gene trait resulting from a variety of molecular mechanisms.
However, there may be more marker genes that are yet unidentified and whose nature may shed light on the pathways leading to drug resistance. A successful strategy employed previously involved the in vitro selection for SbIII or SbV tolerant parasite populations that were subsequently subjected to an expression analysis for candidate genes. Unanticipated resistance genes cannot be detected in this approach. Likewise, genetic linkage analyses cannot detect random variations of gene expression within populations and isolates. Other, non-hypothesis-driven strategies need to be implemented, such as quantitative genomics and functional cloning.
Leishmania parasites are unicellular protozoa of the order Kinetoplastida. Typical of this order is the lack of gene-specific transcription regulation (Clayton, 2002), a unique feature among the Eukaryota. Most Leishmania genes are transcribed as multi-cistronic pre-mRNAs that are subject to subsequent processing through trans-splicing and polyadenylation. Generally, protein coding genes are not interrupted by introns. This fact makes gDNA cosmid libraries the tool of choice for functional cloning in Old World Leishmania (Clos and Choudhury, 2006). By contrast, the use of selectable episomes is not feasible in the related Trypanosoma brucei (Clayton, 1999), possibly due to the existence of an RNA silencing system (Ullu et al., 2004). Recent evidence indicates that L. (Viannia) braziliensis also possesses the genes encoding key proteins of RNA silencing (Peacock et al., 2007), and is susceptible to RNA interference (Lye et al., 2010). This was unknown at the onset of our study.
We have recently used a functional cloning strategy (Clos and Choudhury, 2006) to search for genetic markers of drug resistance in Leishmania infantum. Promastigotes were transfected with a shuttle cosmid library of L. infantum genomic DNA and challenged with the antileishmanial drug, Miltefosine. The two cosmids selected overlapped in a single gene which was shown subsequently to confer increased Miltefosine and SbIII resistance upon overexpression (Choudhury et al., 2008).
Expanding on the same strategy, we attempted to identify genes that may differ between antimony resistant and antimony sensitive Leishmania clinical isolates. Antimony resistance might arise in a stepwise manner, first to SbV and then to SbIII (Yardley et al., 2006; Rijal et al., 2007). This is supported by our observation of three combinations of in vitro antimony susceptibility phenotypes among field isolates: (i) parasites sensitive or (ii) tolerant to both SbV and SbIII, and (iii) parasites tolerant to SbV only (the majority) (Yardley et al., 2006; Rijal et al., 2007). In this study, we used the SbIII- and SbV-resistant isolate MHOM/PE/02/PER104 of L. braziliensis (Yardley et al., 2006; Adaui et al., 2011a) as donor for a genomic DNA cosmid library, to transfect SbIII s/SbV r clones of isolate MHOM/PE/01/PER002 and to challenge the recombinant population with antimonyl tartrate (SbIII). Five independent screens using two libraries from resistant and sensitive isolates, combined with two L. braziliensis clones and one L. peruviana acceptor strain, yielded three cosmids that overlapped in the same region of the L. braziliensis chromosome 20. The region harbours a gene, LbrM20.0210, which upon overexpression in L. infantum increases the IC50 for SbIII up to threefold and increases parasite survival inside macrophages under SbV pressure.
2. Materials and methods
2.1. Parasite strains and isolates
L. braziliensis isolates MHOM/PE/2002/PER104 (PER104) and MHOM/PE/2001/PER002 (PER002) have been described (Soto et al., 2005; Llanos-Cuentas et al., 2008). Of the latter, two clones were isolated with confirmed SbIII s/SbV r phenotype. L. peruviana strain MHOM/PE/??/ LC2434, clone 5, was a gift from Lenea Campino, Lisbon. L. infantum strain MHOM/FR/91/LEM2259 was described (Garin et al., 2001; Choudhury et al., 2008).
2.2. Parasite cultivation and selection procedures
Promastigotes were cultivated at 25 °C, pH 7.0 in Locke’s Solution (0.8% NaCl; 0.02% KCl; 0.03% KH2PO4; 0.01% MgSO4 × 7H2O; 0.1% NaHCO3; 0.25% glucose; all w/v) over Tobie’s blood agar (20% rabbit blood, defibrinated; 1.5% agar; 1.5% Bacto-Tryptose; 0.4% NaCl; 0.04% KCl; 0.5% Na3PO4 × 12H2O) and with 1% l-glutamine/Penicillin/Streptomycin (Sigma–Aldrich #D3462). G418 (Geneticin sulfate, Carl Roth) was added to 12 μg ml−1 for recombinant cell populations. Cell density was determined using Neubauer chambers with 0.025 mm width.
Alternatively, L. braziliensis and L. peruviana were cultivated at 25 °C, pH 7.0 in Schneider’s Insect Medium supplemented with 10% (v/v) heat-inactivated fetal calf serum, 2% (v/v) sterile filtered human urine, 0.04% (w/v) NaHCO3, 0.079% (w/v) CaCl2 × 2H2O, and 25 μg ml-1 gentamycin. L. infantum was cultivated in supplemented Medium 199 (Krobitsch et al., 1998). G418 was added to 50 μg ml−1 for recombinant cell populations. Cell density was monitored using a Schaerfe System CASY® cell counter. For growth experiments, promastigotes were seeded at 5 × 105 cells ml−1 in modified M199 without G418 and cell density was monitored for 3 days.
2.3. In vitro infection
In vitro infections were performed as described (Reiling et al., 2010). Bone marrow-derived macrophages (BMMs) were isolated from femurs of C57BL/6 mice and incubated in Iscove‘s Modified Dulbecco‘s Medium (IMDM) supplemented with 10% heat inactivated FCS, 5% horse serum, and 30% L929 cell supernatant containing macrophage colony-stimulating factor (MCSF), modified after (Racoosin and Swanson, 1989). After differentiation, BMMs were harvested, washed, and seeded into 8-well chamber slides (Nunc) at a density of 4 × 105 cells/well. Macrophages were incubated for 48 h at 37 °C and 9% CO2 to permit adhesion. BMMs were then infected using stationary phase promastigotes (Racoosin and Beverley, 1997) at 10 parasites per macrophage. After 4 h of incubation at 37 °C in modified Medium 199 (Krobitsch et al., 1998), free parasites were washed off with PBS. Incubation was continued for another 72 h in IMDM without or with 160 μg ml-1 of Pentostam® at 37 °C and 9% CO2. The medium was removed. The cells were washed twice in PBS and subsequently fixed in ice-cold methanol. Infection rates were assessed by nuclear staining with DAPI (1.25 μg ml−1, Sigma) and fluorescence microscopy. Pentostam® (formulated for clinical use, GlaxoSmithKline) was a gift from the Bundeswehr Krankenhaus Hamburg, Tropical Medicine Unit.
2.4. Genomic DNA preparation and cosmid library construction
Construction of a Leishmania genomic DNA cosmid library in the vector pcosTL (Kelly et al., 1994) has been described (Hoyer et al., 2001). Briefly, cosmid libraries of L. braziliensis strains PER104 and PER002 (Yardley et al., 2006; Adaui et al., 2011b) were prepared by cleavage of the shuttle cosmid vector pcosTL with SmaI and BamHI and ligation with size-selected Sau3AI partial digest products of genomic DNA prepared by glass rod spooling from an ethanol precipitation (Sambrook and Russell, 2001). After packaging using the Gigapack Gold II kit (Stratagene), the complexities of the libraries were tested, after which the libraries were amplified and stored at −70 °C.
2.5. Electroporation, selection, and recovery of cosmid DNA
Electrotransfection of Leishmania promastigotes was carried out as described (Krobitsch et al., 1998). Promastigotes were harvested during late log phase of growth, washed twice in ice–cold PBS, once in pre-chilled electroporation buffer and suspended at a density of 1 × 108 ml−1 in electroporation buffer (Laban and Wirth, 1989; Kapler et al., 1990). 50 μg of circular DNA in an electroporation cuvette was mixed on ice with 0.4 ml of the cell suspension. The mixture was immediately subjected to electroporation using a Bio-Rad Gene Pulser apparatus. Electrotransfection of DNA was carried out by three pulses at 2.750 V/cm and 25 μF in a 4 mm electroporation cuvette. Mock transfection was performed in identical fashion, however without plasmid or cosmid DNA, to obtain negative control strains for antibiotic selection. Following electroporation, cells were kept on ice for 10 min before they were transferred to 10 ml drug-free medium. G418 (12–50 μg ml−1) was added after 24 h for selection of recombinant cells.
