Abstract
Here we present a quantitative mechanism-based investigation aimed at comparing the cell uptake, intracellular trafficking, endosomal escape and final fate of lipoplexes and lipid–protamine/deoxyribonucleic acid (DNA) (LPD) nanoparticles (NPs) in living Chinese hamster ovary (CHO) cells. As a model, two lipid formulations were used for comparison. The first formulation is made of the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and the zwitterionic lipid dioleoylphosphocholine (DOPC), while the second mixture is made of the cationic 3β-[N-(N,N-dimethylaminoethane)-carbamoyl] cholesterol (DC-Chol) and the zwitterionic helper lipid dioleoylphosphatidylethanolamine (DOPE). Our findings indicate that lipoplexes are efficiently taken up through fluid-phase macropinocytosis, while a less efficient uptake of LPD NPs occurs through a combination of both macropinocytosis and clathrin-dependent pathways. Inside the cell, both lipoplexes and LPD NPs are actively transported towards the cell nucleus, as quantitatively addressed by spatio-temporal image correlation spectroscopy (STICS). For each lipid formulation, LPD NPs escape from endosomes more efficiently than lipoplexes. When cells were treated with DOTAP–DOPC-containing systems the majority of the DNA was trapped in the lysosome compartment, suggesting that extensive lysosomal degradation was the rate-limiting factors in DOTAP–DOPC-mediated transfection. On the other side, escape from endosomes is large for DC-Chol–DOPE-containing systems most likely due to DOPE and cholesterol-like molecules, which are able to destabilize the endosomal membrane. The lipid-dependent and structure-dependent enhancement of transfection activity suggests that DNA is delivered to the nucleus synergistically: the process requires both the membrane-fusogenic activity of the nanocarrier envelope and the employment of lipid species with intrinsic endosomal rupture ability.
Keywords: Lipoplex, Lipid nanoparticle, Transfection barrier, Gene delivery
1. Introduction
Lipid nanocarriers have attracted much attention because of being biodegradable, nontoxic and nonimmunogenic delivery vectors for drugs and genes [1–3]. Among all the common lipid systems, lipoplexes and core–shell-type lipid nanoparticles (CSLNPs) have emerged as the two main classes of nanocarriers [4]. Lipoplexes are typically formed by bulk mixing between cationic liposomes and DNA solutions and are arranged as multilayer structures in which DNA is intercalated between alternating lipid bilayers [8–12] (Fig. 1). Historically, lipoplexes have been used for decades [5–7], but limitations concerning their physical and chemical stability, batch-to-batch reproducibility and low transfection efficiency (TE) have been thoroughly described [8–12]. Conversely, CSLNPs were originally synthesized with a lipid shell and a core composed of inorganic materials such as silica [13], or organic materials such as polymer microgels [14]. Based on the concept of core–shell architecture, lipid NPs composed of two functional building blocks were synthesized [15,16]. The first building block is a core of DNA complexed with protamine or other polycations (Fig. 1). The core of lipid/protamine–DNA (LPD) NPs acts as the carrier for the gene payload, provides mechanical stability, controlled morphology, narrow size distribution and reduces the amount of lipid needed for complete drug encapsulation. The second building block is the lipid shell that protects DNA from degradation, imparts biocompatibility and improves stability in biological fluids. Optimization of the core and the shell of LPD NPs can result in tunable and sustained drug release profiles. A recent suggestion is that LPD NPs can exhibit superior performances in the transfection potency compared to their lipoplex counterpart (i.e. lipoplexes with the same lipid composition and lipid/DNA charge ratio) [17], but the precise reason remains unclear. It is generally accepted that TE is rate limited by intracellular processes such as cellular uptake, intracellular trafficking and endosomal escape [18–24]. Therefore, a systematic, quantitative and mechanism-based investigation (and comparison) of the transfection process of lipoplexes and LPD NPs in live cells is needed if we want to substantially improve the effectiveness of lipid vectors. The acquired information would also enable us to recognize to what extent and how specific intracellular barriers affect the vector transfection activity. Thus, here we propose a quantitative mechanism-based investigation of the cell uptake, intracellular trafficking, endosomal escape and final fate of lipoplexes and LPD NPs in Chinese hamster ovary (CHO) living cells. In the present study, two lipid formulations were used for comparison since they are diversely potent and widely used [25–27]. The first formulation is made of the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and the zwitterionic lipid dioleoylphosphocholine (DOPC). The second mixture is made of the cationic 3β-[N-(N,N-dimethylaminoethane)-carbamoyl] cholesterol (DC-Chol) and the zwitterionic helper lipid dioleoylphosphatidylethanolamine (DOPE). Our findings indicate that fluid-phase macropinocytosis is the main internalization pathway of lipoplexes, while the uptake of LPD NPs occurs equally through macropinocytosis and the classical clathrin-associated endocytosis. Inside the cell, both lipoplexes and LPD NPs are actively transported towards the cell nucleus. For each lipid formulation, LPD NPs escaped from endosomes more efficiently than lipoplexes, most likely due to their distinctive core–shell-type nanostructure. The largest fraction of DOTAP–DOPC-containing systems reaches the lysosome compartment, suggesting that poor endosomal escape and extensive lysosomal degradation are the most relevant barriers in DOTAP–DOPC-mediated transfection. On the other side, escape from endosome is large in DC-Chol–DOPE-containing systems most likely due to the use of DOPE and cholesterol-like molecules, which can cause rupture of endosomes. Collectively, our mechanism-based investigation shows that efficient gene delivery requires both the membrane-fusogenic activity of the core–shell-type nanostructure and the use of lipid species with a tendency to destabilize endosomal membrane.
Fig. 1.
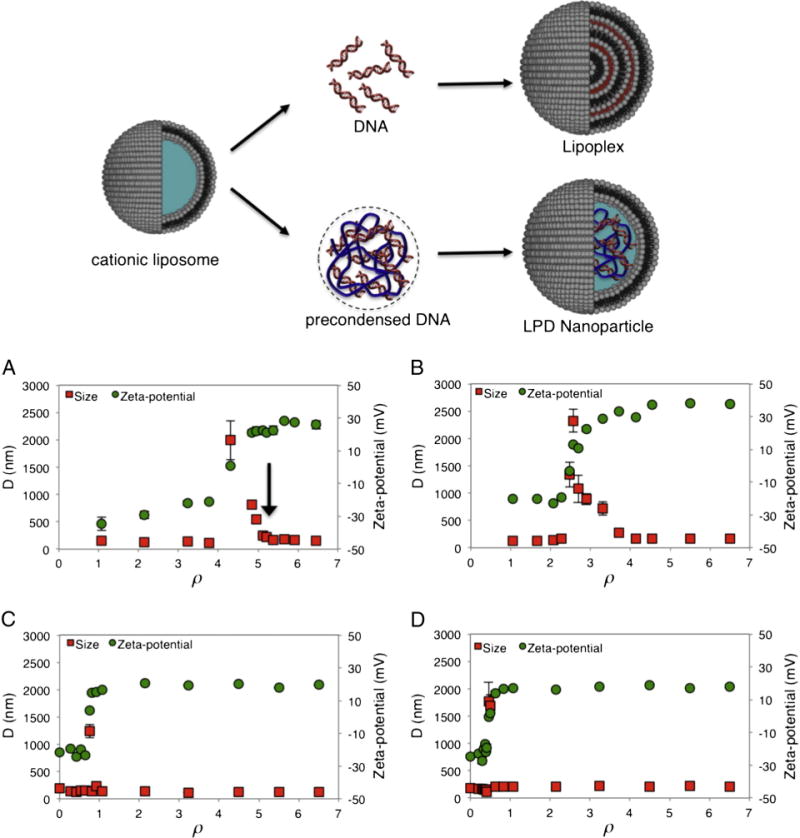
(Cartoon) Mechanism of formation of lipoplexes and lipid nanoparticles: lipoplexes are typically formed by bulk mixing between cationic liposomes and DNA solutions and are arranged as multilayer structures in which DNA is intercalated between alternating lipid bilayers. Lipid NPs are composed of a core of DNA complexed with protamine and covered by a lipid shell that protects DNA from degradation, imparts biocompatibility and improves stability in biological fluids. The hydrodynamic radius, D, and the zeta-potential of DOTAP–DOPC/DNA (panel A), DC-Chol–DOPE/DNA (panel B) lipoplexes and DOTAP–DOPC/P-DNA (panel C), DC-Chol–DOPE/P-DNA (panel D) nanoparticles, as a function of the cationic lipid/DNA charge ratio, ρ.
