Abstract
Objective
CYP2B6 variation predicts pharmacokinetic characteristics of its substrates. Consideration for underlying genetic structure is critical to protect against spurious associations with the highly polymorphic CYP2B6 gene.
Design
The effect of CYP2B6 variation on response to its substrates, nonnucleoside reverse transcriptase inhibitors (NNRTIs), was explored in the Women's Interagency HIV Study.
Methods
Five putative functional polymorphisms were tested for associations with virologic suppression within one year after NNRTI initiation in women naïve to antiretroviral agents (n=91). Principal components (PCs) were generated to control for population substructure. Logistic regression was used to test the joint effect of rs3745274 and rs28399499, which together indicate slow, intermediate, and extensive metabolizers.
Results
Rs3745274 was significantly associated with virologic suppression (OR=3.61, 95% CI 1.16-11.22, p trend=0.03); the remaining polymorphisms tested were not significantly associated with response. Women classified as intermediate and slow metabolizers were 2.90 (95% CI 0.79-12.28) and 13.44 (95% CI 1.66-infinity) times as likely to achieve virologic suppression compared to extensive metabolizers after adjustment for PCs (p trend=0.005). Failure to control for genetic ancestry resulted in substantial confounding of the relationship between the metabolizer phenotype and treatment response.
Conclusion
The CYP2B6 metabolizer phenotype was significantly associated with virologic response to NNRTIs; this relationship would have been masked by simple adjustment for self-reported ethnicity. Given the appreciable genetic heterogeneity that exists within self-reported ethnicity, these results exemplify the importance of characterizing underlying genetic structure in pharmacogenetic studies. Further follow-up of the CYP2B6 metabolizer phenotype is warranted given the potential clinical importance of this finding.
Keywords: CYP2B6, population substructure, women, NNRTIs, confounding
Introduction
Genetic variation in pathways of drug absorption, delivery, metabolism and excretion (ADME) contributes to the variability in pharmacokinetic parameters of drug levels and clearance in plasma or cells over time, such as half life or area under the curve (AUC) [1-2]. Pharmacokinetic characteristics are often associated with clinical endpoints, such as fasting glucose levels after exposure to anti-diabetic medications or viral suppression after exposure to antiretroviral agents [1,2]. One of the most robust examples of a pharmacokinetic association is observed with genetic variation in the cytochrome p450 2B6 gene (CYP2B6) [1-2], whose protein product is an enzyme that metabolizes an array of medications [3], including bupropion, tamoxifen, propofol, and the non-nucleoside reverse transcriptase inhibitors (NNRTI) efavirenz and nevirapine that are used to treat human immunodeficiency virus (HIV) infection.
Several CYP2B6 loss-of-function alleles are associated with pharmacokinetic characteristics of NNRTIs [4,5]. Particularly, consistent associations of two single nucleotide polymorphisms (SNP), rs3475274 and rs28399499, have been observed individually with plasma and intracellular NNRTI levels [4-14] and to a lesser degree with clinical response [10-15]. The CYP2B6 metabolizer phenotype, derived from the composite genotype for rs3745274 and rs28399499, categorizes individuals as extensive, intermediate, and slow metabolizers of CYP2B6 substrates [16-18]. The metabolizer phenotype predicts efavirenz [16,17] and nevirapine [16] plasma concentrations. Regarding clinical response, an association between the metabolizer phenotype and virologic failure, defined as two consecutive HIV viral load measures ≥ 200 copies/ml, in African-Americans has been suggested [18].
Confounding by race/ethnicity is a concern in all genetic association studies and the two variants comprising the CYP2B6 metabolizer phenotype may be particularly susceptible to this type of confounding. Substantial differences in minor allele frequencies for these two SNPs occur across racial/ethnic groups [5-8] and race/ethnicity has often been associated with pharmacokinetic parameters and treatment responses [19]. Gross confounding by race/ethnicity (population stratification) can be addressed by adjusting statistical models for self-reported ethnicity, but cryptic relatedness and admixture require more detailed genetic ancestry data to guard against false positive or false negative results [20-24]. Such cryptic relatedness occurs in seemingly homogenous populations within the same self-reported race/ethnicity group [24]. In theory, affected individuals (cases) may be more related to one another than controls and as a result the allelic distribution among cases may be skewed [24].
Spurious associations can also occur in a study population that contains individuals from a single ethnic group that has experienced recent genetic admixture [25,26]. Varying proportions of ancestry from distinct ‘parental’ populations contribute to a population's genome; admixture describes this mosaic of genetic ancestry. Admixture is particularly evident in African-American and Hispanic-American populations [27-29]. For example, among African-Americans from different geographical regions in the United States (U.S.), European ancestry varies between 11.6% and 22.5%; the remaining proportion includes African and Native American ancestry [27]. Thus, addressing population substructure in genetic association studies beyond adjustment for self-reported race/ethnicity is critical to protect against potentially spurious results.