Dose-inhibition curves for antimonyl tartrate (Sigma–Aldrich #383376) were established by seeding the promastigotes at 5 × 105 ml−1 in medium containing various concentrations of a drug, but lacking the selection antibiotic G418. For primary screens, the selection was performed in the presence of G418, to select against spontaneous drug resistance.
Recombinant populations of promastigotes transfected either with cosmid library DNA or with the unmodified vector, pcosTL, were seeded in G418-containing medium with SbIII at a concentration that inhibits >95% of the growth. This was determined empirically for each batch of antimonyl tartrate and parasite strain. When a cell density of 1 × 107 cells ml−1 was reached, the cells were suspended in fresh medium containing the drug. Cultivation was continued for several days. Conditions were deemed stringent if the control population transfected with the vector pcosTL did not show growth. Surviving parasites were harvested for cosmid isolation.
Cosmid DNA was prepared from Leishmania promastigotes by alkaline lysis, following the protocol for plasmid DNA mini-preparation (Sambrook and Russell, 2001). After phenol/chloroform/isoamylalcohol (25:24:1) extraction, cosmid DNA was precipitated by adding 0.7 vol. 2-propanol, washed once with 70% ethanol and dissolved in Tris/EDTA (pH 8.0) buffer.
2.6. Cosmid characterisation
Approximately 100 ng of cosmid DNA was mixed on ice with Library Efficiency DH5α Competent Cells (Invitrogen). Transfection was performed following the manufacturer’s protocol, and recombinant bacteria were plated on LB agar under ampicillin selection.
Cosmid DNA mini-preparations were performed from ⩾50 individual colonies, and the isolated cosmid DNA was subjected to analytical digest with the restriction enzymes EcoRV and XbaI. The pattern of restriction fragments was analysed by field inversion gel electrophoresis (Reiling et al., 2010).
2.7. Construction of cosmid derivates
The cosmid pcosC1.6 was cut with AflII and re-ligated to create derivative (a). Digest with BamHI and subsequent re-ligation yielded construct (b). A combined digest with NdeI and BamHI followed by Klenow enzyme fill-in reaction and ligation yielded construct (c). Excising the XbaI fragment and fusing it into the XbaI site of pcosTL resulted in construct (d). To produce construct (e), pcosC1.6 was digested with XbaI and SpeI, and the resulting 5200 bp fragment was ligated into XbaI-opened pcosTL vector. Using BclI instead of SpeI in the second step produced construct (210), while cutting construct (e) with XbaI and BclI followed by Klenow enzyme treatment and ligation resulted in construct (220). Schematic representations can be found in Fig. 1C and in Supplementary data 2.
Fig. 1.
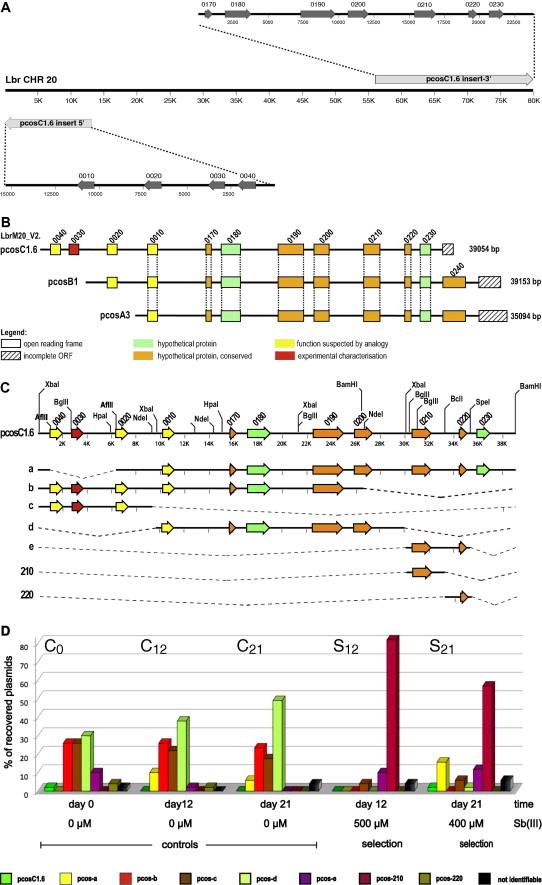
(A) Sequence alignment between the experimentally determined insert sequence of pcosC1.6 and L. braziliensis chromosome 20. The 5′ ∼15 kb of the insert align in reverse complimentary direction with the 5′ end of chromosome 20, whilst the C′-terminal 24 kb align in colinear fashion with chromosome 20 sequences between positions 56 and 80 kb. (B) Schematic representation of the overlapping genomic DNA inserts of cosmid C1.6, A3 and B1. Eight open reading frames (ORFs) are present in all three cosmid inserts. The four digit gene numbers correspond to the systematic numbers for chromosome 20 (LbrM20.____) derived from version 2 of the L. braziliensis genome project. The total length of each insert (to the right in [bp]) was derived from end-sequencing. Green boxes symbolise ORFs for hypothetical proteins, orange boxes stand for conserved hypothetical proteins, and yellow boxes signify coding regions for proteins with predicted functions. Hatched boxes stand for incomplete ORFs. Vertical dotted lines connect identical ORFs found in all 3 cosmids. Note that the sequence was incomplete in the genome project, causing the apparent non-linear numbering of gene candidates. (C) Truncation of cosmid insert C1.6. The inserts of seven deletion constructs derived from the cosmid pcosC1.6 are schematically shown. The deletions are represented by the dashed lines. The positions of AflII, BamHI, BclI, SpeI and XbaI restriction sites are shown on insert C1.6. (D) Recovery rates [%] of cosmid pcosC1.6 and its truncated derivatives after selection in L. infantum. Promastigotes transfected with pcosC1.6 and any of the truncated variants (pcos-a to pcos-e, pcos210, pcos220) were mixed at equal ratio (C0). Two control cultures cultivated without SbIII for 12 days (C12) or 21 days (C21) were analysed along with the cultures selected at 500 μM SbIII for 12 days (S12) or at 400 μM SbIII for 21 days (S21). Episomes from surviving parasites were isolated and used for transformation of E. coli. From each transformation, the cosmid DNAs from 50 bacterial colonies were characterised by RFLA, and the share [%] of each cosmid type is displayed.
2.8. cDNA synthesis and qPCR
Real-time qPCR was performed essentially as described (Choudhury et al., 2008). Gene-specific primers were 20.0210.F2 (5′-TGATGATGAAGGTGACCGTGACG-3′) and 20.0210.B3 (5′-AAGGAGGGTGTAGACGACGCTCTC-3′). LbrM20.0210 mRNA abundance was calculated relative to the actin signal.
2.9. In silico DNA analysis and data handling
Significance was assessed by the Mann–Whitney U-test (Mann and Whitney, 1947). All statistical analyses were performed using the Prism Software (GraphPad). Sequencing was performed by a commercial provider (AGOWA, Berlin). We used the MacVector™ suite software (Versions 10.5–12.7) for in silico sequence analysis. BLAST searches were performed at standard settings, using the TriTryp web site (http://tritrypdb.org/tritrypdb//). Open reading frames were identified both by onsite analysis, using MacVector™, and by mining of the TriTryp Genome Databases. Sequence alignments were done in ClustalW using the MUSCLE algorithm included with the MacVector® software package. Transmembrane domain predictions were performed at the TMpred website (http://www.ch.embnet.org/software/TMPRED_form.html) (Hofmann and Stoffel, 1993). Figures were compiled using the Intaglio® vector graphics software.
2.10. Animal ethics
The isolation of bone marrow-derived macrophages from sacrificed mice was done in accordance with the German animal protection laws and regulations.