2. Materials and methods
2.1. Cationic liposomes
Cationic 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and (3β-[N-(N′,N′-dimethylaminoethane)-carbamoyl])-cholesterol (DC-Chol), the neutral helper lipids dioleoylphosphatidylethanolamine (DOPE) and dioleoylphosphocholine (DOPC), and the fluorescent neutral lipids 1-oleoyl-2-[12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino] dodecanoyl]-sn-glycero-3-phosphocholine (PC-NBD) and 1-oleoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoethanolamine (PE-NBD) were purchased from Avanti Polar Lipids (Alabaster, AL, USA) and used without further purification.
Cationic liposomes (CLs) were prepared according to standard protocols. In brief, the proper amount of lipids was dissolved in chloroform and the solvent was evaporated under vacuum for at least 24 h. The obtained lipid films were hydrated with the appropriate amount of Tris–HCl buffer solution (10−2 M, pH 7.4) to achieve the desired final concentration (1 mg/ml). Liposome dispersions were sonicated to clarity to prepare small unilamellar vesicles (SUVs). Experiments aimed at clarifying the mechanism of endosomal escape were performed by using CLs prepared with PC-NBD and PE-NBD mixed with unlabeled DOPC and DOPE to obtain labeled liposomes.
2.2. Lipoplexes
When adequate amounts of the DNA solutions were mixed with suitable volumes of CL liposome dispersions, self-assembled CL/DNA lipoplexes at desired cationic lipid/DNA charge ratio, ρ, were obtained. For size and zeta-potential experiments, calf thymus DNA was used. For confocal fluorescence microscopy experiments, Cy3-labeled 2.7 kbp plasmid DNA (Mirus Bio Corporation, Madison, WI, USA) was used.
2.3. Lipid-protamine/DNA nanoparticles
Protamine sulfate salt (P) from salmon (MW = 5.1 kDa) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Positively charged P/DNA microspheres at RW = 0.5 were mixed with SUVs at the desired charge ratio, ρ.
2.4. Size and zeta-potential measurements
Size and zeta-potential distributions of CLs, LPD nanoparticles, and lipoplexes were measured at 25 °C by a Malvern NanoZetaSizer spectrometer equipped with a 5 mW HeNe laser (wavelength λ = 632.8 nm) and a digital logarithmic correlator. The normalized intensity autocorrelation functions were analyzed by using the CONTIN method, which analyzes the autocorrelation function through an inverse Laplace transform [28] in order to obtain the distribution of the diffusion coefficient D of the particles. This coefficient is converted into an effective hydrodynamic radius RH by using the Stokes–Einstein equation RH = KBT/(6πηD), where KBT is the thermal energy and η the solvent viscosity. The electrophoretic mobility measurements were carried out by means of the laser Doppler electrophoresis technique, by the same apparatus used for size measurements. The mobility u was converted into the zeta-potential using the Smoluchowski relation zeta-potential = uη/ε, where η and ε are the viscosity and the permittivity of the solvent phase, respectively.
2.5. Transfection efficiency experiments
Cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with Glutamax-1 (Invitrogen, Carlsbad, CA, USA) supplemented with 1% penicillin–streptomycin (Invitrogen) and 10% fetal bovine serum (Invitrogen) at 37 °C and 5% CO2 atmosphere, splitting the cells every 2–4 days to maintain monolayer coverage. For luminescence analysis, Chinese hamster ovary (CHO) cells were transfected with pGL3 control plasmid (Promega, Fitchburg, WI, USA). The day before transfection, cells were seeded in 24-well plates (150,000 cells per well) using medium without antibiotics. Cells were incubated until they were 75–80% confluent, which generally took 18–24 h. For TE experiments, LPD nanoparticles and lipoplexes were prepared in Optimem (Invitrogen) by mixing for each well of 24-well plates 10 μl of sonicated lipid dispersions (1 mg/ml) with 0.5 μg of plasmid (lipoplexes) or 0.5 0.5 μg of plasmid pre-complexed with 0.25 μg of protamine. Complexes were left for 20 min at room temperature before adding them to the cells. On the day of transfection, the growth medium was replaced with 400 μl of Optimem and the cells were incubated for 30 min at 37 °C, before adding 100 μl of lipoplexesor LPD nanoparticles in Optimem. Cells were incubated at 37 °C for an additional 4 h to permit transient transfection. Finally, to avoid internalization of complexes that could remain bound to the cell surface after medium replacement, the cells were extensively washed 3× with phosphate buffered saline (PBS) before DMEM medium supplemented with 10% fetal bovine serum at 37 °C was added. After 48 h, cells were analyzed for luciferase expression using Luciferase Assay System from Promega. Briefly, cells were washed in PBS and harvested in 200 μl 1 × reporter lysis buffer (Promega). Of the cell suspension, 20 μl was diluted in 100 μl luciferase reaction buffer (Promega) and the luminescence was measured 10 s using a Berthold AutoLumat luminometer LB-953 (Berthold, Bad Wildbad, Germany). Results were expressed as relative light units per mg of cell proteins as determined by Bio-Rad Protein Assay Dye Reagent (Bio-Rad, Hercules, CA). Each condition was performed in quadruple and repeated three times.
2.6. Flow cytometric analysis
CHO cells were seeded in 24-well plates (450,000 cells/well) using medium without antibiotics. After 24 h, cells were incubated for 3 h with LPD NPs or lipoplexes prepared with Cy3-labeled DNA. Cells were then dissociated and suspended in PBS (1 × 106 cells/sample). Fluorescence was analyzed by FACScalibur with CellQuest and FLowJo software (Becton Dickinson, Mountain View, CA, USA).
2.7. Cell culture, transfection, colocalization assay
CHO-K1 cells were purchased from American Type Culture Collection (CCL-61 ATCC) and were grown in Ham’s F12K medium supplemented with 10% of fetal bovine serum at 37 °C and in 5% CO2. For transfection experiments, lipoplexes were prepared in PBS by mixing 1 μl of Cy3-labeled DNA with 10 μl of sonicated lipid dispersions. These complexes were left for 20 min at room temperature before adding them to the cells. In order to identify the endocytic vesicles involved in lipoplex internalization, we performed colocalization assays in living cells. CHO-K1 cells were coincubated with lipoplexes and different endocytic fluorescent markers: 1 mg/ml 70 kDa dextran–fluorescein isothiocyanate (FITC) conjugate at 37 °C for 30 min to label macropinosomes, 50 mM Lysosensor for 30 min to label lysosomes, 2 μg/ml transferrin-Alexa488 conjugate for 30 min to label recycling and sorting endosomes. Transfection of caveolin-E1GFP was carried out using lipofectamine reagent (Invitrogen) according to the manufacturer’s instruction. Colocalization of green and red signals was analyzed by the ImageJ software (NIH Image; http://rsbweb.nih.gov/ij/) [29].
To provide quantitative validation of endosomal escape, doubly labeled complexes were prepared with Cy3-labeled DNA (red) and NBD-labeled lipids (green). Cells were treated with complexes as for transfection, and the localization of the complexes monitored by time-lapse imaging. An estimate of the escape efficiency can be given by the fraction of cytoplasmic DNA, F(cyt), calculated as the pixel area corresponding to the red signal divided by the total pixel area of red and yellow signals.
2.8. Laser scanning confocal microscopy
Laser scanning confocal microscopy (LSCM) experiments were performed with the Olympus Fluoview 1000 (Olympus, Melville, NY) confocal microscope interfaced with a 405 nm diode laser, a 488 nm Argon laser, and 543 nm helium–neon laser. Glass bottom Petri dishes containing transfected cells were mounted in a temperature-controlled chamber at 37 °C and 5% CO2 and viewed with a 60 × 1.25 numerical aperture (NA) water immersion objective. The following collection ranges were adopted: 500–540 nm (EGFP, Alexa488-transferrin and FITC-dextran 70 kDa), 555–655 nm (Cy3), and 460–530 (Lysosensor). Images were collected in sequential mode to eliminate emission cross talk between the various dyes.