The Women's Interagency HIV Study (WIHS) is a multi-ethnic cohort study of HIV-infected and uninfected women who have been followed every six months since 1995. We report here the association between genetic variation in CYP2B6 and virologic response to initial antiretroviral therapy regimens containing a NNRTI. Simultaneously, population substructure was characterized using a panel of ancestry informative markers (AIM) selected to distinguish between European, West African, Asian, and Native American ancestry [30]. CYP2B6 genetic associations were used to demonstrate that residual confounding may exist when underlying genetic structure is ignored.
Materials and Methods
WIHS Study Design
The WIHS is a prospective cohort study of HIV-infected women and a comparison group of HIV-uninfected women. Participants were recruited from six sites across the U.S., located in Bronx/Manhattan, New York; Brooklyn, New York; Washington, D.C.; Los Angeles, California; San Francisco/Bay Area, California; and Chicago, Illinois. Enrollment was conducted in two phases; 2 054 HIV seropositive and 569 seronegative women were enrolled in 1994-1995 and 737 HIV seropositive and 406 seronegative women were enrolled from 2000-2001. A more detailed description of the WIHS cohort has been published elsewhere [31,32].
Inclusion and Exclusion Criteria
HIV-infected women who consented to genetic studies, initiated antiretroviral therapy containing a NNRTI during study follow-up, maintained a three drug HAART regimen at the subsequent visit, had HIV RNA measurements at the visit immediately prior to, the visit of, and the visit immediately after initiating a NNRTI-containing regimen were eligible for this study. Women who were treated with antiretroviral drugs prior to reporting treatment with a NNRTI were excluded to limit the inclusion of women infected with a virus that might have acquired resistance to antiretroviral agents. A total of 100 subjects met these study criteria and were genotyped on CYP2B6 SNPs.
Virologic Response
HIV viral loads were quantified with an assay with a lower limit of detection of 80 HIV RNA copies/ml. A positive virologic response (“responders”) was defined as achievement of an undetectable viral load at the visit during which the NNRTI regimen was first reported or at the visit subsequent to it, which corresponds to a maximum of 54 weeks of treatment. Participants not achieving an undetectable viral load at the visit of first reported NNRTI regimen use (in the previous six months) or subsequent visit were classified as “non-responders”. This definition conforms to clinical guidelines for treatment with antiretroviral regimens [33].
SNP Selection
CYP2B6 pharmacogenetic association studies in the literature were queried to obtain a list of polymorphisms located in the exons, 3′ UTR, 5′ UTR and splice sites (i.e. putative functional polymorphisms). From this list, SNPs with a minor allele frequency (MAF) of 5% or higher were scored for assay performance on Illumina's GoldenGate® genotyping platform (San Diego, CA). A total of 11 CYP2B6 SNPs were selected: rs3211370, rs3211397, rs36079186, rs34128717, rs3211374, rs3211371, rs8192709, rs34097093, rs1042389, rs3745274, and rs28399499.
Genotyping and Quality Control
Data for the eleven CYP2B6 SNPs were generated as a subset of a larger genotyping panel (n=384 SNPs). Four of the eleven CYP2B6 SNP assays (rs3211370, rs3211397, rs36079186, rs34128717) did not generate three distinct genotypes and were deemed failures. Of the remaining seven SNPs that were successfully genotyped, rs3211374 was monomorphic (no variants observed) in all race/ethnic groups and excluded from the analysis. In total, six CYP2B6 polymorphisms (rs3211371, rs8192709, rs34097093, rs1042389, rs3745274, and rs28399499) passed the QC criterion of successful genotyping on 89.5% of the samples. Five of the 100 subjects genotyped (5%) did not pass the QC criterion of <10% missing genotype data and were excluded from the analysis. QC for the entire panel is described in supplementary information.
Characterization of Genetic Ancestry in the WIHS
Selection of AIMs, Genotyping and Quality Control
A subset of markers that were originally identified by Smith and colleagues was utilized to distinguish between West African, European, East Asian, and Native American populations [30]. All subjects in the WIHS cohort who consented to genetic studies and had DNA available were characterized for genetic ancestry as these markers are being used for all genetic association studies being conducted in the WIHS. A total of 185 AIMs were selected as part of a larger panel (n=384); 168 (91%) AIMs passed the QC criterion of successful genotyping on 89.5% of the samples Four of 100 eligible subjects did not pass the QC criterion of <10% missing genotype data and were excluded from the analysis. QC for the entire panel is described in supplementary information.
Estimation of Genetic Ancestry Components
Individual genetic ancestry proportions for 2 318 WIHS women were inferred using the software package STRUCTURE 2.3.1 [22,34], which employs a Bayesian Markov chain Monte Carlo clustering algorithm. A 30 000 repetition burn-in period and 10 000 subsequent iterations for different values of k (number of assumed subpopulations, k=3-6) were initiated under the admixture model with independent allele frequencies. Four independent simulations for each value of k were performed to ensure that estimates were consistent across runs. The admixture model with the greatest log likelihood for each value of k was selected. HapMap2 and HapMap3 [35] reference population data on 168 AIMs and 105 AIMs, respectively, were included in the STRUCTURE analyses to increase the accuracy of admixture estimation [36]. Results were formatted and graphically displayed using the Distruct 1.1 software package [37].