3. Results
3.1. Library construction and transfection
The genomic DNA from two L. braziliensis isolates, the SbIII resistant MHOM/PE/02/PER104 (PER104) and the SbIII sensitive MHOM/PE/01/PER002 (PER002), were used for construction of two cosmid libraries, pcos104 and pcos002 respectively. The cosmid library DNAs were then used for stable transfection of L. braziliensis PER002, clone 7, promastigotes. In addition, the cosmid library pcos104 was electro-transfected into L. peruviana promastigotes, to test whether the outcome of the screen was influenced by the acceptor species. The number of recombinant parasites per electroporation reaction was determined by limiting dilution analysis (not shown). Results varied between 960 and 1900 clones per reaction. We therefore pooled 6 electroporation reactions for each library to ensure the necessary number of >4200 clones, that is needed to cover the L. braziliensis genome with a confidence of >99%.
3.2. First selection (S1) of PER002cl7 parasites bearing the pcos104 library
First, we established the sensitivity of PER002 promastigotes against SbIII in biphasic medium. Dose-inhibition curves suggested that growth of a control strain bearing the empty cosmid vector, PER002 [pcosTL], was reduced by >95% at 90 μM antimonyl tartrate (data not shown). We decided to perform selections at two concentrations, 25 and 90 μM.
We next selected the cosmid library from the SbIII resistant isolate PER104 in the SbIII sensitive PER002 background. PER002cl7[pcos104] promastigotes were seeded in vitro at 1 × 107 cells ml−1 in biphasic medium and challenged with antimonyl tartrate. PER002cl7[pcosTL] controls were cultivated in parallel. The challenge was maintained for up to 33 days, or until the library-transfected populations showed significant growth. In total, three selections (S1.1–S1.3) were performed, one (S1.1) at 25 μM and two (S1.2 and S1.3) at 90 μM SbIII.
The cosmid DNA from the surviving parasite populations was isolated and then used for transformation of competent Escherichia coli DH5α cells. The cosmid DNA from ⩾50 resulting E. coli clones was isolated and subjected to restriction fragment length analysis (RFLA) (Supplementary data 1). One cosmid, designated pcosC1.6, was dominant in all three selections (Table 1). In selection S1 under low antimony pressure (25 μM SbIII), pcosC1.6 was identified in 66% of the bacterial clones. In selections S1.2 and S1.3 (both at 90 μM SbIII), the same cosmid was recovered from 90% to 98% respectively of the analysed clones. Another cosmid that appeared frequently turned out to be a plasmid contamination, likely contracted during E. coli transformation (Supplementary data 1). The fact that only one cosmid species was recovered after 90 μM SbIII selection experiments implicates a strong selective pressure towards a/the gene/s harboured in cosmid pcosC1.6.
Table 1.
Outcomes for 6 independent selection screens.
| Selection | Acceptor strain | Donor strain | SbIII (μM) | Time (d) | Selected cosmids (%) | Chromosome/region |
|---|---|---|---|---|---|---|
| S1.1 | PER002 | PER104 | 25 | 18 | C1.6 (66) | 20/903 984–946 647 |
| S1.2 | PER002 | PER104 | 90 | 33 | C1.6 (90) | 20/903 984–946 647 |
| S1.3 | PER002 | PER104 | 90 | 15 | C1.6 (98) | 20/903 984–946 647 |
| S2.1 | PER002 | PER002 | 75 | 17 | A3 (10) | 20/913 015–951 701 |
| S2.2 | PER002 | PER002 | 75 | 17 | A3 (96) | 20/913 015–951 701 |
| S3 | L. peruviana | PER104 | 15 | 30 | B1 (40) | 20/908 315–951 075 |
Screens S1.1–S1.3 were performed using the SbIII sensitive strain PER002 and a cosmid library derived from the SbIII resistant strain PER104; screens S2.1.1 and S2.1.2 were consecutively performed using PER002 both as donor and acceptor, and screen 3 was performed using L. peruviana as receptor and strain PER104 as donor. SbIII concentrations (column 3) and selection time (column 4) varied depending on antimony batch variations. The dominant selected cosmids and their share of the overall recovered cosmids are given in column 5, while the corresponding chromosomal regions are given in column 6.
Partial 5′- and 3′ sequence analysis of the genomic DNA insert of pcosC1.6 was performed and used for an alignment with the L. braziliensis genome project. The region bracketed by the partial sequences contained two large gaps. Using a primer walking strategy, we filled those sequence gaps to obtain a complete sequence of the pcosC1.6 insert (Supplementary data 3). Alignment of the complete insert sequence with the L. braziliensis chromosome 20 (CHR20) sequence (as of Sep 20, 2013) results in a split alignment (Fig. 1A). The 5′ ∼15 kb align in reverse complimentary direction to the telomere end of CHR20 (positions ∼50–14,000) whilst the 3′ 24 kb of the cosmid insert align in colinear fashion to CHR20 sequences between positions 56,000 and 80,000. This can be either due to a ligation artefact during library construction joining two smaller DNA fragments or to a faulty contig building for CHR20. The cosmid insert thus comprises 39 kb of genomic DNA and includes eleven complete and one partial open reading frames (Fig. 1B, Supplementary data 2).
3.3. Quantification of the effect by pcosC1.6
To quantify the effect of the pcosC1.6 episomes, we performed a dose-inhibition experiment. The SbIII susceptibilities of acceptor and library donor parasites, PER002cl7 and PER104, are different. We found PER104 (Fig. 2A, full circles) to have a >2-fold higher IC50 (50%-inhibitory concentration) than PER002cl7 (Fig. 2A, open circles). The vector control strain PER002cl7[pcosTL] showed an IC50 of 7 μM antimonyl tartrate, whilst the PER002cl7[pcosC1.6] parasites displayed an IC50 of 16 μM. This 2.3-fold difference caused by the pcosC1.6 transgene is therefore measurable, reproducible (n = 5, p = 0.002) and in the range of the donor strain PER104.
Fig. 2.
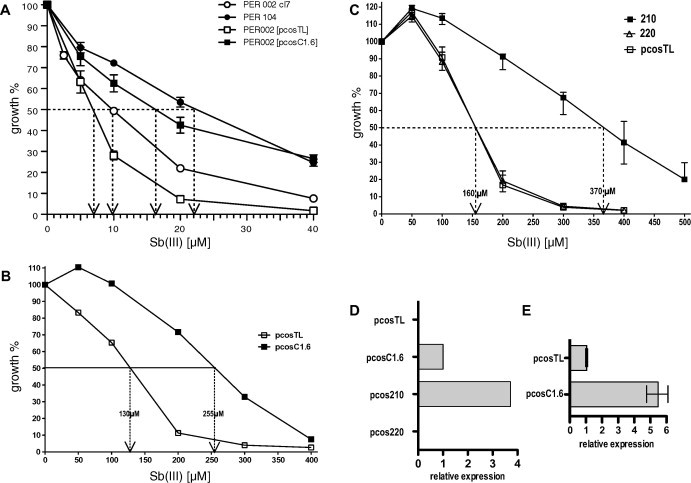
(A) Dose–effect of antimonyl tartrate (SbIII) on the growth of L. braziliensis strains PER104 and PER002 transfected with cosmid pcosTL or pcosC1.6 respectively. Growth at 0 μM SbIII was defined as 100%. The dashed arrows indicate the 50% inhibiting concentration (IC50). Significance (IC50): p = 0.002, n = 6. (B) Effect of pcosC1.6 in L. infantum. Empty vector pcosTL and cosmid pcosC1.6 were transfected into L. infantum and selected under G418. Cells of both recombinant populations (5 × 105 cells ml−1) were grown under the indicated SbIII concentrations for 72 h. The diagram shows the cell density after 72 h relative to the control culture (0 μM SbIII). Numerical IC50 values are indicated. Significance (IC50) p = 0.016, n = 5. (C) Constructs pcos-210, pcos-220, and the empty vector pcosTL were transfected into L. infantum and selected under G418. Cells of each recombinant population (5 × 105 cells ml−1) were challenged with the indicated SbIII concentrations for 72 h. The diagram shows the cell density after 72 h relative to the control culture (0 μM SbIII). The bars indicate the mean error. Significance (IC50) p = 0.008, n = 5. (D) Control of LbrM20.0210 expression by qPCR. Expression mediated by cosmid pcosC1.6 was given the value 1. Note that L. infantum expresses a homologous gene that is not amplified by the species-specific primers. Thus, the degree of overexpression cannot be determined. (E) Quantification of LbrM20.0210 expression in L. braziliensis strains PER002[pcosTL] and PER002cl7[pcosC1.6]. Expression in the control strain PER002cl7[pcosTL] was assigned the value 1. n = 6. The standard error of means is depicted.
