Abstract
Cardiac gene expression is precisely regulated and its perturbation causes developmental defects and heart disease. Leucine-rich repeat containing 10 (Lrrc10) is a cardiac-specific factor that is crucial for proper cardiac development and deletion of Lrrc10 in mice results in dilated cardiomyopathy. However, the mechanisms regulating Lrrc10 expression in cardiomyocytes remain unknown. Therefore, we set out to determine trans-acting factors and cis-elements critical for mediating Lrrc10 expression. We identify Lrrc10 as a transcriptional target of Nkx2-5 and GATA4. The Lrrc10 promoter region contains two highly conserved cardiac regulatory elements, which are functional in cardiomyocytes but not in fibroblasts. In vivo, Nkx2-5 and GATA4 endogenously occupy the proximal and distal cardiac regulatory elements of Lrrc10 in the heart. Moreover, embryonic hearts of Nkx2-5 knockout mice have dramatically reduced expression of Lrrc10. These data demonstrate the importance of Nkx2-5 and GATA4 in regulation of Lrrc10 expression in vivo. The proximal cardiac regulatory element located at around −200 bp is synergistically activated by Nkx2-5 and GATA4 while the distal cardiac regulatory element present around −3 Kb requires SRF in addition to Nkx2-5 and GATA4 for synergistic activation. Mutational analyses identify a pair of adjacent Nkx2-5 and GATA binding sites within the proximal cardiac regulatory element that are necessary to induce expression of Lrrc10. In contrast, only the GATA site is functional in the distal regulatory element. Taken together, our data demonstrate that the transcription factors Nkx2-5 and GATA4 cooperatively regulate cardiac-specific expression of Lrrc10.
Keywords: LRRC10, Nkx2-5, GATA4, transcriptional regulation, cardiac gene expression
1. Introduction
Leucine-rich repeat containing 10 (LRRC10) is a cardiac-specific factor in mice, zebrafish and humans [1–4], which exhibits robust expression in the developing and adult heart [1, 4]. Lrrc10 plays critical roles in cardiac development and function in zebrafish [3] and LRRC10 interacts with actin and actinin in the heart [5], suggesting a prominent function for LRRC10 in cardiac physiology and a potential role in human heart disease.
Tissue-specific expression of genes requires precise coordinated molecular controls to confer specificity. Given the remarkable cardiac-specific expression of Lrrc10 from the precardiac region to the adult heart and its important roles in cardiac function [1, 3–5], it is critical to investigate the molecular mechanisms that regulate expression of Lrrc10. Lrrc10 expression is dynamically regulated during development, with an approximately four-fold increase in mRNA levels at birth [1]. Lrrc10 homozygous knockout mice exhibit prenatal cardiac functional deficits and progressive dilated cardiomyopathy in postnatal life [5], indicating that proper regulation of Lrrc10 expression is essential for normal cardiac function. The promoter region of Lrrc10 contains multiple DNA binding motifs for key cardiac transcription factors [4, 6] and LacZ transgenic reporter mice containing 7 Kb upstream of the Lrrc10 sequence recapitulate its cardiac-specific expression in vivo [4]. However, the critical transacting factors and cis-elements that regulate cardiac Lrrc10 expression remain to be elucidated.
Nkx2-5 is a cardiac-restricted transcription factor essential for proper cardiac development [7–9] and conduction system function [10–13]. Mutations of Nkx2-5 result in congenital heart disease, electrophysiological abnormalities, and sudden death in animal models [7, 8, 10–13] and humans [14]. Nkx2-5 regulates cardiac transcription often in conjunction with other transcriptional cofactors, including GATA4 [15–19], SRF (serum response factor) [17, 20, 21], Tbx5 [12, 22], and Jarid2 [23]. GATA4 is a zinc finger transcription factor that is required for early cardiac development and adult cardiac function [24–27]. GATA4 regulates cardiac gene expression by forming complexes with transcriptional factors, including Nkx2-5 [15–19], NFAT (nuclear factor of activated T cells) [28], Tbx5 [25, 29], SRF [17, 30, 31], Smad1/4 [32] and Jarid2 [23]. Furthermore, mutations of GATA4 have been shown to cause cardiac septal defects in humans [25]. Cardiac-specific deletion of Gata4 [33] or perinatal knockout of Nkx2-5 [10] in mice results in compromised cardiac function and dilated cardiomyopathy, suggesting a prominent role for Nkx2-5- and GATA4-mediated transcription in adult cardiac function and disease.