Genetic ancestry components were also evaluated with principal components analysis on the WIHS genotype data for 168 AIMs (n=2 318) following the method used with the EIGENSTRAT software [38,39]. Adjusting for PCs is the preferred method to control for population substructure, as the model does not depend on an assumption of the number of source populations [38,39]. PCs were used in the models examining the association between CYP2B6 genotypes and virologic response to therapy.
Statistical Analysis
The final dataset consisted of 91 subjects meeting study inclusion and exclusion criteria and with complete data for CYP2B6 and AIM SNPs. Logistic regression was used to test associations between each CYP2B6 polymorphism and virologic response. Odds ratios (OR) per allele and 95% CIs were estimated by modeling the genotypes as an ordinal variable, where common allele homozygotes, heterozygotes and minor allele homozygotes were coded as 0, 1, and 2, respectively. This log-additive model provides a p-value for corresponding test of the trend for increased probability of virologic response per allele.
CYP2B6 metabolizer phenotypes were constructed using two polymorphisms, rs3745274 and rs28399499, to test the association between the metabolizer phenotype and virologic response. Women who were common allele homozygotes at rs3745274 and rs28399499 (GG and TT, respectively) were coded as 0 “extensive metabolizers”. Women with one heterozygote genotype and one common allele homozygote genotype at either polymorphism were coded as 1 “intermediate metabolizers”. Women with a total of two minor alleles (one minor allele homozygote genotype, or two heterozygote genotypes) across both SNPs were coded as 2 “slow metabolizers”. No women carried one minor allele at one SNP and two minor alleles at the other SNP, or four minor alleles across the two SNPs.
Metabolizer phenotype-specific ORs and 95% CIs for intermediate metabolizers and slow metabolizers compared with extensive metabolizers, were estimated with exact logistic regression, since there were zero non-responders with the slow metabolizer phenotype. Additionally, the metabolizer phenotype was treated as an ordinal variable to obtain the exact p for trend. Nominal p-values are reported throughout the manuscript.
To assess the potential confounding effects of population substructure, models were fit unadjusted, adjusted for self-reported race/ethnicity (Non-Hispanic White, African American, Hispanic, and Asian/Other), and adjusted for genetic ancestry principal components. The three most important PCs that accounted for the largest change in the main effect β in the individual SNP analyses were included in the metabolizer phenotype model.
Self-reported adherence was also evaluated as a potential confounder (change in the genotype main effect β of 10% or more was considered confounding). Adherence data were taken at the visit at which the participant achieved the virologic response outcome since the adherence variable at this visit reflects treatment adherence in the six months leading up to the visit in which the outcome was achieved. For modeling purposes, adherence was dichotomized as ≥ 95% adherent and < 95% adherent from original categories as collected (100%, 95-99%, 75-95%, <75%).
Results
Detection of population substructure
Figure 1 illustrates the individual ancestry proportions for 2 318 WIHS participants (n=1 796 HIV seropositive and n=522 seronegative) by self-reported ethnic groups for k=4 cluster STRUCTURE models (results for self-reported Asian, Native American and ‘other’ groups are not illustrated due to small numbers). In summary, the WIHS women likely descended from four source populations, European, Asian, Yoruban, and non-Yoruban African ancestry. Admixture was estimated by including reference population data from HapMap2 (Fig. 1A) and HapMap3 (Fig. 1B). Assuming k=4 subpopulations (log likelihood= -449 517.3), the fourth subpopulation (yellow) was represented in WIHS African-American and Hispanic women, but was not substantially present in Yoruban, European, or East Asian reference populations (Fig. 1A). The k=4 model suggests that there is ancestry in the WIHS that is not represented in the HapMap2 populations. Assuming k=4 subpopulations (log likelihood= -358 970.2) with HapMap3 data, the fourth subpopulation (orange) was over-represented in the Maasai Kenyans, in which the average proportion of ancestry attributed to the fourth subpopulation was 79% (Fig. 1B). Since the Maasai population show admixture with Yoruban ancestry (12%), the fourth component was labeled as non-Yoruban African ancestry. The HapMap3 Mexican-American population shows a considerable amount of admixture represented by the Asian subpopulation, which was inferred to represent Native American ancestry as Native American ancestry is correlated with East Asian ancestry [40].
Figure 1.


Individual ancestry proportions for self-reported WIHS ethnic groups and HapMap populations.