3.4. Second selection (S2) of PER002cl7 parasites bearing the pcos002 library
The SbIII resistance conferred by pcosC1.6 may be caused by a sequence variation in the responsible gene or by the overexpression from an episome. To distinguish between these possibilities, we also performed a selection using the recombinant population PER002cl7[pcos002]. This was designed to identify gene(s) that induce SbIII resistance by gene over-expression only, as donor and acceptor strain gDNAs are identical. The parasites were seeded in supplemented Schneider‘s insect medium at 5 × 105 cells ml−1 and challenged with 75 μM SbIII, the empirically determined IC95 (95%-inhibitory concentration) for that batch of antimonyl tartrate. After 17 days of selection, cosmid DNA was isolated from the surviving, recombinant parasites. Competent E. coli XL1-blue cells were transformed with the recovered cosmid DNA. RFLA was performed for 100 bacterial clones (data not shown). Five cosmid species (pcosA1-5) were detected after the initial screen (S2.1). Parasites bearing the cosmids pcosA1-5 at equal ratios were then subjected to a second screen (S2.2) under the same conditions. After the secondary selection, 96% of the recovered cosmids corresponded to pcosA3.
Cosmid pcosA3, like pcosC1.6, contains genomic DNA derived from chromosome 20 and overlaps the genomic DNA insert of pcosC1.6 (Fig. 1B). This indicates that a quantitative effect is underlying the increased SbIII tolerance, meaning that both pcosA3 and pcosC1.6 act through an increase of the relevant gene copy number(s).
3.5. Third selection (S3) of L. peruviana parasites bearing the pcos104 library
To test whether the closely related L. peruviana yields a similar selection result, a recombinant population of L. peruviana [pcos104] was cultivated in vitro in supplemented Schneider‘s insect medium and then seeded at 1 × 105 cells ml−1 under a challenge of 15 μM SbIII, due to that species’s much lower tolerance for antimonyl tartrate. After 30 days of in vitro cultivation, cosmid DNA was isolated from the selected population and transformed into competent E. coli XL1-blue cells. RFLA of 50 bacterial clones was performed (data not shown), and a dominant cosmid, pcosB1 was recovered from 40% of the bacterial clones.
Again, the selected cosmid, pcosB1, corresponds to sequences on chromosome 20 and overlaps the inserts of both pcosC1.6 and pcosA3 (Table 1, Fig. 1B). Thus, five independent in vitro selections under antimonyl tartrate implicate the same genomic region on chromosome 20. The gene(s) responsible for the selective advantage must therefore code for (a) dominant, dose-dependent resistance marker(s) located there.
Furthermore, the isolation of 3 different cosmids with synthenic gene arrays covering essentially the same region on CHR20 argues against ligation artefacts as the basis for the split sequence alignment between cosmid inserts and the current CHR20 sequence.
3.6. Functional deletion analysis of pcosC1.6
To identify the candidate gene(s) responsible for SbIII-resistance, the cosmid pcosC1.6 was truncated by digestion with different restriction enzymes and subsequent re-ligation, resulting in seven constructs that represent different regions of pcosC1.6 (Fig. 1C). These constructs were then transfected into L. braziliensis PER002 clone 7. The recombinant strains were then mixed and selected under SbIII. We could not observe the selection of specific truncated pcosC1.6 derivates in those selections, and dose-inhibition experiments for selected transgenic parasite strains were also inconclusive (data not shown).
At that time, studies of the L. braziliensis genome revealed that the components of RNA interference (RNAi) pathways exist in the subgenus Viannia (Peacock et al., 2007) and that gene regulation due to homologous dsRNA may occur. This was since confirmed (Lye et al., 2010). This led us to suspect that dsRNA may interfere with the overexpression from the truncated cosmid episomes. Our constructs and experimental set-up did not allow for an analysis of dsRNA-mediated RNA breakdown by qPCR. Nevertheless, we decided to test the constructs in another species known for the absence of RNAi (Beverley, 2003).
3.7. Selection for the resistance-mediating gene using L. infantum as acceptor species
We decided to use the Old World Leishmania species, L. infantum, which lacks genes for key components of the RNAi machinery, such as Argonaute and Dicer, and for which we had experience regarding experimental SbIII resistance (Choudhury et al., 2008). First, we confirmed that the L. braziliensis-derived cosmid pcosC1.6 conferred elevated IC50 for SbIII to L. infantum. We performed dose-inhibition growth experiments with L. infantum bearing either the empty vector, pcosTL, or the selected cosmid pcosC1.6. Indeed, the IC50 of L. infantum [pcosC1.6] was approximately twice that of L. infantum [pcosTL] (Fig. 2B).
We next performed two independent SbIII screens with L. infantum strains bearing the cosmid pcosC1.6 and its truncated derivatives (Fig. 1C) and mixed at equal ratios (culture C0). We seeded two cultures each at 4 × 104 cells ml−1 and selected them either for 12 days at 500 μM SbIII (S12) or for 21 days at 400 μM SbIII (S21) μM SbIII. Two groups (C12 and C21) were also cultivated for 12 and 21 days respectively, but without antimony pressure. After 12 days (C12 and S12) or 21 days (C21 and S21) of in vitro cultivation, cosmid DNA was isolated from the resulting populations. E. coli XL1-blue cells were transformed with the re-isolated cosmids. The cosmids from 50 bacterial clones for each selection or control culture were prepared, and RFLA was performed on them to determine the prevalence of truncated derivatives of pcosC1.6.
Cosmid pcos-210, harbouring only the gene LbrM20.0210, was the most dominant of the recovered transgenes and recovered from 82% (S12) to 57% (S21) respectively of the bacterial clones (Fig. 1D). Constructs (a and e), both also harbouring LbrM20.0210, were recovered at lesser rates. This means that 92% (S12) and 86% (S21) of the re-isolated constructs carry the LbrM20.0210 gene. Since the recovery rates for these cosmids from the non-selected populations were ⩽10%, the cause for LbrM20.0210 selection must be an in vitro SbIII resistance mediated by this gene.
3.8. Analysis of the L. braziliensis resistance gene in L. infantum
The effect of LbrM20.0210 was further verified and quantified by performing individual SbIII dose-inhibition experiments for L. infantum carrying the constructs pcosTL (vector control), pcos210 (suspected resistance gene), and pcos-220 (negative control). While Lin[pcosTL] and Lin[pcos-220] show indistinguishable dose-inhibition curves (Fig. 2C), Lin[pcos-210] increases the IC50 for SbIII 2.3-fold.
We also confirmed the expression of LbrM20.0210 in L. infantum transfected with pcosC1.6 and with pcos-210, using real-time qPCR (Fig. 2D). Expression was ∼3-fold higher from the pcos-210 construct compared to the original cosmid, pcosC1.6. As expected, L. infantum controls did not express the L. braziliensis gene. This result explains the preferential selection of pcos-210 over pcosC1.6 as shown in Fig. 1D. To determine the general level of overexpression from pcosC1.6 over the L. braziliensis wild type PER002cl7, we performed another qPCR with cDNA derived from PER002[pcosTL] and PER002[pcosC1.6]. Fig. 2E shows the result. The pcosC1.6 transgene gives rise to a 5.5-fold elevated LbrM20.0210 RNA level. Although not directly applicable to the L. infantum system, this indicates a sizeable overexpression of LbrM20.0210 in the recombinant parasites.