Nkx2-5 is expressed very early in the precardiac region when Lrrc10 expression is first detected [1] and regulates the expression of a number of transcriptional targets in the heart, including endothelin-converting enzyme-1 [34], Jarid2 [35], and β-catenin [36]. GATA4 also controls the transcription of several important cardiac genes, such as carnitine palmitoyltransferase Iβ [31], troponin I [37], troponin C [38], brain natriuretic peptide [39–41], and α-myosin heavy chain [42]. Moreover, Nkx2-5 and GATA4 physically interact [15, 16, 18] and have been shown to cooperatively regulate the expression of essential cardiac target genes, including ANF [15, 18, 19, 43], T- and L-type Ca2+ channels [44], connexin 40 [22], α-actin [16, 17, 20, 21], and Id2 [45].
Here, we provide evidence that Lrrc10 is a novel transcriptional target of Nkx2-5 and GATA4. In vivo, Nkx2-5 and GATA4 occupy highly conserved cardiac regulatory regions of the Lrrc10 genomic locus in the heart and deletion of Nkx2-5 in mice results in dramatically reduced Lrrc10 expression. In vitro, we identify proximal and distal cardiac regulatory elements (PCE and DCE, respectively) near the Lrrc10 promoter by reporter gene assays. Nkx2-5 and GATA4 synergistically activate the Lrrc10 promoter containing the PCE and respective binding sites are required for Nkx2-5- and GATA4-mediated activation. The DCE contains a highly conserved GATA site critical for GATA4-mediated activation and is cooperatively activated by SRF, Nkx2-5, and GATA4.
2. Materials and Methods
2.1. Animals
All procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (NIH) and the University of Wisconsin Research Animal Resource Center policies. Procedures were approved by a University of Wisconsin-Madison Institutional Animal Care and Use Committee. Genotyping of Nkx2-5−/− embryos was described elsewhere [9].
2.2. Cell Culture and Luciferase Reporter Gene Assays
Mouse neonatal cardiomyocytes were cultured as described previously [1]. Lrrc10 reporter plasmids were constructed by cloning various regions of the Lrrc10 genomic locus by PCR and ligating the PCR product into the pGL3 basic vector carrying a firefly luciferase gene (Promega). Transient transfection assays were performed as described [23, 46]. Briefly, cardiomyocytes were cotransfected with Lrrc10 reporter constructs and a β-galactosidase-CMV vector using Lipofectamine LTX (Invitrogen). Luciferase assays were performed 48 hours post transfection using the Luciferase Assay System (Promega) and normalized to β-galactosidase activity. Assays were repeated at least three times in duplicate. Reporter gene assays were also performed in which 10T1/2 cells were cotransfected with plasmids encoding the cardiac transcription factors Nkx2-5, GATA4, and/or MEF2A in the pcDNA3.1-myc vector (Promega) [23, 47] and/or pCGN-SRF [48]. Nkx2-5 binding elements (NKEs) and/or GATA binding sites within the −178 bp Lrrc10 promoter-enhancer reporter were mutated as depicted in Figure 4B using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). All new constructs were subjected to diagnostic digestion and confirmed by DNA sequencing. The oligonucleotides used for constructing Lrrc10 mutant reporter genes are as follows with mutated nucleotides in bold and NKEs and GATA factor sites underlined:
Figure 4.