A. k=4, 168 AIMs, HapMap2
B. k=4, 105 AIMs, HapMap3
Figure A. Labels of blocks from top to bottom
White, Non-Hispanic
Hispanic
African-American
CEPH European (HapMap2)
Yoruban, Nigeria (HapMap2)
Chinese Han, Beijing (HapMap2)
Japanese, Tokyo (HapMap2)
Figure B. Labels of blocks from top to bottom
White, Non-Hispanic
Hispanic
African-American
CEPH European (HapMap3)
Yoruban, Nigeria (HapMap3)
Chinese Han, Beijing (HapMap3)
Japanese, Tokyo (HapMap3)
Toscani, Italy (HapMap3)
Luhya tribe, Kenya (HapMap3)
Maasai tribe, Kenya (HapMap3)
African-American (HapMap3)
Chinese-American (HapMap3)
Mexican-American (HapMap3)
Gujarti Indian (HapMap3)
Legend
Green: European ancestry
Purple: East Asian ancestry
Pink: Yoruban ancestry
Yellow: Non-Yoruban ancestry
Orange: Non-Yoruban ancestry
The estimated admixture proportions in self-reported African-American, Hispanic, and non-Hispanic White women were evaluated by the geographic location of the WIHS sites (results not shown). In contrast to African-Americans and Whites, ancestry proportions in Hispanics varied across sites. There was a gradient of decreasing European ancestry and increasing Asian ancestry in Hispanics from East to West. Additionally, African admixture (Yoruban and non-Yoruban) was substantial in Hispanics in New York (0.13-0.19) but minimal among Los Angeles Hispanics (0.03-0.04).
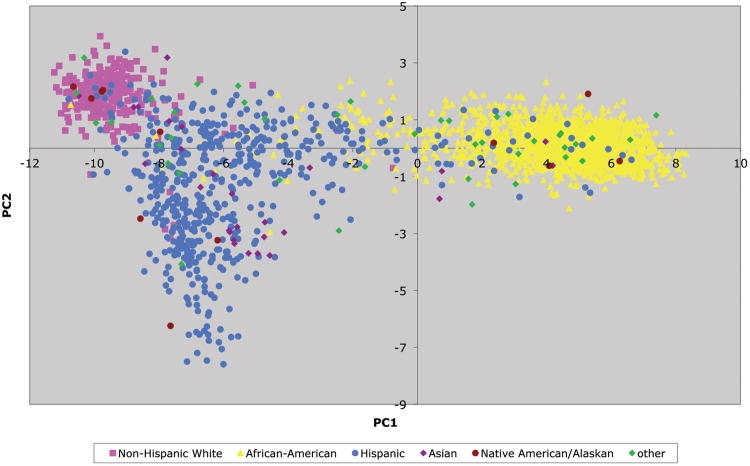
While estimating ancestry proportions using STRUCTURE is beneficial to labeling subpopulations for ease of interpretation, PCs are preferred as ancestry covariates in statistical models. Graphical display of the PCs illustrate the separation of broad ethnic groups and the dispersion of individuals along the axes confirms the variation that exists within self-reported ethnic groups as seen in Figure 1. Figure 2 shows the plot of PC1 vs. PC2 with color-coded self-reported ethnic groups. PC1 (x-axis) separates African vs non-African ancestry and PC2 (y-axis) separates Hispanic vs non-Hispanic ancestry.
Figure 2.
Principal component (PC) 1 vs. PC 2 for WIHS women (n=2 318) from analysis on 168 ancestry informative markers. Self-identified race/ethnicity groups are color-coded.
Legend
Pink: self-reported Non-Hispanic White
Yellow: self-reported African-American
Blue: self-reported Hispanic
Purple: self-reported Asian
Maroon: self-reported Native American/Alaskan
Green: self-reported “Other”
CYP2B6 associations with response to NNRTIs and the impact of population substructure
Nine out of 21 virologic non-responders (43%) had a nadir CD4+ count lower than 200 cells/ml prior to reporting a NNRTI-based regimen, a difference that was not statistically significantly different from responders (N=24/70, 34%, p=0.47). The median viral load measured at the visit prior to first report of a NNRTI was 28 000 copies/ml (IQR 6 300-110 000) for responders and 45 000 copies/ml (IQR 13 000-69 000) for non-responders (Kruskal-Wallis test p=0.57). Nearly all of the non-responders (n=18/21) reported having African-American race/ethnicity and the remaining three women reported having Hispanic race/ethnicity. Among the responders (N=70), 53% self-reported as African-American, 31% as White, and 7% as Hispanic.
The genotype frequencies by responder status and MAF by self-reported ethnicity are given in Table 1 for the six SNPs that were successfully genotyped. There were no non-responders among carriers of the minor allele for rs3211371 and therefore effect estimates could not be calculated. The six SNPs were not in linkage disequilibrium (r2 ≥ 0.80) in any of the WIHS self-reported racial/ethnic groups (Supplementary Figure 1).
Table 1.
Frequency counts for genotypes by virologic responder status1 and minor allele frequencies by self-reported race/ethnic group in 91 HIV-infected women treated with NNRTIs (efavirenz or nevirapine).