3.9. Overexpression of LbrM20.0210 protects intracellular amastigotes against antimony (V)
Antimony (V) is reduced to antimony (III) in Leishmania amastigotes and in macrophages, with the trivalent form being the cytotoxic principle. SbIII resistant strains are usually SbV-resistant, but not necessarily vice versa. Therefore, we next tested whether overexpression of LbrM20.0210 also protects the relevant life cycle stage, intracellular amastigotes, against an SbV-based drug formulation, namely Pentostam®.
L. infantum promastigotes transfected either with the vector pcosTL or with pcos210 were grown to stationary phase in vitro and used to infect bone marrow-derived murine macrophages in vitro. After 4 h, the free parasites were removed and the infected cells were overlayered with medium without or with 160 μg ml−1 SbV. Fig. 3A shows the accumulated results from 4 separate infection experiments. In the absence of SbV, the infection rates of L. infantum [pcosTL] and L. infantum [pcos-210] show only insignificant variation. Under SbV treatment of infected macrophages we observe significant (p = 0.029) differences. While L. infantum [pcosTL] infection rates drop by 90%, the effect on the L. infantum [pcos-210] parasites is less severe. This indicates that LbrM20.0210 can partially protect intracellular amastigotes of L. infantum against the effect of an SbV-containing drug. We therefore assigned the moniker ARM58 (antimony resistance marker of 58 kDa) to LbrM20.0210 and its product.
Fig. 3.
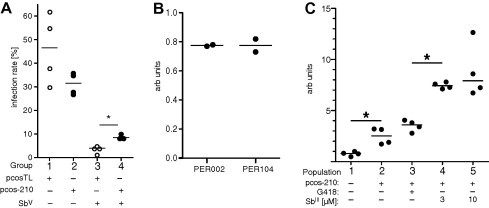
(A) Infection rate in bone marrow-derived macrophages under SbV challenge. BMMs were infected with stationary growth phase L. infantum transfected either with vector pcosTL or with pcos-210. After removal of free parasites, the infected macrophages were incubated in the absence or presence of 160 μg ml−1 Pentostam®. After 72 h, the cells were fixed with ice–cold methanol, stained with DAPI, and subjected to fluorescence microscopy. For each dataset, 100 macrophages were examined for the presence of parasites. Experiments were performed in quadruplicate and on separate days. The median infection rates are indicated by horizontal bars. Significance (*p = 0.029) was tested using the Mann–Whitney ranking test. n = 4. (B) Basic expression rate of ARM58 in two clinical isolates of L. braziliensis, PER002 (SbIII sensitive) and PER104 (SbIII resistant), measured by qPCR. Experiments were done in duplicate, arbitrary units. (C) ARM58 RNA expression in L. braziliensis populations under various selective pressure. Clone PER002cl7[pcos-210] was subjected to in vitro passage for 4 weeks with twice-weekly medium changes. The populations were kept without selection (2), under 50 μg ml−1 G418 (3), under 3 μM SbIII (4), or under 10 μM SbIII (5). The PER002cl7 wild type was used for normalisation (1). Cultures were grown in duplicate, qPCR was done in duplicate for each culture (n = 4). The bars indicate the medians. Asterisks indicate significance (∗p ⩽ 0.05).
3.10. ARM58 in Sb resistant and sensitive isolates
To assess whether ARM58 gene copy number and/or RNA abundance vary between strains, we performed a comparative qPCR analysis to quantify ARM58 RNA in L. braziliensis strains PER002 (SbIII-sensitive acceptor strain in this study) and PER104 (natural SbIII-resistant donor strain for gDNA library). At least based on the qPCR results (Fig. 3B), we cannot detect variations in ARM58 mRNA levels in the two isolates. Since the strains show different SbIII sensitivities in vitro, we can exclude variant mRNA levels as cause.
We also assessed the stability of ARM58 overexpression under different conditions. L. brasiliensis PER002[pcos-210] were kept without selection, under G418 (50 μg ml−1), or under SbIII (3 or 10 μM) for 4 weeks, with twice weekly medium changes. Following these in vitro passages, we collected the selected parasites, isolated RNAs from each population, and performed cDNA synthesis and qPCR on them. Fig. 3C shows the results, with wild type L. braziliensis PER002cl7 as control. We observe no difference between the unselected population bearing the transgene and the population selected under G418, indicating that the transgene is stable for >4 weeks without selection. The mRNA levels are 3-fold elevated compared with the wild type control population. Under SbIII selection, however, ARM58 mRNA levels increase significantly to 800% of wild type expression. This is good indication that the ARM58 gene is indeed selected under SbIII pressure. Lacking an ARM58-specific antibody, we can only assume that those changes translate into higher protein levels.
Lastly, we analysed whether there may be sequence polymorphisms in ARM58 causing different functionality in isolates PER002 and PER104. We amplified the ARM58 open reading frame DNA from genomic DNA of either isolate and subcloned the products into plasmid pBluescript II KS+. Three plasmid clones each were subjected to sequence analysis. Indeed, both sequences differed in one base pair from the sequence published in the TriTryp database, resulting in a S235A amino acid exchange (data not shown). We also observed two base pair changes between isolates PER002 and PER104 (data not shown). However, both polymorphisms are silent as neither base pair change translates into an amino acid exchange. From these data, we conclude that ARM58 exerts its role as resistance marker in a solely quantitative manner.
3.11. ARM58 is unique to the Leishmaniae
The chromosomal region harbouring ARM58 is syntenic for at least 5 Leishmania species (Fig. 4A), although usually found on larger chromosomes, i.e. CHR33 in the New World species L. mexicana and CHR34 in the Old World Leishmaniae L. infantum, L. major, and L. tarentolae. Upstream of ARM58, a structurally related gene, LbrM20.0200, is found that does not confer antimony resistance (Fig. 1D, construct (d)). We dubbed this gene ARM58rel (ARM58-related). While ARM58 is notably absent from the corresponding T. brucei region, ARM58rel is present. Thus, ARM58 is unique to Leishmania spp.
Fig. 4.
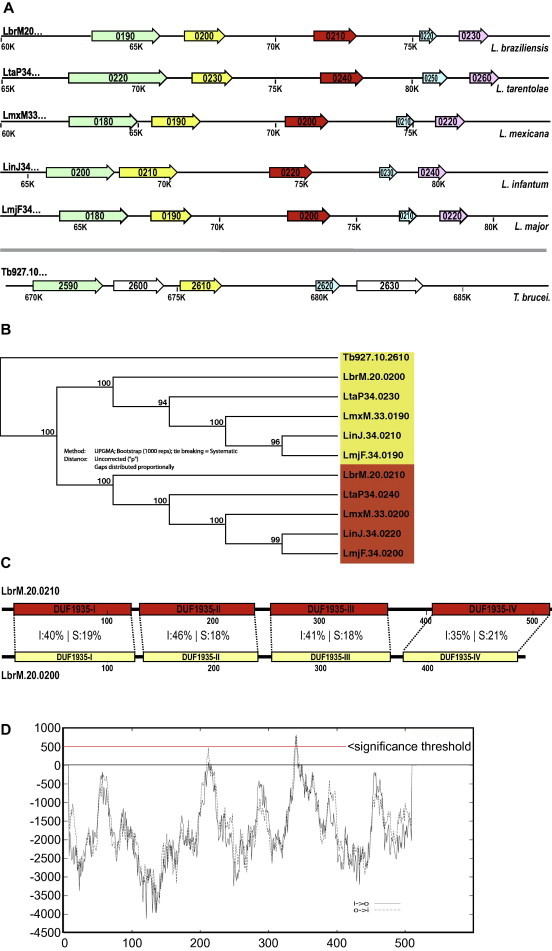
(A) ORF maps of the region around LbrM20.0210 for 5 Leishmania species and Trypanosoma brucei. (B) Phylogeny tree for LbrM20.0210 and LbrM20.0200. UPGMA algorithm, bootstrap analysis (1000 reps), uncorrected. Gene numbers are underlaid with colours red (for LbrM20.0210 orthologs) or yellow (LbrM20.0200 orthologs). (C) Domain structures for LbrM20.0210 and -0200. DUF = domain of unknown function. Domains are numbered sequentially in Roman numerals (I–IV) from N terminus to C terminus. Sequence conservation for each domain is indicated (I = identity, S = side chain similarity). Length markers in [amino acids] are given below. (D) Transmembrane domain prediction for LbrM20.0210 using the TMpred algorithm (http://www.ch.embnet.org/software/TMPRED_form.html). Note the peak (score 792/819 i>o/o>i) corresponding to domain DUF1935-III and exceeding the threshold score of 500.