2.9. Spatio-temporal image correlation spectroscopy
Confocal microscopy experiments were carried out using a Fluoview FV-1000 (Olympus, Tokyo, Japan) microscope, with the HeNe laser at 543 nm to excite the fluorescently labeled pDNA Following the practical guidelines by Kolin and Wiseman [30], the experiments were carried out at 37 °C and were controlled by a data acquisition software (FV10-ASW, Olympus, Tokyo, Japan). For each region of interest (ROI), a temporal stack of at least 50 raster scanned images of 256 × 256 pixels was acquired. The pixel size was set equal to 0.1–0.25 μm/pixel, while the pixel dwell time was set equal to 8–20 μs/pixel. Due to the slow dynamics of the complexes, the time resolution (i.e. the distance in time between two subsequent frames) was set equal to the frame acquisition time plus a delay time on the order of 1–5 s (Δt in the following). Data were analyzed by a custom-made data acquisition software (SimFCS, Laboratory for Fluorescence Dynamics, Irvine, CA, USA – downloadable from www.lfd.uci.edu) and an additional custom-made Matlab (The MathWorks, Natick, MA) program. STICS is a powerful tool that allows us to obtain dynamic parameters of particles within different ROIs in a confocal two-dimensional image. From the raw temporal stack, a generalized discrete spatio-temporal correlation function can be defined [31]:
| (1) |
where δi(x, y, t) = i(x, y, t) − <i(x, y, t)>t is the intensity fluctuation at image pixel position (x, y) and time t, the angular brackets in the denominator represent spatial ensemble averaging over images at time kΔt and kΔt + τ in the time series, X and Y are the number of pixels spanning the region being analyzed (e.g. X = Y = 16, 32, 64, 128, 256,… pixels), N is the number of images in the image series (e.g. N = 50 in our experimental data), k is a dummy variable, Δt is the time resolution and δx = δy is the pixel size. In order to minimize the contribution of immobile fluorescent cell structures, an immobile filtering is applied to the image intensities prior to use Eq. (1) [30]. At zero-lag time (τ = 0), the autocorrelation function g(ξ,η,0) could be fitted to a 2D Gaussian centered in (ξ,η) = (0,0). If flow is present, the peak position moves in the ξ–η plane as a function of τ (ξ(τ) = −νxτ and η(τ) = − νyτ). Therefore, it is possible to obtain the velocity in the x–y plane from a linear fitting of the peak position. This first step allows us to distinguish types of motion, i.e. normal diffusion from flow and diffusion, without any a priori knowledge or assumption. In order to obtain the diffusion coefficient D, the temporal correlation function g(0,0,τ) could be fitted to the analytical model equation for normal diffusion (Eq. (2)) or flow and diffusion (Eq. (3)), according to the motion categorization achieved in the previous step.
| (2) |
where g(0,0,0) is the zero-lag amplitude, g∞ is the longtime offset, is the ratio between the average radial and axial beam radii, and the characteristic diffusion time .
| (3) |
where |ν| is the two-dimensional velocity, that in principle should be equal to the one obtained by tracking the moving peak of the STICS correlation function.
3. Results
3.1. Size and zeta-potential
DOTAP–DOPC and DC-Chol–DOPE CLs were found to be positively charged (zeta-potential = 50.3 ± 1.6 and 60.0 ± 2.5 mV, respectively) with a hydrodynamic diameter of D = 110 ± 8 and 98 ± 6 nm, respectively. To identify the most appropriate cationic lipid/DNA charge ratio (ρ) for transfection, size and zeta-potential of both lipoplexes and LPD NPs were measured. In Fig. 1A the hydrodynamic diameter (D) and the zeta-potential of DOTAP–DOPC/DNA lipoplexes are plotted against ρ. For 0.5 < ρ < 4, complexes are negatively charged and both their size and zeta-potential increase with increasing ρ. At ρ ~ 5 DOTAP–DOPC/DNA lipoplexes become neutrally charged (zeta-potential ~ 0 mV) and exhibit the maximum size (D ~ 2 μm). Further increase of the lipid content (ρ > 5) induces, as expected, charge inversion and re-entrant condensation of complexes [32,33]. In the case of DC-Chol–DOPE/DNA lipoplexes, the same phenomenology was observed, as reported in Fig. 1B. In more detail, charge inversion and re-entrant condensation of DC-Chol–DOPE/DNA lipoplexes start at ρ ~ 2.5. In Fig. 1C, D size and zeta-potential of DOTAP–DOPC/P-DNA and DC-Chol–DOPE/P-DNA NPs are reported. It is worth noting that both charge and size saturation were found to occur at ρ values much smaller than those observed for lipoplexes. This finding indicates that LPD NPs are definitely more efficient than lipoplexes in encapsulating DNA To compare the biological activity of different formulations, the same cationic lipid/DNA ratio must be chosen. To accomplish this requirement, the charge ratio ρ = 5 was chosen.
3.2. Transfection efficiency
The transfection efficiency of lipoplexes prepared in a serum-free cell culture medium was tested in vitro using the luciferase expression assay in CHO cells, as described in the Materials and methods section. Fig. 2 shows that DC-Chol–DOPE/DNA lipoplexes and DC-Chol–DOPE/P-DNA NPs are about three orders of magnitude more efficient than their DOTAP–DOPC counterpart. At the same time, TE of DC-Chol–DOPE/P-DNA NPs is about 2-fold times higher than that of DC-Chol–DOPE/DNA lipoplexes. This quantitative picture of the overall transfection process confirms the potential superior efficacy of LPD NPs as compared with lipoplexes. To understand which intracellular barriers and/or processes are responsible for the different transgene expressions, we performed a quantitative mechanism-based investigation comparing the cell uptake, intracellular trafficking, endosomal escape and final fate of the four formulations in the selected cell line.
Fig. 2.
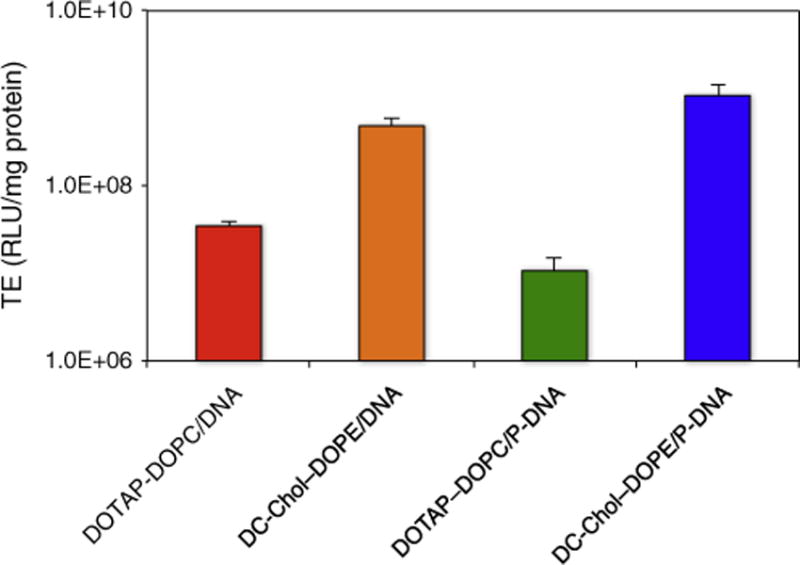
Transfection efficiency (TE) in RLU per mg of cellular proteins of lipoplexes and lipid nanoparticles.
3.3. Cell uptake
In Fig. 3 we report the amount of fluorescent positive cells after 3 h of incubation with lipoplexes and LPD NPs at 37 °C, measured by flow cytometry. These data support two main conclusions: i) cellular uptake of lipoplexes is higher than that of their LPD NP counterparts; ii) for both lipoplexes and LPD NPs cellular uptake of the DOTAP–DOPC-based complexes is higher than that of the DC-Chol–DOPE-based ones. It is remarkable that, upon CHO treatment with the less efficient formulation of DOTAP–DOPC/DNA lipoplexes, cell uptake reached a level as high as ~40%, anyhow superior to the most efficient formulation of DC-Chol–DOPE/P-DNA NPs (less than 10%). Although in the high-fluorescence population (less than 3%), the average fluorescence intensity of LPD NPs was larger than that of lipoplexes (data not reported), we are prompted to conclude that cellular uptake does not correlate with the measured transgene expression (Fig. 2).
Fig. 3.
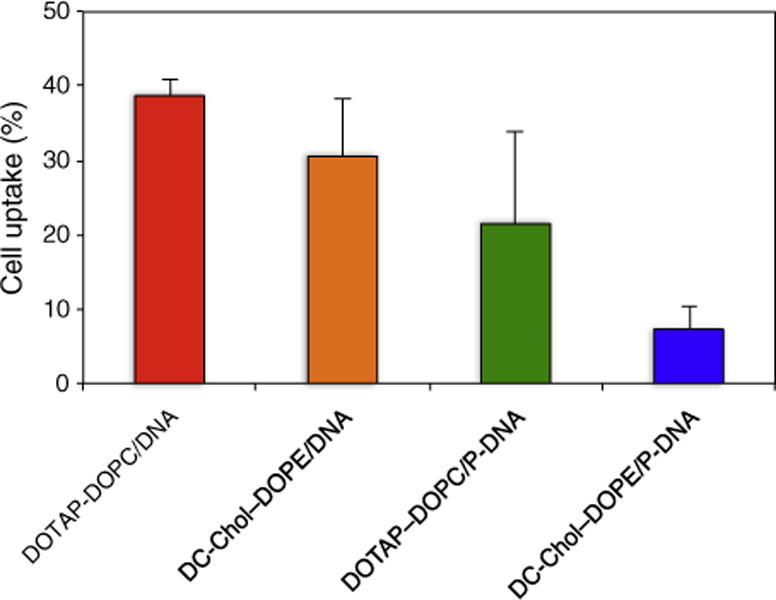
Cell uptake of lipoplexes and lipid nanoparticles given as percentage of fluorescent positive cells after 3 h of incubation with at 37 °C.