| Minor Allele Frequency | ||||||
|---|---|---|---|---|---|---|
| CYP2B6 SNP2 | Responders (%) | Non-responders (%) | African American (n=55) | White (n=22) | Hispanic (n=8) | Other3 (n=6) |
| rs3211371 | 0.03 | 0.07 | 0 | 0 | ||
| CC | 63 (90) | 21 (100) | ||||
| CT | 6 (10) | 0 | ||||
| TT | 0 | 0 | ||||
| missing | 1 | 0 | ||||
| rs3409703 | 0.22 | 0 | 0 | 0.08 | ||
| TT | 52 (76) | 14 (74) | ||||
| TC | 16 (24) | 4 (21) | ||||
| CC | 0 | 1 (5) | ||||
| missing | 2 | 2 | ||||
| rs1042389 | 0.25 | 0.14 | 0.21 | 0.08 | ||
| TT | 46 (66) | 13 (62) | ||||
| TC | 21 (30) | 7 (33) | ||||
| CC | 3 (4) | 1 (5) | ||||
| rs8192709 | 0.06 | 0 | 0.07 | 0 | ||
| CC | 64 (91) | 20 (95) | ||||
| CT | 6 (9) | 1 (5) | ||||
| TT | 0 | 0 | ||||
| rs3745274 | 0.28 | 0.24 | 0.21 | 0.33 | ||
| GG | 32 (46) | 14 (67) | ||||
| GT | 34 (48) | 7 (33) | ||||
| TT | 4 (6) | 0 | ||||
| rs28399499 | 0.10 | 0 | 0 | 0 | ||
| TT | 62 (88) | 19 (90) | ||||
| TC | 7 (10) | 2 (10) | ||||
| CC | 1 (2) | 0 | ||||
Virologic response defined as achievement of undetectable viral load up to 54 weeks after self- reported initiation of NNRTI-based regimen
SNP= single nucleotide polymorphism; all SNPs are missense with the exception of rs1042389 which is located in the 3′ untranslated region
Asian, Native American, Other
The associations between the remaining five SNPs (rs3409703, rs1042389, rs8192709, rs3745274 and rs28399499) and response to a NNRTI-based regimen are shown in Table 2. Overall, no statistically significant associations between these five SNPs and virologic response were observed after adjustment for self-reported race/ethnicity (second column). However, a high magnitude of effect on virologic response was observed for the two SNPs which comprise the metabolizer phenotype (rs3745274 and rs28399499) after adjustment for genetic ancestry PCs and the effect of rs3745274 reached statistical significance (p-trend =0.03).
Table 2.
Associations between CYP2B6 single nucleotide polymorphisms (SNP) and with virologic response1 to NNRTI-based regimens in women who were naïve to antiretroviral drugs (N=91).
| Crude | Adjusted for SR Race/Ethnicity3 | Adjusted for top three PCs4 | |
|---|---|---|---|
| CYP2B6 SNP2 | OR (95% CI) | OR (95% CI) | OR (95% CI) |
| rs34097093 | |||
| per C allele | 0.70 (0.25-1.99) | 1.21 (0.39-3.77) | 1.25 (0.37-4.22) |
| p trend | 0.50 | 0.74 | 0.72 |
| rs1042389 | |||
| per C allele | 0.88 (0.38-2.03) | 1.14 (0.46-2.84) | 1.13 (0.43-2.94) |
| p trend | 0.76 | 0.77 | 0.80 |
| rs81927095 | |||
| CC | 1.00 | 1.00 | 1.00 |
| CT | 1.87 (0.21-16.52) | 3.35 (0.38-29.90) | 2.96 (0.30-28.87) |
| rs3745274 | |||
| per T allele | 2.42 (0.93-6.30) | 2.49 (0.90-6.88) | 3.61 (1.16-11.22) |
| p trend | 0.07 | 0.08 | 0.03 |
| rs28399499 | |||
| per C allele | 1.32 (0.30-5.91) | 2.23 (0.47-10.61) | 3.63 (0.69-19.18) |
| p trend | 0.71 | 0.31 | 0.13 |
Virologic response was defined as achievement of undetectable viral load up to 54 weeks after self-reported initiation of NNRTI-based regimen
rs3211371 could not be analyzed due to lack of non-responders with the TC genotype
Adjusted for self-reported (SR) race/ethnicity (White, Hispanic, African American, Asian/Native American/Other)
Adjusted for the top three genetic ancestry principal components (PCs)
No carriers of the TT genotype observed
When considering the joint effects of the two SNPs, intermediate and slow metabolizers were 2.90 (95% CI 0.79-12.28) and 13.44 (95% CI 1.66-infinity) times as likely to respond to NNRTIs compared to extensive metabolizers after adjustment for genetic ancestry PCs (p-trend =0.005; Table 3). Substantial evidence of confounding is present with the metabolizer phenotype when comparing the crude, self-reported ethnicity-adjusted and genetic ancestry PCs-adjusted estimates. After adjustment for self-reported race/ethnicity, the OR for slow metabolizers compared to extensive metabolizers increased by 54%. After adjustment for genetic ancestry PCs, the confounding effect of population substructure was further reduced; the OR for slow metabolizers compared to extensive metabolizers increased by 169% over the unadjusted estimate and by 75% over the self-reported race/ethnicity estimate. PC1, which separates African vs. non-African ancestry, explained most of the confounding, in which the crude estimate changed by 55% after including PC1 in the model. Adjustment for PC9 increased the PC1-adjusted estimate by 19% and PC7 accounted for an additional 4% increase thereafter. The level of confounding attributable to population substructure is not surprising given the appreciable heterogeneity that exists within each self-reported race/ethnicity group (Figures 1 and 2).
Table 3.