ARM58 and ARM58rel form distinct lineages (Fig. 4B) and show considerable sequence divergence. The gene products, ARM58 and ARM58rel, consist of four similar domains of unknown function, DUF1935. This type of domain is also found singly in various calpain-like cysteine peptidases, also described as SMPs (Tull et al., 2004). However, an arrangement of four DUF1935 is exclusive to ARM58 and ARM58rel. Comparing the two putative amino acid sequences domain by domain, there is no obvious candidate domain for the drug resistance functionality. The degree of amino acid conservation in the DUF1935 varies between 56% and 64%, with the fourth DUF1935 being the least conserved (Fig. 4C). Also, an insertion between DUF1935-III and DUF1935-IV is unique to ARM58. Moreover, a search for possible transmembrane domains (Hofmann and Stoffel, 1993) identified a candidate TMD within DUF1935-III (Fig. 4D).
4. Discussion
Resistance against antimony-based antileishmanial drugs was first noted in Northern India in the 1970s and has since reached alarming numbers. In high-endemicity areas the percentage of non-responders can exceed 60% rendering this mainstay treatment unusable (Croft et al., 2006).
Several molecules have been reported as effectors of antimony tolerance. The multi-drug resistance (MDR) loci that encode membrane-associated P-glycoproteins were shown to play a role in laboratory induced resistance (Legare et al., 2001; Ouellette et al., 2001). Gene copy number and the resulting increased expression of P-glycoproteins rendered promastigotes more resistant against SbV. Abundance of aquaglyceroporins is inversely correlated with antimony resistance (Gourbal et al., 2004). Also, expression levels of the cytosolic chaperones Hsp70 and Hsp90 were reported to correlate with antimony tolerance (Brochu et al., 2004; Vergnes et al., 2007) suggesting that the protective effects of chaperones may ameliorate toxicity. A very large protein, P299, confers SbIII and Miltefosine resistance when overexpressed in L. infantum (Choudhury et al., 2008). Yet another resistance mechanism seems to involve the parasite-mediated activation of host P-glycoproteins that will extrude SbV before it can be reduced to the toxic form, SbIII (Mookerjee Basu et al., 2008).
Our in vitro selection screens using two cosmid libraries of genomic DNA from resistant or sensitive L. braziliensis isolates identified a single genomic region from chromosome 20 of L. braziliensis. Episomal transgenes derived from this region confer SbIII resistance not only to L. braziliensis, but also to other species such as L. (Viannia) peruviana and a European isolate of L. infantum.
It is most interesting that in five selection screens using three host parasite strains and two libraries, only LbrM20.0210-containing cosmids dominated the surviving populations. We have no evidence that any of the established or suspected antimony resistance markers was selected in our screens. Given the rather stringent selection protocol using SbIII at 90 μM (>95% growth inhibition), our screen may not reflect the typical protocol for raising spontaneous drug resistant strains by gradually increasing the drug concentration (Haimeur et al., 2000; Brochu et al., 2003). It is possible that some of the known markers were not well represented in the library, but it would be unlikely that none of them were present in the cosmid libraries since the calculated representation of genes exceeded 99% in the recombinant populations. Therefore, ARM58 appears to be a dominant marker of SbIII resistance conferring unparalleled protection under stringent selection.
The product of ARM58 confers increased tolerance to both SbIII and SbV, regardless of whether the gene originated in strain PER104 or PER002. This was in keeping with the results of a sequence comparison between the ARM58 homologues from PER104 and PER002 which yielded identical putative amino acid sequences and ruled out ARM58 sequence polymorphisms as cause for varying resistance phenotypes.
On the other hand, we did not see any variation of ARM58 expression, at least at the RNA level. Nevertheless, selection of the ARM58 transgenic parasites under SbIII resulted in an increased expression, presumably by selection for higher transgene copy numbers (Fig. 3C). In the field, gene copy numbers can also vary, even within isolates, based on a highly variable chromosome ploidy in Leishmania spp. (Ubeda et al., 2008; Leprohon et al., 2009; Rogers et al., 2011; Mannaert et al., 2012). Therefore, our results do not rule out a natural amplification of chromosome 20 or parts thereof under antimony challenge.
ARM58 functionality is not restricted to SbIII-sensitive L. braziliensis acceptor strains either. It could also confer SbIII and SbV resistance to L. infantum. While there is a high degree of synteny in this genomic region between Leishmania species (Fig. 4A), the amino acid sequence identity between the L. braziliensis and L. infantum orthologs is only 81%, with 7% similar amino acids (data not shown), underscoring the evolutionary distance between sensu stricto Leishmaniae and the Viannia subgenus.
We have tried to obtain information regarding the subcellular localisation of ARM58. We produced polyclonal antibodies against the protein after expression in E. coli. The antibodies recognise the recombinantly expressed protein with good specificity and sensitivity. However, no signals were obtained in Western blots of Leishmania cell lysates (Schäfer, unpublished data), suggesting post-translational modification of ARM58 in Leishmania. Another Leishmania protein sharing a single DUF1935, SMP-1 (Small Myristoylated Protein), forms a β-sandwich structure (Gooley et al., 2006; Tull et al., 2010), bears post-translational modifications such as myristoylation (Tull et al., 2004) and associates with the flagellar membrane (Tull et al., 2010, 2012). It is conceivable that ARM58 with its four DUF1935 might also be membrane associated. Indeed, a ARM58::mCHERRY fusion protein localises close to the flagellar pocket in L. infantum (Schäfer et al., 2013).
A dedicated deletion mutational analysis is required to find out whether one or more of the DUF1935 in ARM58 is mediating antimony resistance and whether the insertion sequence between DUF1935-III and DUF1935-IV plays any role. Given the discouraging experience with L. braziliensis as acceptor organisms for short episomal transgenes, such analyses will be carried out in a better suited model, i.e. L. infantum. Indeed, the L. infantum ortholog of ARM58, LinJ34.0220, is also capable of conferring antimony resistance and has been subject to deletion mutagenesis as well as cell biological analysis (Schäfer et al., 2013).
A remaining question is whether expression rates and/or gene copy numbers for ARM58 or its orthologs in other Leishmaniae varies between natural drug sensitive and drug resistant isolates. This should be investigated by using isolates from hot spots of drug resistance, such as Northern India (Croft et al., 2006), and using L. donovani.
Our experience shows that the Viannia subgenus (i.e. L. braziliensis complex) is less accommodating for functional cloning than Old World Leishmania or L. mexicana complex members. While the selection of full-length cosmids appeared to work reliably – 5 independent screens identified the same genomic region – we encountered problems with decreasing length gDNA inserts. This problem persisted even when different vectors were used (not shown) and irrespective of the number of in vitro passages to which the recombinant strains were subjected. It is possible that shorter gDNA sections are more likely to give rise to antisense RNA production which would then interfere with the stability of sense transcripts (Peacock et al., 2007; Lye et al., 2010). Considering our limited experience we propose to restrict functional cloning in L. braziliensis to the primary screens with full length cosmids. For subsequent steps, Old World Leishmania spp. can be used as acceptor cells. Alternatively, gene integration strategies may be employed similar to the situation with T. brucei (Clayton, 1999).
5. Conclusion
In spite of a strategy aimed at identifying structurally distinct drug resistance markers from resistant versus sensitive L. braziliensis isolates, the screen yielded a marker gene that acts in a gene dose-dependent manner. The gene codes for a hypothetical protein of 58 kD comprising four structurally related DUF1935 domains, with one possible trans-membrane domain. The gene and its product are functionally conserved in Old World and New World Leishmania spp., but not beyond. ARM58 overexpression protects both free living promastigotes and intracellular amastigotes against the effects of SbIII and SbV respectively. More must be learned about the impact of ARM58 in vivo, its possible role in clinical antimony treatment failure, its function in antimony tolerance and the significance of its tetra-partite domain structure.