3.4. Uptake mechanism
To define the endocytotic route of lipid-mediated DNA delivery, CHO cells were treated with Cy3-DNA (red) and co-labeled with specific markers of endocytic pathways (green): clathrin-mediated endocytosis (Alexa488-labeled transferrin), caveolae-mediated endocytosis (caveolin-E1GFP) and macropinocytosis (70 kDa dextran). Colocalization of red and green fluorescence gave rise to visible yellow/orange punctate structures (Fig. 4). In addition to a qualitative inspection of confocal images, we performed a quantitative analysis of the fluorescence signal biodistribution by applying an object-based colocalization analysis protocol previously described [34]. The colocalization values are given as the fraction of cell-associated NPs colocalizing with fluorescently labeled endocytotic structures. At N = 20 cells, the colocalization parameters reached a plateau (not reported), suggesting that such a number of measurement repetitions was enough for a general conclusion to be valid. Therefore, in the following, each analyzed group is composed of at least 20 cells. At 1 h of treatment, DOTAP–DOPC/DNA and DC-Chol–DOPE/DNA lipoplexes do not colocalize with caveolin-E1GFP (Fig. 4A, B). This result clearly indicates that lipoplexes do not use this pathway to enter CHO cells. Similar conclusions can be drawn at t = 30 min for clathrin dependent endocytosis, in view of the lack of co-localization of the Cy3-DNA with Alexa488-labeled transferrin. Lastly, we used a fluorescently labeled 70 kDa dextran to visualize fluid phase macropinocytosis. Since macropinocytosis is slower than other endocytosis pathways, measurements were performed after 3 h of incubation. At this time point we observe that both lipoplex formulations considerably colocalize with the dextran marker, indicating macropinocytosis as the preferential pathway of lipoplex entry into CHO cells. By contrast, the internalization of LPD NPs is more heterogeneous in nature, as an increased number of NPs at any time would be able to occupy labeled endocytic vesicles (Fig. 4C, D), compared to the case with lipoplexes. Similarly to what is observed for lipoplexes, a fraction of NPs shows colocalization with macropinosomes. By contrast, however, image analysis reveals a positive contribution of clathrin-mediated endocytosis, as demonstrated by the significant overlap of the fluorescence signals from Cy3-DNA and Alexa488-labeled transferrin. It is worth noting that by summing the fractional contributions of all the mechanism involved in NP uptake, we obtain a number above 1. This is not surprising, in light of the well known structural overlapping between the different intracellular endocytotic pathways (e.g. the same endosome can be populated by molecules taken up by macropinocytosis or clathrin-mediated endocytosis). In summary, LSCM shows that, while lipoplexes enter CHO cells exclusively through fluid-phase macropinocytosis, LPD NPs can use two different endocytosis pathways: macropynocitosis and clathrin-mediated endocytosis.
Fig. 4.
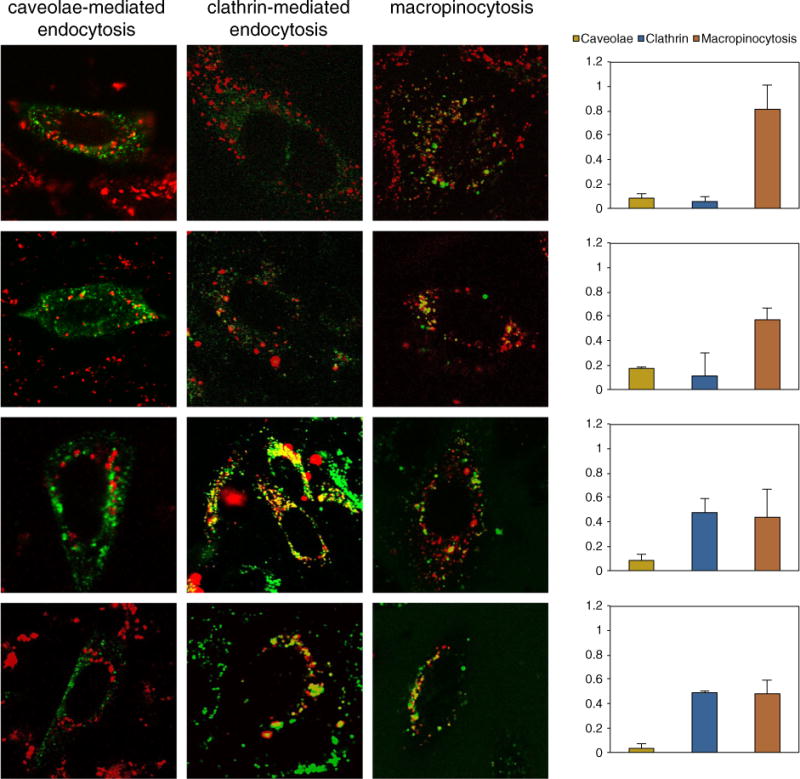
Colocalization of DOTAP–DOPC/DNA (panel A), DC-Chol–DOPE/DNA (panel B) lipoplexes and DOTAP–DOPC/P-DNA (panel C), DC-Chol–DOPE/P-DNA (panel D) nanoparticle signals (red) with specific markers of endocytic pathways (green): clathrin-mediated endocytosis (Alexa488-labeled transferrin), caveolae mediated endocytosis (caveolin-E1GFP) and macropinocytosis (70 kDa dextran). The histograms on the left show an object-based colocalization analysis protocol. The colocalization values are given as the fraction of cell-associated nanoparticles colocalizing with fluorescently labeled endocytic structures.
3.5. Intracellular trafficking
To date, most of the studies have focused on identifying the way of entry and final destination of nanocarriers, while completely missing their intracellular trafficking and interactions. To address this challenge, some of us recently introduced a quantitative method based on STICS that allows for characterizing the mode of motion of nanocarriers and for quantifying their transport parameters as they move through the cytosol in a living cell [36]. Fig. 5 introduces the fundamental steps of this analysis for a representative time series of DC-Chol–DOPE-P/DNA NPs. Fig. 5A is the superimposition of DC-Chol–DOPE-P/DNA NP fluorescence and the Nomarski image of the corresponding cell. By Eq. (1) we calculate the STICS function and fit it to a 2D Gaussian. The ROI size is first set to the image size, i.e. 256 × 256 pixels; this allows us to extract the average information about the intracellular dynamics motion of lipoplexes and LPD nanoparticles at the single-cell level. Without any previous assumption, it is possible to follow the 2D Gaussian peak motion from Fig. 5B in the x–y plane. From a linear interpolation of the peak position (Fig. 5C), the x- and y-components of the velocity can be retrieved as well as the particle velocity, . According to Herbert et al. [31], when vSTICS < 0.009 μm/s, the motion can be categorized as Brownian diffusion, while a value of vSTICS > 0.009 μm/s is clear indication of an active mode of motion. After having categorized the motion as combination of flow and diffusion, one can obtain the diffusion coefficient, DTICS, and the velocity, vTICS, by fitting g(0,0,τ), to Eq. (3). By its nature, the STICS technique measures an ensemble of particles and can report average values but not their distribution, as can be done by using single particle tracking (SPT). However, due to the ability of correlation techniques to analyze distinct ROIs in a confocal image, we could investigate the motion of particles within different subcellular regions. By segmenting the single-cell images into boxes as small as 64 × 64 pixels we revealed the presence of distinct local dynamics. In more detail, we observe that a large fraction of both lipoplexes and LPD nanoparticles is actively transported, while, in some less frequent cases, they mainly display a diffusive motion. Table 1 lists the average dynamic parameters obtained by applying this analysis to all temporal stacks acquired for lipoplexes and LPD nanoparticles. With respect to the mode of motion neither the kind of the delivery system nor the lipid formulation affords a reasonable explanation for the observed differences in TE.
Fig. 5.