Estimates for the association between the CYP2B6 metabolizer phenotype (using rs3745274 and rs28399499) with virologic response1 to NNRTI-based HAART regimens in women who were naïve to antiretroviral drugs (N=91).
| Metabolizer Phenotype2 | Responders (%) | Non-responders (%) | Crude | Adjusted for SR Race/Ethnicity3 | Adjusted for top three PCs4 |
|---|---|---|---|---|---|
| OR (95% CI) | OR (95% CI) | OR (95% CI) | |||
| Extensive | 28 (40) | 12 (57) | 1.00 | 1.00 | 1.00 |
| Intermediate | 33 (47) | 9 (43) | 1.56 (0.52-4.88) | 1.66 (0.47-6.11) | 2.90 (0.79-12.28) |
| Slow | 9 (13) | 0 | 5.00 (0.70-inf) | 7.69 (1.04-inf) | 13.44 (1.66-inf) |
| p trend exact | 0.09 | 0.04 | 0.005 |
Virologic response defined as achievement of undetectable viral load up to 54 weeks after self- reported initiation of NNRTI-based regimen
Extensive = no minor alleles at either rs3745274 or rs28399499 Intermediate = one minor allele at either rs3745274 or rs28399499 Slow = two minor alleles at either rs3745274 or rs28399499, or one minor allele at both polymorphisms
Adjusted for self-reported race/ethnicity (White, Hispanic, African American, Asian/Other)
Adjusted for the top three genetic ancestry principal components (PCs)
Discussion
We observed a statistically significant association between the CYP2B6 metabolizer phenotype (joint effect of rs3745274 and rs28399499) and virologic response to NNRTI-based antiretroviral regimens (p-trend =0.005). This association was strongly confounded by underlying population substructure for which adjustment of self-reported race/ethnicity was not a sufficient control. Our findings confirm prior report of the role of these SNPs in NNRTI metabolism and further highlight the importance of controlling for population substructure using genetic ancestry estimates, such as PCs, in genetic association studies.
It is possible that both genetic ancestry estimates and self-reported race/ethnicity may confound the association between SNPs and outcome, as would be seen if the additional factors captured by self-reported ethnic group (e.g. socio-economic status) are also associated with the SNP of interest. However, adjusting for both PCs and self-reported race/ethnicity did not change the estimated effect of CYP2B6 metabolizer phenotype on virologic response to NNRTIs compared to PCs alone. These findings suggest that PCs alone capture the underlying population substructure and the additional factors related to self-identification in a particular ethnic group are not associated with these SNPs.
To our knowledge, we are the first to report a statistically significant joint effect of rs3745274 and rs28399499 (metabolizer phenotype) on virologic response to NNRTIs in a multi-ethnic observational study of HIV-infected women. Previously, a study of participants randomized to efavirenz in the AIDS Clinical Trial Group (ACTG), of which 19% were female and 34% were African-American, explored the effect of the metabolizer phenotype on virologic failure [18]. The incidence of virologic failure adjusted for the competing event of efavirenz discontinuation was lower among self-reported African American participants with the slow metabolizer phenotype (p=0.02) [18]. However, this effect was not seen in Whites or Hispanics, which is likely an artifact of the lower allele distribution in non-African Americans. The authors reported that the results were similar after incorporation of genome-wide association study (GWAS) PCs, though it is unclear whether this model was tested in all participants.
The two polymorphisms that contribute to the CYP2B6 metabolizer phenotype are functionally relevant. The minor allele (T) at rs3745274 causes an aberrant splicing mechanism that decreases expression and activity of CYP2B6 in the liver [41]. The minor allele (C) at rs28399499 has been consistently associated with reduced CYP2B6 expression [1,7], though the mechanism is unclear [41]. Additionally, it is well established that both SNPs predict several short-term NNRTI pharmacokinetic parameters, such as AUC, oral clearance, and half-life [4-18,42]. In a separate WIHS study, carriers of the TT genotype compared to GG and GT genotypes at rs3745274 experienced an over 3-fold increase (p=1.0 × 10-10) in efavirenz concentration in hair specimens after adjustment for non-genetic predictors of long-term efavirenz exposure (e.g. adherence and consumption of orange juice) [39]. The minor allele C at rs28399499 was associated with a 1.70 fold increase of efavirenz levels in hair (p=0.02) [43]. The independent associations of these SNPs with long-term exposure to efavirenz lends support to our results of increased probability of virologic suppression in intermediate and slow metabolizers through the mechanism of sustained increased drug levels.
A limitation of this study is the absence of viral genotyping on pre-NNRTI specimens from each participant, which would allow us to limit exclusion of treatment-experienced NNRTI users to those who are resistant to NNRTIs. There is no reason to believe that the exclusion of treatment-experienced women who reported initial treatment with a NNRTI would bias the results away from the null. Another limitation of our study is the small sample size, however, we observed a robust and statistically significant association that survived a Bonferroni correction for the number of tests we performed (corrected α=0.05/6 = 0.008). The functional role of the SNPs comprising the metabolizer phenotype and associations within and external to the WIHS with pharmacokinetic parameters is compelling enough to assign a high prior probability that this association is real.