Acknowledgements
We acknowledge helpful suggestions from the members of the LeishEpiNetSA consortium. This work was funded by EU Grant INCO-CT2005-015407-LeishEpiNet. AN was a fellow of the Deutscher Akademischer Austauschdienst (DAAD) for parts of the project. We also acknowledge expert technical assistance from Manfred Krömer (†2009) and Anne Macdonald. We thank Laura Jade Lee for a careful reading of the manuscript. We are not aware of conflicts of interest for any of the authors.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Appendix A. Supplementary data
References
- Adaui V., Castillo D., Zimic M., Gutierrez A., Decuypere S., Vanaerschot M., De Doncker S., Schnorbusch K., Maes I., Van der Auwera G., Maes L., Llanos-Cuentas A., Arevalo J., Dujardin J.C. Comparative gene expression analysis throughout the life cycle of Leishmania braziliensis: diversity of expression profiles among clinical isolates. PLoS Negl. Trop. Dis. 2011;5:e1021. doi: 10.1371/journal.pntd.0001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adaui V., Maes I., Huyse T., Van den Broeck F., Talledo M., Kuhls K., De Doncker S., Maes L., Llanos-Cuentas A., Schonian G., Arevalo J., Dujardin J.C. Multilocus genotyping reveals a polyphyletic pattern among naturally antimony-resistant Leishmania braziliensis isolates from Peru. Infect. Genet. Evol. 2011;11:1873–1880. doi: 10.1016/j.meegid.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Adaui V., Schnorbusch K., Zimic M., Gutierrez A., Decuypere S., Vanaerschot M., DE Doncker S., Maes I., Llanos-Cuentas A., Chappuis F., Arevalo J., Dujardin J.C. Comparison of gene expression patterns among Leishmania braziliensis clinical isolates showing a different in vitro susceptibility to pentavalent antimony. Parasitology. 2011;138:183–193. doi: 10.1017/S0031182010001095. [DOI] [PubMed] [Google Scholar]
- Alvar J., Velez I.D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., den Boer M. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo J., Ramirez L., Adaui V., Zimic M., Tulliano G., Miranda-Verastegui C., Lazo M., Loayza-Muro R., De Doncker S., Maurer A., Chappuis F., Dujardin J.C., Llanos-Cuentas A. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J. Infect. Dis. 2007;195:1846–1851. doi: 10.1086/518041. [DOI] [PubMed] [Google Scholar]
- Azeredo-Coutinho R.B., Mendonca S.C., Callahan H., Portal A.C., Max G. Sensitivity of Leishmania braziliensis promastigotes to meglumine antimoniate (glucantime) is higher than that of other Leishmania species and correlates with response to therapy in American tegumentary leishmaniasis. J. Parasitol. 2007;93:688–693. doi: 10.1645/GE-1031R.1. [DOI] [PubMed] [Google Scholar]
- Beverley S.M. Protozomics: trypanosomatid parasite genetics comes of age. Nat. Rev. Genet. 2003;4:11–19. doi: 10.1038/nrg980. [DOI] [PubMed] [Google Scholar]
- Brochu C., Wang J., Roy G., Messier N., Wang X.Y., Saravia N.G., Ouellette M. Antimony uptake systems in the protozoan parasite Leishmania and accumulation differences in antimony-resistant parasites. Antimicrob. Agents Chemother. 2003;47:3073–3079. doi: 10.1128/AAC.47.10.3073-3079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochu C., Haimeur A., Ouellette M. The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite leishmania. Cell Stress Chaperones. 2004;9:294–303. doi: 10.1379/CSC-15R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury K., Zander D., Kube M., Reinhardt R., Clos J. Identification of a Leishmania infantum gene mediating resistance to miltefosine and SbIII. Int. J. Parasitol. 2008;38:1411–1423. doi: 10.1016/j.ijpara.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Clayton C.E. Genetic manipulation of kinetoplastida. Parasitol. Today. 1999;15:372–378. doi: 10.1016/s0169-4758(99)01498-2. [DOI] [PubMed] [Google Scholar]
- Clayton C.E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos J., Choudhury K. Functional cloning as a means to identify Leishmania genes involved in drug resistance. Mini Rev. Med. Chem. 2006;6:123–129. doi: 10.2174/138955706775476028. [DOI] [PubMed] [Google Scholar]
- Croft S.L., Sundar S., Fairlamb A.H. Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 2006;19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin Y.J., Sulahian A., Pratlong F., Meneceur P., Gangneux J.P., Prina E., Dedet J.P., Derouin F. Virulence of Leishmania infantum is expressed as a clonal and dominant phenotype in experimental infections. Infect. Immun. 2001;69:7365–7373. doi: 10.1128/IAI.69.12.7365-7373.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooley P.R., Mertens H.D., Tull D., McConville M.J. 1H, 13C and 15N resonance assignments of SMP-1: a small myristoylated protein from Leishmania major. J. Biomol. NMR. 2006;36(Suppl. 1):26. doi: 10.1007/s10858-006-0010-1. [DOI] [PubMed] [Google Scholar]
- Gourbal B., Sonuc N., Bhattacharjee H., Legare D., Sundar S., Ouellette M., Rosen B.P., Mukhopadhyay R. Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J. Biol. Chem. 2004;279:31010–31017. doi: 10.1074/jbc.M403959200. [DOI] [PubMed] [Google Scholar]
- Grondin K., Haimeur A., Mukhopadhyay R., Rosen B.P., Ouellette M. Co-amplification of the gamma-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene pgpA in arsenite-resistant Leishmania tarentolae. EMBO J. 1997;16:3057–3065. doi: 10.1093/emboj/16.11.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimeur A., Guimond C., Pilote S., Mukhopadhyay R., Rosen B.P., Poulin R., Ouellette M. Elevated levels of polyamines and trypanothione resulting from overexpression of the ornithine decarboxylase gene in arsenite-resistant Leishmania. Mol. Microbiol. 1999;34:726–735. doi: 10.1046/j.1365-2958.1999.01634.x. [DOI] [PubMed] [Google Scholar]
- Haimeur A., Brochu C., Genest P., Papadopoulou B., Ouellette M. Amplification of the ABC transporter gene PGPA and increased trypanothione levels in potassium antimonyl tartrate (SbIII) resistant Leishmania tarentolae. Mol. Biochem. Parasitol. 2000;108:131–135. doi: 10.1016/s0166-6851(00)00187-0. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Stoffel W. TMBASE – a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hoyer C., Mellenthin K., Schilhabel M., Platzer M., Clos J. Leishmania and the Leishmaniases: Use of genetic complementation to identify gene(s) which specify species-specific organ tropism of Leishmania. Med. Microbiol. Immunol. 2001;190:53–56. doi: 10.1007/s004300100077. [DOI] [PubMed] [Google Scholar]
- Kapler G.M., Coburn C.M., Beverley S.M. Stable transfection of the human parasite Leishmania major delineates a 30-kilobase region sufficient for extrachromosomal replication and expression. Mol. Cell. Biol. 1990;10:1084–1094. doi: 10.1128/mcb.10.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.M., Das P., Tomás A.M. An approach to functional complementation by introduction of large DNA fragments into Trypanosoma cruzi and Leishmania donovani using a cosmid shuttle vector. Mol. Biochem. Parasitol. 1994;65:51–62. doi: 10.1016/0166-6851(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Krobitsch S., Brandau S., Hoyer C., Schmetz C., Hübel A., Clos J. Leishmania donovani heat shock protein 100: characterization and function in amastigote stage differentiation. J. Biol. Chem. 1998;273:6488–6494. doi: 10.1074/jbc.273.11.6488. [DOI] [PubMed] [Google Scholar]
- Laban A., Wirth D.F. Transfection of Leishmania enriettii and expression of chloramphenicol acetyltransferase gene. Proc. Natl. Acad. Sci. USA. 1989;86:9119–9123. doi: 10.1073/pnas.86.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legare D., Cayer S., Singh A.K., Richard D., Papadopoulou B., Ouellette M. ABC proteins of Leishmania. J. Bioenergy Biomembr. 2001;33:469–474. doi: 10.1023/a:1012870904097. [DOI] [PubMed] [Google Scholar]