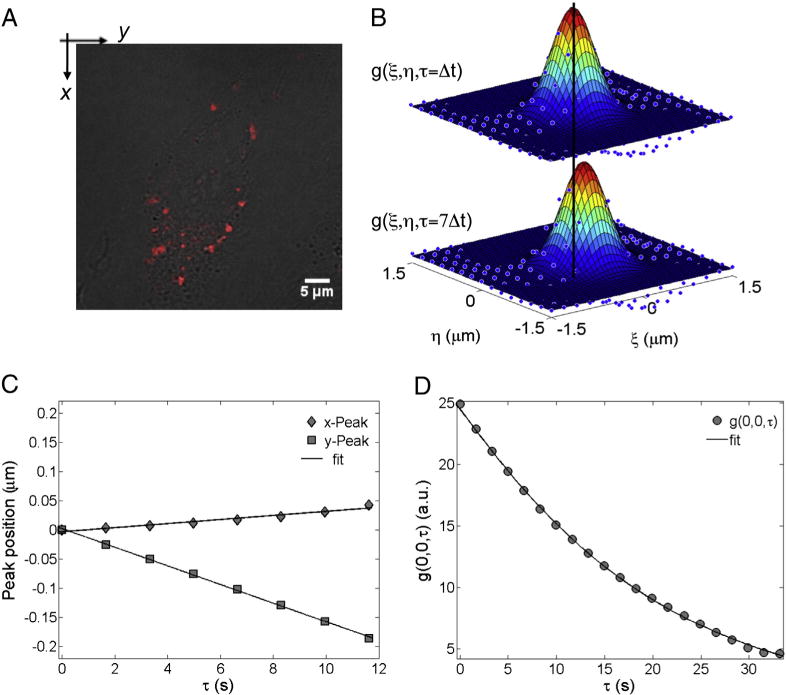
Steps of the STICS analysis: (A) superimposition of the confocal fluorescence image (the first image of the temporal stack) on top of the corresponding CHO-K1 cell Nomarski image; (B) the STICS functions at τ = Δt and 7Δt (the vertical black line helps to follow the motion of the STICS peak position); (C) the x- and y-coordinates (filled diamonds and squares, respectively) of the 2D Gaussian peak position as a function of the delay time τ. The solid lineis the best linear fit to the data. From the slope of the linear fits, the velocities (vSTICS) along the x- and y-directions could be calculated; (D) the raw temporal image correlation function (circles) and the related fit (solid line) using Eqs. (2) and (3) (depending on the type of motion) to obtain the dynamic parameters, i.e. the velocity vTICS and the diffusion coefficient DTICS.
Table 1.
Average dynamical parameters of lipoplexes and LPD NPs as obtained by the peak movement of the STICS function (vSTICS) and by fitting the temporal image correlation function using Eqs. (2) and (3) (vTICS and DTICS).
| vSTICS [μm/s] | vTICS [μm/s] | DTICS [μm2/s] | |
|---|---|---|---|
| DOTAP–DOPC/DNA | 0.0104 ± 0.0088 | 0.0225 ± 0.0106 | 0.0010 ± 0.0008 |
| DOTAP–DOPC-P/DNA | 0.0178 ± 0.0145 | 0.0219 ± 0.0123 | 0.0013 ± 0.0012 |
| DC-Chol–DOPE/DNA | 0.0298 ± 0.0149 | 0.0216 ± 0.0069 | 0.0010 ± 0.0007 |
| DC-Chol–DOPE/P-DNA | 0.0190 ± 0.0150 | 0.0192 ± 0.0115 | 0.0015 ± 0.0013 |
3.6. Endosomal escape
As endosomes maturate and fuse (and travel to the perinuclear region, the location of the lysosomal compartment), several particles may share the same endosomal compartment and become indistinguishable by LSCM as their separation is smaller than the resolution limit of the microscope. This effect should be more pronounced for particles that remain trapped in endosomes, which indeed look like larger, brighter spots in the images. As Fig. 6 shows, this is clearly the case of DOTAP–DOPC-based nanocarriers. Conversely, DC-Chol–DOPE-based complexes that appear as small, point-like particles remain spatially separated and are possibly more prone to escape from endosomal compartments. To provide quantitative validation of this suggestion, doubly labeled complexes were prepared with Cy3-labeled DNA (red) and NBD-labeled lipids (green in Fig. 6). Cells were treated with complexes as for transfection, and the localization of the complexes monitored by time-lapse imaging. After 4 h, cells show a punctuate fluorescence pattern: yellow clusters, as indicated by yellow arrows, are proof of the colocalization of the pDNA (red) and shell lipids (green), i.e. proof of the complex integrity. Isolated red clusters (red arrows) can be reasonably defined as cytoplasmic DNA. According to literature [37], each red cluster consists of a several DNA molecules, not a single one, and the pixel areas reflect the amount of the DNA in each cluster. Large (>1 μm) patches of red fluorescence appear indicating that DC-Chol–DOPE-based complexes largely escaped the endosome pathway. In this reasoning, an estimate of the escape efficiency can be given by the fraction of cytoplasmic DNA, F(cyt), calculated as the pixel area corresponding to the red signal divided by the total pixel area of red and yellow signals [37]. After transfection with DC-Chol–DOPE/DNA for 3 h, we selected up to 50 clusters randomly (in not less than 20 cells) to calculate the fraction of DNA in the cytoplasm. As Fig. 6E shows, values of F(cyt) were in the following order: DOTAP–DOPC/DNA < DOTAP–DOPC-P/DNA < DC-Chol–DOPE/DNA < DC-Chol–DOPE-P/DNA. Our results suggest that escape from lipid vesicles is responsible for the levels of transgene expression in TE experiments.
Fig. 6.

Laser scanning confocal microscopy on live CHO cells using doubly labeled complexes prepared with Cy3-labeled DNA (red) and NBD-labeled lipids (green): (panel A) DOTAP–DOPC/DNA lipoplexes, (panel B) DC-Chol–DOPE/DNA lipoplexes, (panel C) DOTAP–DOPC/P-DNA nanoparticles, (panel D) DC-Chol–DOPE/P-DNA nanoparticles. Yellow clusters, as indicated by yellow arrows, represent the colocalization of the pDNA (red) and lipids (green). Other red clusters not colocalized with green structure (as indicated by red arrows) were defined as cytoplasmic pDNA. The histogram on the left shows the fraction of cytoplasmic DNA, F(cyt), calculated as the pixel area of the red signal divided by the total pixel area of the red and yellow signals [37].
3.7. Lysosomal degradation
In previous section we have demonstrated that endosomal escape efficiency does correlate with TE. It cannot be excluded, however, that the fraction of DNA entrapped in vesicles 4 h after transfection (1-F(cyt)) might escape from endosomes on longer timescales. As a consequence, this fraction may be further divided into two subfractions: the fraction of DNA complexed with cationic lipid vesicles that is contained in the lysosome compartment (F(lys)) (destined to degradation) and the fraction of DNA that stays in vesicles that are not transported to lysosomes (F(lip)). Initially, we labeled the CHO cells with Lysosensor, a well-known lysosome marker (Fig. 7). Consequently, the cy3-DNA signal colocalized with the green Lysosensor signal (yellow clusters) was interpreted as the fraction of DNA in lysosomes, F(lys). After 3 h of incubation with DOTAP–DOPC/DNA lipoplexes, a marked colocalization of red and green fluorescence in large perinuclear vesicular structures was observed (Fig. 7A). This observation suggests that DOTAP–DOPC/DNA lipoplexes are largely destined to metabolic degradation in the lysosomes and provides a reasonable explanation for their low TE (Fig. 2). Strikingly, when CHO cells are transfected with DC-Chol–DOPE/DNA lipoplexes, a substantially lower degree of colocalization of Cy3-DNA and Lysosensor is detected (Fig. 7B). Fig. 7C, D also shows that labeled LPD NPs exhibit a further lower colocalization with lysosomes than their lipoplex counterparts. Thus, by applying the object-based colocalization analysis protocol described above, we quantitatively addressed the fraction of DNA complexed with cationic lipid vesicles that is contained in lysosomes. This analysis (Fig. 7E) shows that the degree of colocalization of nanocarriers with Lysosensor was in the order: DOTAP–DOPC/P-DNA > DOTAP–DOPC/DNA > DC-Chol–DOPE/DNA > DC-Chol–DOPE/P-DNA. This further clarifies why DC-Chol–DOPE-mediated DNA delivery could provide a much greater transfection efficiency than that of DOTAP–DOPC/DNA complexes.
Fig. 7.