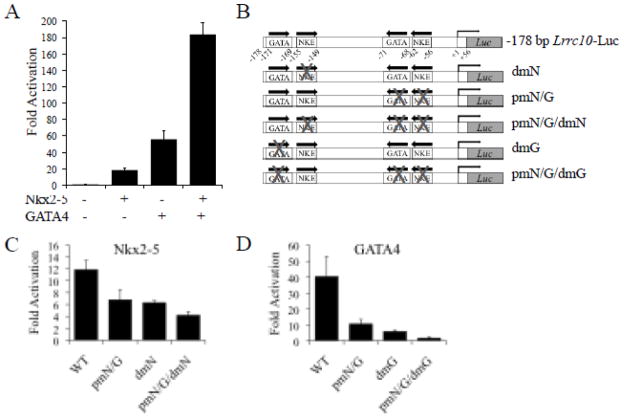
The proximal cardiac regulatory element (PCE) of Lrrc10 contains functional Nkx2-5 and GATA4 binding sites. (A) A −178 bp Lrrc10 reporter was cotransfected with Nkx2-5 and/or GATA4, indicating that Nkx2-5 and GATA4 synergistically induce Lrrc10 expression. (B) Schematic representation of −178 bp Lrrc10 reporter genes containing mutations in NKEs and GATA binding sites. Mutation of (C) NKEs or (D) GATA binding sites within the PCE of Lrrc10 diminishes Nkx2-5- and GATA4-mediated activation, respectively. 10T1/2 cells were cotransfected with a −178 bp Lrrc10 WT reporter or a mutant reporter containing one or both of the NKEs or GATA sites mutated and Nkx2-5 or GATA4. Activation is expressed relative to pcDNA3.1 vector control.
Proximal mNKE/GATA forward (F)
5′GCCTGTCACCAGACACTCAAGCTGCCACCAGGAGTATTCGATTCTGTCAAAGTGACTGGCTTT′
Distal mNKE
5′GGAGAGTAGATAAGAGATCAGGGCTTTTGCCATTAAAGACAAATATGTGAGTTGGAC′
Distal mGATA F
5′GCCCTTGTCGAGGAGAGTAACTAAGAGATCAGGGCTTAAG′
2.3. Quantitative Real Time PCR
(qRT-PCR) qRT-PCR was performed as described previously [5]. Data are normalized to 18S expression. The following primers were designed using Primer Express 1.0 (ABI Prism):
Lrrc10 F 5′GCCACCTCCTCCCCTTCTT′; Lrrc10 R 5′GCCTTCAACCTCAGCTGTTCA′;
18S F 5′CGCCGCTAGAGGTGAAATTCT′; 18S R 5′CGAACCTCCGACTTTCGTTCT′
2.4. Quantitative Chromatin Immunoprecipitation (qChIP)
qChIP experiments were performed as described previously [49, 50] on two pooled wild type mouse hearts at postnatal day three with Nkx2-5 (Supplemental Fig 1), GATA4 (Santa Cruz), or SRF (Santa Cruz) specific antibodies. Preimmune serum or nonspecific IgG (Sigma) was used as a negative control. Primers were designed to amplify 50- to 150-bp amplicons. Products were measured using SYBR Green fluorescence (FastStart SYBR Green Master, Roche; iCycler, BioRad) and normalized to a standard curve of input chromatin. The following primers were used for amplification of the conserved regions of the Lrrc10 locus:
5.5 Kb F 5′ACAAACAATTTGTTCCGCTGAA′; -5.5 Kb R 5′CAAGGTTGAGGAGCCCTTAGC′;
3 Kb F 5′CCATTTGTGGTCACCACTGG′; -3 Kb R 5′TCGCGCGGTCTTACCTCTAT′;
2.3 Kb F 5′TTTACTCTCACACAAGGATGTATGCA′
2.3 Kb R 5′ CCAAGCAATGTTCAGACAATTCTATAAT′
1.7 Kb F 5′TGGCAGCACTTTGGGTCAT′; -1.7 Kb R 5′GGAAGCGTCTGTCCCAGAGTT′
200 bp F 5′AGTGATTAAAGACAAATATGTGAGTTGGA′;
200 bp R 5′AAAGCCAGTCACTTTGACAGAATAAA′
2.5. Anti-Nkx2-5 Antibody Generation, Western Blotting, and In Situ Hybridization
To generate anti-Nkx2-5 antibody, mouse Nkx2-5 was purified from E. coli as a Maltose Binding Protein (MBP) fusion protein and the MBP tag was removed by TEV cleavage leaving a 6 His N-terminal tag on Nkx2-5. The protein was confirmed by SDS-PAGE followed by Coomassie blue staining and was used to immunize rabbit to generate polyclonal anti-Nkx2-5 antibodies (Proteintech Group). Affinity of the antiserum was tested using an enzyme-linked immunosorbent assay and specificities of anti-Nkx2-5 antibodies were confirmed by Western blotting and immunostaining as described previously [5] (Supplemental Fig 1). The LRRC10 antibody was previously described [1]. Whole-mount in situ hybridization of mouse embryos using antisense cRNA probes against Lrrc10 was previously described [1].