- Leprohon P., Legare D., Raymond F., Madore E., Hardiman G., Corbeil J., Ouellette M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucl. Acids Res. 2009;37:1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos-Cuentas A., Tulliano G., Araujo-Castillo R., Miranda-Verastegui C., Santamaria-Castrellon G., Ramirez L., Lazo M., De Doncker S., Boelaert M., Robays J., Dujardin J.C., Arevalo J., Chappuis F. Clinical and parasite species risk factors for pentavalent antimonial treatment failure in cutaneous leishmaniasis in Peru. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2008;46:223–231. doi: 10.1086/524042. [DOI] [PubMed] [Google Scholar]
- Lye L.F., Owens K., Shi H., Murta S.M., Vieira A.C., Turco S.J., Tschudi C., Ullu E., Beverley S.M. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathogens. 2010;6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H.B., Whitney D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947;18:50–60. [Google Scholar]
- Mannaert A., Downing T., Imamura H., Dujardin J.C. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol. 2012;28:370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Mookerjee Basu J., Mookerjee A., Banerjee R., Saha M., Singh S., Naskar K., Tripathy G., Sinha P.K., Pandey K., Sundar S., Bimal S., Das P.K., Choudhuri S.K., Roy S. Inhibition of ABC transporters abolishes antimony resistance in Leishmania infection. Antimicrob. Agents Chemother. 2008;52:1080–1093. doi: 10.1128/AAC.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B., Mukhopadhyay R., Bannerjee B., Chowdhury S., Mukherjee S., Naskar K., Allam U.S., Chakravortty D., Sundar S., Dujardin J.C., Roy S. Antimony-resistant but not antimony-sensitive Leishmania donovani up-regulates host IL-10 to overexpress multidrug-resistant protein 1. Proc. Natl. Acad. Sci. USA. 2013;110:E575–E582. doi: 10.1073/pnas.1213839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M., Haimeur A., Grondin K., Legare D., Papadopoulou B. Amplification of ABC transporter gene pgpA and of other heavy metal resistance genes in Leishmania tarentolae and their study by gene transfection and gene disruption. Methods Enzymol. 1998;292:182–193. doi: 10.1016/s0076-6879(98)92015-8. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Legare D., Papadopoulou B. Multidrug resistance and ABC transporters in parasitic protozoa. J. Mol. Microbiol. Biotechnol. 2001;3:201–206. [PubMed] [Google Scholar]
- Peacock C.S., Seeger K., Harris D., Murphy L., Ruiz J.C., Quail M.A., Peters N., Adlem E., Tivey A., Aslett M., Kerhornou A., Ivens A., Fraser A., Rajandream M.A., Carver T., Norbertczak H., Chillingworth T., Hance Z., Jagels K., Moule S., Ormond D., Rutter S., Squares R., Whitehead S., Rabbinowitsch E., Arrowsmith C., White B., Thurston S., Bringaud F., Baldauf S.L., Faulconbridge A., Jeffares D., Depledge D.P., Oyola S.O., Hilley J.D., Brito L.O., Tosi L.R., Barrell B., Cruz A.K., Mottram J.C., Smith D.F., Berriman M. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racoosin E.L., Swanson J.A. Macrophage colony-stimulating factor (rM-CSF) stimulates pinocytosis in bone marrow-derived macrophages. J. Exp. Med. 1989;170:1635–1648. doi: 10.1084/jem.170.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racoosin E.L., Beverley S.M. Leishmania major: promastigotes induce expression of a subset of chemokine genes in murine macrophages. Exp. Parasitol. 1997;85:283–295. doi: 10.1006/expr.1996.4139. [DOI] [PubMed] [Google Scholar]
- Reiling L., Chrobak M., Schmetz C., Clos J. Overexpression of a single Leishmania major gene is sufficient to enhance parasite infectivity in vivo and in vitro. Mol. Microbiol. 2010;76:1175–1190. doi: 10.1111/j.1365-2958.2010.07130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal S., Yardley V., Chappuis F., Decuypere S., Khanal B., Singh R., Boelaert M., De Doncker S., Croft S., Dujardin J.C. Antimonial treatment of visceral leishmaniasis: are current in vitro susceptibility assays adequate for prognosis of in vivo therapy outcome? Microbes Infect. 2007;9:529–535. doi: 10.1016/j.micinf.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Roberts W.L., Berman J.D., Rainey P.M. In vitro antileishmanial properties of tri- and pentavalent antimonial preparations. Antimicrob. Agents Chemother. 1995;39:1234–1239. doi: 10.1128/aac.39.6.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.B., Hilley J.D., Dickens N.J., Wilkes J., Bates P.A., Depledge D.P., Harris D., Her Y., Herzyk P., Imamura H., Otto T.D., Sanders M., Seeger K., Dujardin J.C., Berriman M., Smith D.F., Hertz-Fowler C., Mottram J.C. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Molecular Cloning. [Google Scholar]
- Schäfer C., Tejera Nevado P., Zander D., Clos J. ARM58 Overexpression Reduces Intracellular Antimony Concentration in Leishmania infantum. Antimicrob Agents Chemother. 2013 doi: 10.1128/AAC.01881-13. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked-Mishan P., Ulrich N., Ephros M., Zilberstein D. Novel Intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J. Biol. Chem. 2001;276:3971–3976. doi: 10.1074/jbc.M005423200. [DOI] [PubMed] [Google Scholar]
- Soto J., Toledo J., Vega J., Berman J. Short report: efficacy of pentavalent antimony for treatment of Colombian cutaneous leishmaniasis. Am. J. Trop. Med. Hyg. 2005;72:421–422. [PubMed] [Google Scholar]
- Sundar S. Treatment of visceral leishmaniasis. Med. Microbiol. Immunol. (Berl) 2001;190:89–92. doi: 10.1007/s004300100088. [DOI] [PubMed] [Google Scholar]
- Tull D., Vince J.E., Callaghan J.M., Naderer T., Spurck T., McFadden G.I., Currie G., Ferguson K., Bacic A., McConville M.J. SMP-1, a member of a new family of small myristoylated proteins in kinetoplastid parasites, is targeted to the flagellum membrane in Leishmania. Mol. Biol. Cell. 2004;15:4775–4786. doi: 10.1091/mbc.E04-06-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tull D., Naderer T., Spurck T., Mertens H.D., Heng J., McFadden G.I., Gooley P.R., McConville M.J. Membrane protein SMP-1 is required for normal flagellum function in Leishmania. J. Cell Sci. 2010;123:544–554. doi: 10.1242/jcs.059097. [DOI] [PubMed] [Google Scholar]
- Tull D., Heng J., Gooley P.R., Naderer T., McConville M.J. Acylation-dependent and-independent membrane targeting and distinct functions of small myristoylated proteins (SMPs) in Leishmania major. Int. J. Parasitol. 2012;42:239–247. doi: 10.1016/j.ijpara.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Ubeda J.M., Legare D., Raymond F., Ouameur A.A., Boisvert S., Rigault P., Corbeil J., Tremblay M.J., Olivier M., Papadopoulou B., Ouellette M. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9:R115. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Tschudi C., Chakraborty T. RNA interference in protozoan parasites. Cell. Microbiol. 2004;6:509–519. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- Vergnes B., Gourbal B., Girard I., Sundar S., Drummelsmith J., Ouellette M. A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol. Cell. Proteomics. 2007;6:88–101. doi: 10.1074/mcp.M600319-MCP200. [DOI] [PubMed] [Google Scholar]
- Yardley V., Ortuno N., Llanos-Cuentas A., Chappuis F., Doncker S.D., Ramirez L., Croft S., Arevalo J., Adaui V., Bermudez H., Decuypere S., Dujardin J.C. American tegumentary leishmaniasis: is antimonial treatment outcome related to parasite drug susceptibility? J. Infect. Dis. 2006;194:1168–1175. doi: 10.1086/507710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