Although patients with CYP2B6 intermediate and slow metabolizer phenotypes are more likely to achieve undetectable viral loads after treatment with NNRTIs in our study, it is possible that the low grade toxicities that are experienced in carriers of the minor allele at rs3745274 [4,9] could impact adherence to NNRTIs over time or result in drug-switching. This is an area of active research in the WIHS. Nevertheless, the association of the CYP2B6 metabolizer phenotype and response to NNRTIs has important potential for clinical decision-making, in which the genetic profile of an individual is taken into account prior to the initiation of a given antiretroviral agent. In fact, this paradigm has already been implemented with the antiretroviral agent, abacavir, in which acquisition of genetic information is highly recommended prior to the institution of this drug, in order to avoid potential hypersensitivity reactions [44]. The body of evidence supporting a role for CYP2B6 SNPs in NNRTI virologic response suggests that further study is warranted to determine whether this information can be used in clinical in decision making related to antiretroviral regimens.
Supplementary Material
Supplementary Figure 1. Linkage disequilibrium blocks (r2) for rs3211371, rs1042389, rs8192709, rs3745274, rs28399499 for White, African American, Hispanic, and Other self-reported race/ethnicity groups using all WIHS women who were genotyped for the 384 SNP panel which included these CYPB26 SNPs (n=1,188).
Acknowledgments
Data in this manuscript were collected by the Women's Interagency HIV Study (WIHS) Collaborative Study Group with centers (Principal Investigators) at New York City/Bronx Consortium (Kathryn Anastos); Brooklyn, NY (Howard Minkoff); Washington DC, Metropolitan Consortium (Mary Young); The Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt); Los Angeles County/Southern California Consortium (Alexandra Levine); Chicago Consortium (Mardge Cohen); Data Coordinating Center (Stephen Gange).
The WIHS is funded by the National Institute of Allergy and Infectious Diseases (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (UO1-HD-32632). The study is co-funded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders. Funding is also provided by the National Center for Research Resources (UCSF-CTSI Grant Number UL1 RR024131). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest: The authors have no conflict of interest to report.
Author contributions: Analysis: Melissa Frasco, Wendy Mack, David Conti, and C. Leigh Pearce
Writing the manuscript: Melissa Frasco, Wendy Mack, C. Leigh Pearce, and Bradley Aouizerat
WIHS study design: Ruth Greenblatt, Kathryn Anastos, and Mardge Cohen
Substudy design: C. Leigh Pearce, David Conti, Kathryn Anastos
Critical review of manuscript: all authors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zanger UM, Turpeinen M, Klein K, Schwab M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal Bioanal Chem. 2008;392:93–108. doi: 10.1007/s00216-008-2291-6. [DOI] [PubMed] [Google Scholar]
- 2.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic, and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Wang HB, Tompkins LM. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab. 2008;9:598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphisms on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics. 2005;15:1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Klein K, Lang T, Saussele T, Barbosa-Sicard E, Schunck WH, Eichelbaum M, et al. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequency variants and possible implications for anti-HIV therapy with efavirenz. Pharmacogenetic Genomics. 2005;15:861–73. doi: 10.1097/01213011-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Sonnenberg A, Rane A, Josephson F, Lundgren S, Stahle L, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genomics. 2006;16:191–198. doi: 10.1097/01.fpc.0000189797.03845.90. [DOI] [PubMed] [Google Scholar]
- 7.Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Decosterd L, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81:557–66. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 8.Wyen C, Hendra H, Vogel M, Hoffmann C, Knechten H, Brockmeyer NH, et al. Impact of CYP2B6 983T>C polymorphisms on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61:914–8. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: An Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 10.Haas DW, Smeaton LM, Shafer RW, Robbins GK, Morse GD, Labbe L, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: An Adult AIDS Clinical Trials Group Study. J Infect Dis. 2005;192:1931–1942. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 11.Motsinger AA, Ritchie MD, Shafer RW, Robbins GK, Morse GD, Labbe L, et al. Multilocus genetic interactions and response to efavirenz-containing regimens: An adult AIDS Clinical Trials Group study. Pharmacogenetics and Genomics. 2006;16:837–845. doi: 10.1097/01.fpc.0000230413.97596.fa. [DOI] [PubMed] [Google Scholar]
- 12.Saitoh A, Sarles E, Capparelli E, Aweeka F, Kovacs A, Burchett SK, et al. CYP2B6 genetic variants are associated with nevirapine pharmacokinetics and clinical response in HIV-1 infected children. AIDS. 2007;21:2191–9. doi: 10.1097/QAD.0b013e3282ef9695. [DOI] [PubMed] [Google Scholar]
- 13.Habtewold A, Amogne W, Makonnen E, Yimer G, Riedel K, Ueda N, et al. Long-term effect of efavirenz autoinduction on plasma/peripheral blood mononuclear cell drug exposure and CD4 count is influenced by UGT2B7 and CYP2B6 genotypes among HIV patients. J Antimicrob Chemother. 2011;66:2350–2361. doi: 10.1093/jac/dkr304. [DOI] [PubMed] [Google Scholar]