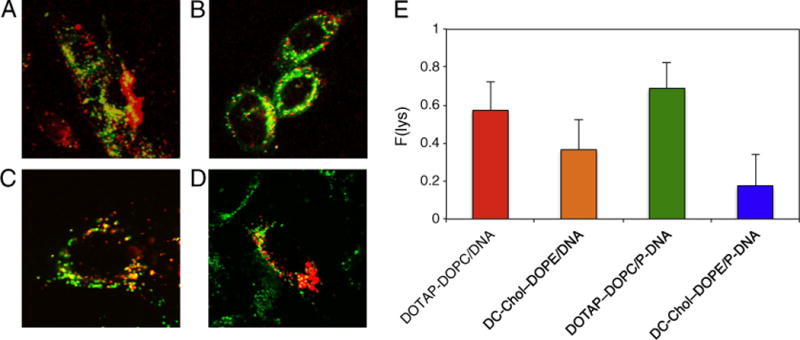
Colocalization of DOTAP–DOPC/DNA (panel A), DC-Chol–DOPE/DNA (panel B) lipoplexes and DOTAP–DOPC/P-DNA (panel C), DC-Chol–DOPE/P-DNA (panel D) nanoparticle signals (red) with Lysosensor (lysosome marker, green). (panel E) Fraction of DNA in the lysosomes, F(lys), calculated by using an object-based colocalization protocol.
Discussion
The successful design of a nonviral gene nanocarriers requires a deep understanding of the mechanisms involved in their interaction with the target cells. However, in the majority of the studies, the final transfection efficiency has been used as the only parameter to define their success. Consequently, it is difficult to predict which of the intracellular barriers represent the main rate-limiting factor in the transfection process. At each intracellular barrier, the nanocarrier has a certain chance of success (i.e. overcome the barrier and go forward in the process). The cumulative probability of success for the entire journey is revealed by the specific TE of the given system. In the present study, we compare the intracellular distribution of exogenous DNA transfected by multilamellar lipoplexes and core–shell type LPD NPs in CHO living cells. We were aimed at increasing our overall understanding of the sequence of events in lipid-mediated DNA delivery. Thus, we propose a combined experimental approach that is able to quantitatively and separately address each step of transfection process. At first, we investigated the formation of complexes by simultaneous determination of size and zeta-potential. As expected, both lipoplexes and LPD NPs exhibited charge inversion and re-entrant condensation, although with a pronounced higher DNA compaction ability by NPs as compared to lipoplexes. Overall, this characterization allowed us to determine the optimum cationic lipid/DNA charge ratio for TE experiments. Regarding the latter, we detected that DC-Chol–DOPE/DNA lipoplexes and DC-Chol–DOPE/P-DNA NPs were about three orders of magnitude more efficient than their DOTAP–DOPC-based counterparts. Among them, DC-Chol–DOPE/P-DNA NPs showed 3-fold higher luciferase expression than DC-Chol–DOPE/DNA lipoplexes. To account for those differences in TE, we first quantified the cellular uptake of lipoplexes and LPD NPs in terms of percentage of positive (fluorescent) cells by flow cytometry. Surprisingly, internalization efficiencies showed a clear anti-correlation with respect to the corresponding TE: e.g. DC-Chol–DOPE/P-DNA NPs yielded the highest TE but the lowest cellular uptake. This outcome prompted us to speculate that the cellular uptake of the two formulations may proceed along different routes. It has been well recognized that lipid/DNA complexes are mainly taken up through endocytosis, but different pathways, with distinct intracellular destinies, have been alternatively proposed [20,21,38]. As a result, a promising strategy for increasing the efficiency of non-viral vectors is to target certain uptake pathways that improve the intracellular fate of the particles. Such a plan requires a comprehensive understanding of the different uptake pathways and the subsequent intracellular events in each case. By confocal imaging and colocalization assays, here we define that the cell entry of lipoplexes occurs through macropinocytosis, while that of LPD NPs equally through macropinocytosis and the classical clathrin-associated endocytosis. To explain this phenomenon, a size-dependent differential uptake of the particles could be considered [20]. It appears, in fact, that the size of lipoplexes (generally more than 200 nm) is too large to be taken up via nonacidic vesicular transport such as caveolin-mediated endocytosis (diameter of caveolae ~60–90 nm) and clathrin-dependent endocytosis (diameter of clathrin-coated vesicles ~100–120 nm), while some LPD NPs are small enough to be internalized via clathrin-dependent endocytosis. Irrespective of the specific route taken, once inside the cell complexes need to travel long distances through the cytoplasm to reach the cell nucleus. Since diffusion through the dense meshwork of the cytoplasm is not likely, other mechanisms must exist by which nanocarriers can move towards the cell nucleus. Recently, it became clear that the cytoskeleton does not constitute a barrier for successful gene delivery, but on the contrary, it actively contributes to DNA transport, just as it does for viruses [36,39,40]. Here we addressed the nanocarrier intracellular trafficking by means of a recently introduced method, based on STICS, which enables the particle motion to be quantified in individual cells with sequential images captured by laser scanning confocal microscopy [36]. By this approach we proved that a large fraction of both lipoplexes and LPD nanoparticles is actively transported, with a minor concomitant contribution of a diffusion-like motion. Our results (Table 1) are consistent with previous reports showing that gene vectors are transported with typical velocity varying between ≈0.02 and ≈0.2 μm s−1 depending on several factors such as the lipid species and the particle size. Results from the four formulations reveal very similar dynamic behaviors, with the average velocities and diffusion coefficients being in the same range of values. Thus, we can conclude that active transport plays some important role in plasmid trafficking, but it seems not to be a significant barrier against transfection. Since endosomal escape and lysosomal degradation of plasmid DNA constitute a major barrier for transgene expression, a quantitative evaluation of these barriers would be highly desirable in terms of optimizing a non-viral gene delivery system. Actually, entrapment and degradation can be regarded as two separate barriers, because preventing lysosomal degradation results in an accumulation of genes in intracellular vesicles without enhancing cytosolic release. Therefore, to reach the nucleus, DNA must avoid degradation within lysosomes but concomitantly escape from intracellular vesicles into the cytosol. To evaluate the extent to which this pathway contributes to the cytoplasmic distribution of DNA, we evaluated the escape efficiency through quantification of the fraction of cytoplasmic DNA, F(cyt). 4 h after transfection fluorescent particles are seen inside of the cells in a punctuate pattern (Fig. 6). In the case of the poorly efficient DOTAP–DOPC/DNA lipoplexes (Fig. 6A), most of the DNA is not released from cationic lipid/vesicles and consequently detected in the cytosol (F(cyt) < 0.1), suggesting that poor endosomal escape may be responsible for low transgene expression. By contrast, DC-Chol–DOPE/DNA lipoplexes (Fig. 6C) and DC-Chol–DOPE/P-DNA NPs (Fig. 6D) show a significantly higher value of cytoplasmic DNA (F(cyt) ≈ 0.5 and 0.7 respectively), which clearly indicates that DC-Chol–DOPE-based systems have a superior efficacy in terms of escape into the cytosol. This remarkable property is most probably due to the specific contribution of DOPE, due to its intrinsic ability to promote lamellar-to-hexagonal phase transitions, resulting in endosomal membrane perturbation and/or disruption. Furthermore, the use of cholesterol-like molecules, as DC-Chol is, in the lipoplex formulation has been shown to enhance transfection both in vitro and in vivo. In vitro this could be due, at least in part, to the formation of cholesterol nanodomains in the lipoplex membrane that may play a role either in cellular uptake or in intracellular trafficking [12,41–43]. In addition, the role of cationic lipids deserves consideration. Indeed, DOTAP is a pH-insensitive lipid with a permanent cationic charge, while DC-Chol has a titrable tertiary amine group (pKa = 7.8). This means that at physiological pH DC-Chol is partially deprotonated resulting in reduced surface charge. Reduction of the cationic charge could weaken the lipid/DNA attraction with the result that DNA could be more easily released from lipoplexes. Fig. 6D also shows that, for a given lipid composition, LPD NPs are about 2-fold superior with respect to their lipoplex counterparts in terms of DNA delivery to the cytosol. A possible explanation arises from the different nanostructures of these complexes. It is accepted that, upon nanocarrier–cellular membrane interaction, anionic cellular lipids laterally diffuse into the complex and locally neutralize cationic lipids [44]. Formation of cationic/anionic mixed bilayers is expected to weaken the electrostatic attraction between cationic lipids and anionic DNA molecules. Only when the membrane charge density of cationic membranes is completely neutralized by anionic lipids the DNA does start to escape from complexes in an appreciable manner [45]. While lipoplexes are multilamellar structures made of some tenths of alternating lipid/DNA layers, LPD NPs are made of DNA complexed with a polycation and encapsulated by a lipid envelope made of a few layers. The ability of LPD NPs to escape from endosomes is presumably related to the more favorable interaction between cationic and anionic cellular lipids due to the absence of competing DNA in the lipid envelope and to the lower number of lipid layers to be peeled off. Moreover, some authors have recently shown that the endosomal escape of lipid gene vectors is extremely low [46]. According to their results, core–shell NPs (containing protamine) may show a burst release from endosomes, whereas lipoplexes could exhibit a more gradual release.