2.6. Statistical Analysis
All statistical analysis was performed by a student’s t-test unless otherwise stated. Results are expressed as mean ± standard error of the mean (SEM).
3. Results
Lrrc10 exhibits robust, cardiac-specific expression in the developing and postnatal heart [1] and plays critical roles in cardiac development and function [3, 5]. We have previously shown that embryonic hearts express abundant Lrrc10 mRNA, which is increased significantly at birth and maintained at an elevated expression level in the adult heart [1]. To determine if this expression pattern is recapitulated at the protein level, Western blotting was performed on mouse heart extracts at various ages. LRRC10 protein expression is also abundant in the embryonic heart, significantly increased in the newborn heart, and maintains elevated abundance in the adult (Supplemental Fig 2). These results suggest that a transcriptional mechanism dynamically regulates LRRC10 protein levels during cardiac development and in the postnatal heart.
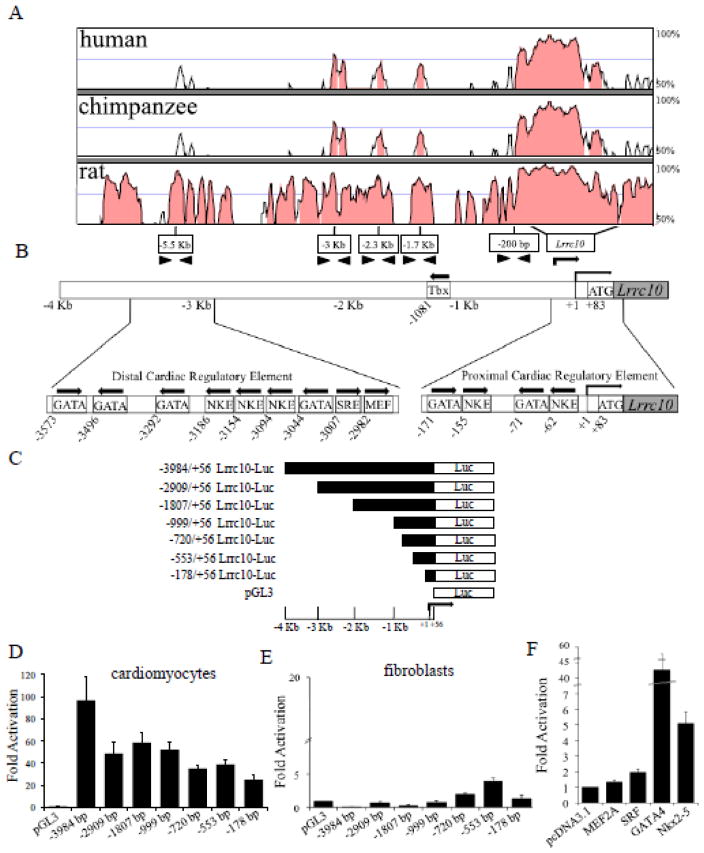
Given the intricate transcriptional regulation of Lrrc10 expression in development, we set out to identify the enhancer/promoter regions of the Lrrc10 genomic locus that confer cardiac-specific expression of Lrrc10 and the corresponding trans-acting factors. A VISTA alignment was performed to identify highly conserved and potential functionally important regions of the Lrrc10 genomic locus (Fig 1A). VISTA alignment of the genomic sequences of mouse, rat, chimpanzee, and human revealed highly conserved regions of 70% or greater sequence homology located at approximately −3 Kb, −2.3 Kb, −1.7 Kb, and −200 bp of the transcription initiation site (TIS). No highly conserved regions were found within 10 Kb downstream of the stop codon (data not shown). Because the Lrrc10 genomic locus contains highly conserved regions within −4 Kb of the TIS (Fig 1A) [4], we analyzed this region for potential transcription factor binding sites using the TFSEARCH program [51] and rVista [52]. Results revealed the presence of multiple DNA binding sites for critical cardiac transcription factors such as Nkx2-5, GATA4, MEF2, and SRF (Fig 1B). Notably, a cluster of three Nkx2-5 binding sites (NKEs) is located around −3.1 Kb of the TIS of Lrrc10, which is flanked by GATA binding sites. Additionally, two NKEs that each lie adjacent to GATA binding sites are found within −200 bp of the TIS (Fig 1B), suggesting a potential role for Nkx2-5 and GATA4 in regulation of Lrrc10 expression.
Figure 1.