- 14.Gozolo C, Gerard L, Loiseau P, Morand-Joubert L, Peytavin G, Molina J, et al. Pharmacogenetics of toxicity, plasma trough concentration and treatment outcome with nevirapine containing regimen in antiretroviral naïve HIV-infected adults: An exploratory study of the TRIANON ANRS 081 trial. Basic Clin Pharmacol Toxicol. 2011;109:513–520. doi: 10.1111/j.1742-7843.2011.00780.x. [DOI] [PubMed] [Google Scholar]
- 15.Glass TR, Rotger M, Telenti A, Decosterd L, Csajka C, Heiner CB, et al. Determinants of sustained viral suppression in HIV-infected patients with self-reported poor adherence to antiretroviral therapy. PLoS One. 2012;7:e29186. doi: 10.1371/journal.pone.0029186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heil SG, van der Ende ME, Schenk PW, van der Heiden I, Lindemans JM, Burger D, et al. Associations between ABCB1, CYP2A6, CYP2B6, CYP2D6, and CYP3A5 alleles in relation to efavirenz and nevirapine pharmacokinetics in HIV-infected individuals. Ther Drug Monit. 2012;34:153–159. doi: 10.1097/FTD.0b013e31824868f3. [DOI] [PubMed] [Google Scholar]
- 17.Haas DW, Gebretsadik T, Mayo G, Menon UN, Acosta EP, Shintani A, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis. 2009;199:872–880. doi: 10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribaudo HJ, Huan L, Schwab M, Schaeffeler E, Eichelbaum M, Motsinger-Reif AA, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetic and treatment response: An AIDS Clinical Trials Group study. J Infect Dis. 2010;202:717–22. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anastos K, Schneider MF, Gange SJ, Minkoff H, Greenblatt RM, Feldman J, et al. The association of race, sociodemographic, and behavioral characteristics with response to highly active antiretroviral therapy in women. J Acquir Immune Defic Syndr. 2005;39:537–544. [PubMed] [Google Scholar]
- 20.Thomas DC, Witte JS. Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- 21.Gorroochurn P, Hodge SE, Heiman G, Greenberg DA. Effect of population stratification on case-control association studies. II False-positive rates and their limiting behavior as number of subpopulations increases. Hum Hered. 2004;58:40–48. doi: 10.1159/000081455. [DOI] [PubMed] [Google Scholar]
- 22.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theor Popul Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- 24.Voight BF, Pritchard JK. Confounding from cryptic relatedness in case-control associations. PloS Genet. 2005;1:e32. doi: 10.1371/journal.pgen.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 26.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parra EJ, Marcini A, Akey J, Batzer MA, Cooper R, Forrester T, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, et al. Genetic admixture and asthma related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29:76–86. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- 29.Halder I, Yang BZ, Kranzler HR, Stein MB, Shriver MD, Gelernter J, et al. Measurement of admixture proportions and description of admixture structure in different U. S. populations. Hum Mutat. 2009;30:1299–1309. doi: 10.1002/humu.21045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1003. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barkan SE, Melnick SL, Preston-Martin S, Weber K, Kalish LA, Miotti P, et al. The Women's Interagency HIV Study. Epidemiology. 1998;9:117–25. [PubMed] [Google Scholar]
- 32.Bacon MC, Von Wyl V, Alden C, Sharp G, Robinson E, Hessol N, et al. The Women's Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol. 2005;12:1013–19. doi: 10.1128/CDLI.12.9.1013-1019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barlett JG, Lane HC. Guidelines for the use of antiretroviral drugs in HIV-1-infected adults and adolescents. In: Infection PCPTHIV, editor. Clinical Guidelines for the treatment and management of HIV Infection. USA: Department of Health and Human Services; 2005. pp. 1–118. [Google Scholar]
- 34.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 36.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78.33. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg NA. Distruct: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- 38.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 39.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, et al. Genetic variation and population structure in Native Americans. PloS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman MH, Blievernicht JK, Klein K, Sausseie T, Schaeffeler E, Schwab M, et al. Abberant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325:284–292. doi: 10.1124/jpet.107.133306. [DOI] [PubMed] [Google Scholar]
- 42.Saitoh A, Spector SA. Effect of host genetic variation on the pharmacokinetics and clinical response of non-nucleoside reverse transcriptase inhibitors. Futur HIV Ther. 2008;2:69–81. doi: 10.2217/17469600.2.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandhi M, Greenblatt RM, Bacchetti P, Jin C, Huang Y, Anastos K, et al. A single nucleotide polymorphism leads to >3-fold increases in efavirenz concentrations in intensive PK curves and hair. J Infect Dis. 2012 doi: 10.1093/infdis/jis508. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaponda M, Pirmohamed M. Hypersensitivity reactions to HIV therapy. Br J Clin Pharmacol. 2011;71:659–671. doi: 10.1111/j.1365-2125.2010.03784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Linkage disequilibrium blocks (r2) for rs3211371, rs1042389, rs8192709, rs3745274, rs28399499 for White, African American, Hispanic, and Other self-reported race/ethnicity groups using all WIHS women who were genotyped for the 384 SNP panel which included these CYPB26 SNPs (n=1,188).