According to literature [37] we evaluated the endosomal escape ability 4 h after transfection. However, DNA might escape from endosomes on longer timescales. On the other side, it is believed that DNA entry into lysosomes is followed by degradation resulting in transfection failure. Thus, to account for differences in TE, final fate of complexes was compared by the fraction of DNA in the lysosomes, F(lys). When CHO cells are treated with DOTAP–DOPC/DNA lipoplexes and DOTAP–DOPC/P-DNA NPs, the majority of DNA-containing vesicles coincides with the lysosome compartment (F(lys) = 0.58 and 0.70 respectively), confirming that metabolic degradation is the rate-limiting factor in DOTAP–DOPC-mediated transfection. On the opposite, the fraction of DNA in lysosomes is much lower for DC-Chol–DOPE/DNA lipoplexes and DC-Chol–DOPE/P-DNA NPs (F(lys) = 0.35 and 0.17 respectively). As a last step towards identifying intracellular barriers responsible for the difference in TE the post-nuclear delivery processes must be investigated [47]. Additionally, to bring optimized nanocarriers to the forefront of gene delivery, understanding how transfection mechanisms are affected by the ‘protein corona’ [48–56] displayed by nanocarriers after exposure to biological fluids is an urgent task.
Conclusions
Collectively, we successfully quantified the cellular uptake, intracellular trafficking and final fate of two formulations of lipoplexes and LPD NPs. As a result, the differences in transgene expression could be explained by the differences in intracellular fate, such as the efficiency of endosomal escape of the DNA and its lysosomal degradation. The lipid-dependent and structure-dependent enhancement of transfection activity suggests that DNA is delivered to the nucleus synergistically: the process requires both the membrane-fusogenic activity of the nanocarrier envelope and lipid species with intrinsic endosomal rupture ability. The acquired knowledge can serve as a guideline for future studies on the optimization of non-viral gene vectors. The last barrier to efficient transfection is the nuclear entry. This mechanism of action is strongly dependent on cell division [57,58] and is currently being investigated in our laboratory.
Acknowledgments
We acknowledge Milka Titin for cell cultivation. This work was partially supported by the Italian Minister for University and Research (MIUR) (Futuro in Ricerca, Grant no. RBFR08TLPO; PRIN Grant no. 2009ACFPN9_002). E.G. and M.A.D. acknowledge support from the National Center for Research Resources (5P41RR003155), the National Institute of General Medical Sciences divisions of the National Institutes of Health (8P41GM103540 and 5P50 GM076516).
References
- 1.Guo X, Huang L. Recent advances in nonviral vectors for gene delivery. Acc Chem Res. 2012;45:971–979. doi: 10.1021/ar200151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balazs DA, Godbey WT. Liposomes for use in gene delivery. J Drug Deliv. 2011;2011:326497. doi: 10.1155/2011/326497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang H, Yeh M. Clinical development of liposome-based drugs: formulation, characterization, and therapeutic efficacy. Int J Nanomedicine. 2012;7:49–60. doi: 10.2147/IJN.S26766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandal B, Bhattacharjee H, Mittal N, Sah H, Balabathula P, Thoma LA, Wood GC. Core–shell-type lipid–polymer hybrid nanoparticles as a drug delivery platform. Nanomedicine: NBM. 2013;9:474–491. doi: 10.1016/j.nano.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Felgner PL, Ringold GM. Cationic liposome-mediated transfection. Nature. 1989;337:387–388. doi: 10.1038/337387a0. [DOI] [PubMed] [Google Scholar]
- 6.Smisterova J, Wagenaar A, Stuart MCA, Polushkin E, Ten Brinke G, Hulst R, Engberts JBFN, Hoekstra D. Molecular shape of the cationic lipid controls the structure of cationic lipid/dioleylphosphatidylethanolamine–DNA complexes and the efficiency of delivery. J Biol Chem. 2001;276:47615–47622. doi: 10.1074/jbc.M106199200. [DOI] [PubMed] [Google Scholar]
- 7.Caracciolo G, Amenitsch H. Cationic liposome/DNA complexes: from structure to interactions with cellular membranes. Eur Biophys J. 2012;41:815–829. doi: 10.1007/s00249-012-0830-8. [DOI] [PubMed] [Google Scholar]
- 8.Marchini C, Pozzi D, Montani M, Alfonsi C, Amici A, Candeloro De Sanctis S, Digman MA, Sanchez S, Gratton E, Amenitsch H, Fabbretti A, Gualerzi CO, Caracciolo G. Role of temperature-independent lipoplex–cell membrane interactions in the efficiency boost of multicomponent lipoplexes. Cancer Gene Ther. 2011;18:543–552. doi: 10.1038/cgt.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozzi D, Caracciolo G, Caminiti R, Candeloro De Sanctis S, Amenitsch H, Marchini C, Montani M, Amici A. Toward the rational design of lipid gene vectors: shape coupling between lipoplex and anionic cellular lipids controls the phase evolution of lipoplexes and the efficiency of DNA release. ACS Appl Mater Interfaces. 2009;10:2237–2224. doi: 10.1021/am900406b. [DOI] [PubMed] [Google Scholar]
- 10.Resina S, Prevot P, Thierry AR. Physico-chemical characteristics of lipoplexes influence cell uptake mechanisms and transfection efficacy. PLoS ONE. 2009;4:e6058. doi: 10.1371/journal.pone.0006058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koynova R, Wang L, MacDonald RC. An intracellular lamellar–nonlamellar phase transition rationalizes the superior performance of some cationic lipid transfection agents. Proc Natl Acad Sci U S A. 2006;103:14373–14378. doi: 10.1073/pnas.0603085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pozzi D, Marchini C, Cardarelli F, Bifone A, Garulli C, Caracciolo G. Transfection efficiency boost of cholesterol-containing lipoplexes. Biochim Biophys Acta. 2012;181:2335–2343. doi: 10.1016/j.bbamem.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Fuller JE, Zugates GT, Ferreira LS, Ow HS, Nguyen NN, Wiesner UB, Langer RS. Intracellular delivery of core–shell fluorescent silica nanoparticles. Biomaterials. 2008;29:1526–1532. doi: 10.1016/j.biomaterials.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Wei B, Gurr PA, Gozen AO, Blencowe A, Solomon DH, Qiao GG, Spontak RJ, Genzer J. Autophobicity-driven surface segregation and patterning of core–shell microgel nanoparticles. Nano Lett. 2008;8:3010–3016. doi: 10.1021/nl802109x. [DOI] [PubMed] [Google Scholar]
- 15.Troutier AL, Ladavière C. An overview of lipid membrane supported by colloidal particles. Adv Colloid Interface Sci. 2007;133:1–21. doi: 10.1016/j.cis.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Zhang L. Lipid–polymer hybrid nanoparticles: synthesis, characterization and applications. Nano LIFE. 2010;1:163–173. [Google Scholar]
- 17.Caracciolo G, Pozzi D, Capriotti AL, Marianecci C, Carafa M, Marchini C, Montani M, Amici A, Amenitsch H, Digman MA, Gratton E, Sanchez SS, Laganà A. Factors determining the superior performance of lipid nanoparticles over lipoplexes. J Med Chem. 2011;54:4160–4171. doi: 10.1021/jm200237p. [DOI] [PubMed] [Google Scholar]
- 18.Zuhorn IS, Hoekstra D. On the mechanism of cationic amphiphile-mediated transfection. To fuse or not to fuse: is that the question? J Membr Biol. 2002;189:167–179. doi: 10.1007/s00232-002-1015-7. [DOI] [PubMed] [Google Scholar]
- 19.Hoekstra D, Rejman J, Wasungu L, Shi F, Zuhorn IS. Gene delivery by cationic lipids: in and out of an endosome. Biochem Soc Trans. 2007;35:68–71. doi: 10.1042/BST0350068. [DOI] [PubMed] [Google Scholar]
- 20.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XX, Allen PG, Grinstaff M. Macropinocytosis is the major pathway responsible for DNA transfection in CHO cells by a charge-reversal amphiphile. Mol Pharm. 2011;8:758–766. doi: 10.1021/mp100366h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini C, Pozzi D, Montani M, Alfonsi C, Amici A, Amenitsch H, Candeloro De Sanctis S, Caracciolo G. Tailoring lipoplex composition to the lipid composition of plasma membrane: a trojan horse for cell entry? Langmuir. 2010;26:13867–13873. doi: 10.1021/la1023899. [DOI] [PubMed] [Google Scholar]