The highly conserved mouse Lrrc10 promoter regions exhibit cardiomyocyte-specific activity. (A) A VISTA alignment was performed on the Lrrc10 locus for mouse, rat, chimpanzee, and human to identify conserved regions. Arrowheads indicate primer sites designed in conserved regions. Rightward arrow indicates transcription initiation site (TIS, +1). Shading denotes regions of greater than 70% conservation. (B) Schematic representation of transcription factor binding sites within conserved regions located approximately −3 Kb and −200 bp of the TIS of Lrrc10. GATA, NKE, MEF, and SRE are putative binding sites for GATA4, Nkx2-5, MEF2, and SRF, respectively. Arrows indicate orientation of putative binding sites. (C–E) Determination of cardiac regulatory elements by reporter gene analyses. Deletion analyses of the Lrrc10 promoter regions were performed in cultured mouse neonatal cardiomyocytes and 10T1/2 cells. (C) Schematic diagram of the 5′ deletions of the Lrrc10 promoter region used for transient transfections. Lrrc10 reporter genes are (D) robustly activated in cardiomyocytes but (E) inactive in 10T1/2 cells. (F) A−3984 bp Lrrc10 luciferase reporter construct is activated in 10T1/2 cells when cotransfected with the cardiac transcription factors Nkx2-5, GATA4, or SRF, but not MEF2A. Fold activation is expressed relative to pcDNA3.1 vector control.
To dissect the cis-elements that mediate cardiac expression, we performed transient transfection assays using reporter genes containing the Lrrc10 promoter region linked to a luciferase reporter. Various serial deletion reporters were constructed (Fig 1C) and luciferase activity was evaluated in primary cardiomyocytes or a fibroblast cell line. The −3984 bp Lrrc10 reporter showed a marked 100-fold activation in cardiomyocytes (Fig 1D), but no activation in fibroblasts (Fig 1E), suggesting that cardiac regulatory elements are located within the −4 Kb region. A marked decrease in luciferase activity was observed from the −3984 bp to −2909 bp reporter constructs in cardiomyocytes but not in fibroblasts (Fig 1D, E). However, further deletion from −2909 bp to −1807 bp constructs did not further decrease cardiac-specific activation, indicating that there is a functionally important cardiac regulatory element present between the −3984 and −2909 bp region. This regulatory region located around −3 Kb (referred to hereafter as the distal cardiac regulatory element, DCE) contains three putative NKEs flanked by GATA binding sites around −3.1 Kb (Fig 1B), which are deleted in the −2909 bp reporter. In contrast, deletion of the −3984 bp to −2909 bp region markedly increased reporter activity in fibroblasts (Fig 1E), suggesting a repressive element functions in fibroblasts between the −3984 bp and −2909 bp region. Therefore, the region between −3984 bp and −2909 bp likely ensures cardiac-specific expression of Lrrc10 by repressing ectopic expression in non-cardiomyocytes.