- 24.Akita H, Kogure K, Moriguchi R, Nakamura Y, Higashi T, Nakamura T, Serada S, Fujimoto M, Naka T, Futaki S, Harashima H. Nanoparticles for ex vivo siRNA delivery to dendritic cells for cancer vaccines: programmed endosomal escape and dissociation. J Control Release. 2010;143:311–317. doi: 10.1016/j.jconrel.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Cardarelli F, Pozzi D, Bifone A, Marchini C, Caracciolo G. Cholesterol-dependent macropinocytosis and endosomal escape control the transfection efficiency of lipoplexes in CHO living cells. Mol Pharm. 2012;9:334–340. doi: 10.1021/mp200374e. [DOI] [PubMed] [Google Scholar]
- 26.Maitani Y, Igarashi S, Sato M, Hattori Y. Cationic liposome (DC-Chol/DOPE = 1:2) and a modified ethanol injection method to prepare liposomes, increased gene expression. Int J Pharm. 2007;342:33–39. doi: 10.1016/j.ijpharm.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 27.Muñoz-Ubeda M, Rodríguez-Pulido A, Nogales A, Martín-Molina A, Aicart E, Junquera E. Effect of lipid composition on the structure and theoretical phase diagrams of DC-Chol/DOPE–DNA lipoplexes. Biomacromolecules. 2010;11:3332–3340. doi: 10.1021/bm1008124. [DOI] [PubMed] [Google Scholar]
- 28.Provencher SW. CONTIN: a general purpose constrained regularization program for inverting noisy linear algebraic and integral equations. Comput Phys Commun. 1982;27:229–242. [Google Scholar]
- 29.Rasband WS. ImageJ. US National Institutes of Health; Bethesda, MD, USA: http://imagej.nih.gov/ij. [Google Scholar]
- 30.Kolin DL, Wiseman PW. Advances in image correlation spectroscopy: measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem Biophys. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- 31.Hebert B, Costantino S, Wiseman P. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys J. 2005;88:3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuzzi S, Cametti C, Onori G. Polyion-induced aggregation of lipidic-coated solid polystyrene spheres: the many facets of complex formation in low-density colloidal suspensions. Langmuir. 2008;24:6044–6049. doi: 10.1021/la8005458. [DOI] [PubMed] [Google Scholar]
- 33.Pozzi D, Marianecci C, Carafa M, Marchini C, Montani M, Amici A, Caracciolo G. Programmed packaging of multicomponent envelope-type nanoparticle system for gene delivery. Appl Phys Lett. 2010;96:183702. doi: 10.1016/j.jconrel.2010.07.063. [DOI] [PubMed] [Google Scholar]
- 34.Sandin P, Fitzpatrick LW, Simpson JC, Dawson KA. High-speed imaging of Rab family small GTPases reveals rare events in nanoparticle tracking in living cells. ACS Nano. 2012;6:1513–1521. doi: 10.1021/nn204448x. [DOI] [PubMed] [Google Scholar]
- 35.Soldati T, Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat Rev Mol Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- 36.Coppola S, Pozzi D, Candeloro De Sanctis S, Digman MA, Gratton E, Caracciolo G. Quantitative measurement of intracellular transport of nanocarriers by spatio-temporal image correlation spectroscopy. Methods Appl Fluoresc. 2013;1:015005. doi: 10.1088/2050-6120/1/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Akita H, Kudo A, Minoura A, Yamaguti M, Khalil IA, Moriguchi R, Masuda T, Danev R, Nagayama K, Kogure K, Harashima H. Multi-layered nanoparticles for penetrating the endosome and nuclear membrane via a step-wise membrane fusion process. Biomaterials. 2009;30:2940–2949. doi: 10.1016/j.biomaterials.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Coppola S, Estrada LC, Digman MA, Gratton E, Caracciolo G. Intracellular trafficking of cationic liposome–DNA complexes in living cells. Soft Matter. 2012;8:7919–7927. doi: 10.1039/C2SM25532D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppola S, Cardarelli F, Pozzi D, Estrada LC, Digman MA, Gratton E, Bifone A, Caracciolo G. The role of cytoskeleton networks on lipid-mediated delivery of DNA. Ther Deliv. 2013;4:191–202. doi: 10.4155/tde.12.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L, Anchordoquy TJ. Effect of cholesterol nanodomains on the targeting of lipid-based gene delivery in cultured cells. Mol Pharm. 2010;7:1311–1317. doi: 10.1021/mp100097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu L, Wempe MF, Anchordoquy TJ. The effect of cholesterol domains on PEGylated liposomal gene delivery in vitro. Ther Deliv. 2011;2:451–460. doi: 10.4155/tde.11.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betker JL, Kullberg M, Gomez J, Anchordoquy TJ. Cholesterol domains enhance transfection. Ther Deliv. 2013;4:453–462. doi: 10.4155/tde.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelphati O, Szoka FC. Mechanism of oligonucleotide release from cationic liposomes. Proc Natl Acad Sci U S A. 1996;93:11493–11498. doi: 10.1073/pnas.93.21.11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caracciolo G, Pozzi D, Amenitsch H, Caminiti R. Interaction of lipoplexes with anionic lipids resulting in DNA release is a two-stage process. Langmuir. 2007;23:8713–8717. doi: 10.1021/la7017665. [DOI] [PubMed] [Google Scholar]
- 46.Hama S, Akita H, Iida S, Mizuguchi H, Harashima H. Quantitative and mechanism-based investigation of post-nuclear delivery events between adenovirus and lipoplex. Nucleic Acids Res. 2007;35:1533–1543. doi: 10.1093/nar/gkl1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ur Rehamn Z, Hoekstra D, Zuhorn IS. Mechanism of polyplex- and lipoplex delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano. 2013;7:3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- 48.Caracciolo G, Callipo L, Candeloro De Sanctis S, Cavaliere C, Pozzi D, Laganà A. Surface adsorption of protein corona controls the cell internalization mechanism of DC-Chol–DOPE/DNA lipoplexes in serum. Biochim Biophys Acta. 2010;1798:536–543. doi: 10.1016/j.bbamem.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Mahon E, Salvati A, Baldelli Bombelli F, Lynch I, Dawson KA. Designing the nanoparticle-biomolecule interface for “targeting and therapeutic delivery”. J Control Release. 2012;161:164–174. doi: 10.1016/j.jconrel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Caracciolo G. The protein corona effect for targeted drug delivery, Bioinspired. Biomimetic and Nanobiomaterials. 2012;2:54–57. [Google Scholar]
- 51.Walkey CD, Chan WCW. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 52.Monopoli MP, Åberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 53.Lesniak A, Salvati A, Santos-Martinez MJ, Radomski MW, Dawson KA, Åberg C. Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency. J Am Chem Soc. 2013;135:438–444. doi: 10.1021/ja309812z. [DOI] [PubMed] [Google Scholar]
- 54.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Baldelli Bombelli F, Hristov DR, Kelly PM, Åberg C, Mahon E, Dawson KA. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat Nanotechnol. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 55.Barrán-Berdón AL, Pozzi D, Caracciolo G, Capriotti AL, Caruso G, Cavaliere C, Riccioli A, Palchetti S, Laganà A. Time evolution of nanoparticle–protein corona in human plasma: relevance for targeted drug delivery. Langmuir. 2013;29:6485–6494. doi: 10.1021/la401192x. [DOI] [PubMed] [Google Scholar]
- 56.Wang F, Yu L, Salvati A, Dawson KA. The biomolecular corona is retained during nanoparticle uptake and protects the cells from the damage induced by cationic nanoparticles until degraded in the lysosomes. Nanomedicine: NBM. 2013 doi: 10.1016/j.nano.2013.04.010. http://dx.doi.org/10.1016/j.nano.2013.04.010. [DOI] [PubMed]
- 57.Kim JA, Åberg C, Salvati A, Dawson KA. Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population. Nat Nanotechnol. 2011;7:62–68. doi: 10.1038/nnano.2011.191. [DOI] [PubMed] [Google Scholar]
- 58.Åberg C, Kim JA, Salvati A, Dawson KA. Theoretical framework for nanoparticle uptake and accumulation kinetics in dividing cell populations. Europhys Lett. 2013;101:38007. [Google Scholar]