Further serial deletion of reporters did not appear to affect cardiac-specific expression of Lrrc10 (Fig 1D). Although there is a Tbx binding site present in the Lrrc10 promoter (Fig 1B), it is in a region that is not highly conserved (Fig 1A) and deletion of the region from −1807 bp to −999 bp containing the Tbx binding site does not affect cardiac-specific activity (Fig 1D). The −178 bp region was sufficient to mediate cardiac-specific activity (Fig 1D), and therefore, this region is hereafter referred to as the proximal cardiac regulatory element, PCE. The PCE contains a pair of adjacent NKEs and GATA binding sites but no SRF DNA binding site (Fig 1B). Because binding sites for the cardiac transcription factors SRF, MEF2, GATA4, and Nkx2-5 are located within 4Kb of the TIS, the ability of these factors to activate the Lrrc10 promoter was evaluated in transient transfection assays. Results indicate that Nkx2-5, SRF, or GATA4 alone activates the −3984 bp Lrrc10 reporter, however, no activation is observed in response to MEF2A (Fig. 1F).
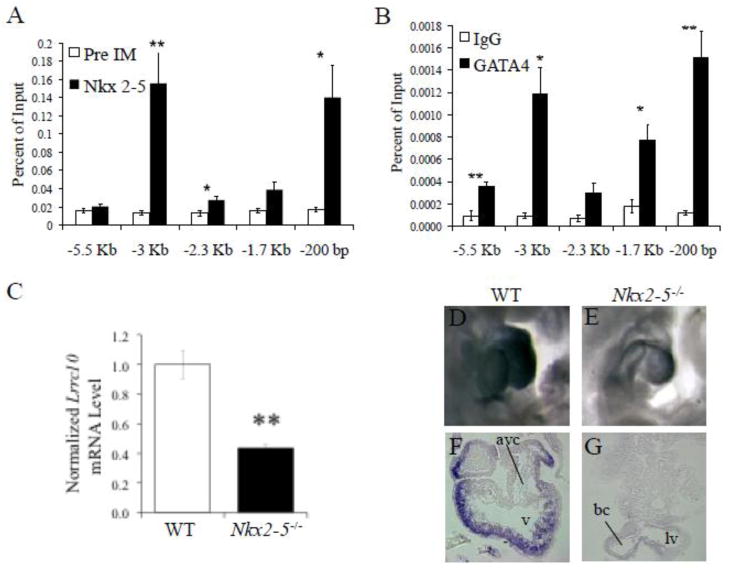
Given the significant activation of the Lrrc10 promoter by Nkx2-5 and GATA4, we hypothesized that Nkx2-5 and GATA4 are key mediators of the cardiac-specific expression pattern of Lrrc10. To determine if Nkx2-5 endogenously occupies conserved regions of the Lrrc10 locus (Fig 1A) in the heart, qChIP analyses were performed on postnatal mouse heart tissue using primers specific for these conserved regions and an Nkx2-5 antibody. Significant occupancy of Nkx2-5 was observed at the highly conserved regions −3 Kb, −2.3 Kb, and −200 bp relative to the TIS of Lrrc10 (Fig 2A), which all contain NKEs (Fig 1B, data not shown). The most substantial accumulation of Nkx2-5 occurs at two conserved regions at −3 Kb and −200 bp (Fig 2A), corresponding to the DCE and PCE, respectively (Fig 2A, 1B). Minor accumulation of Nkx2-5 occurs at the -2.3 Kb region (Fig 2A). However, truncation of the NKE within this region (at −2285/−2279 bp) does not reduce transcriptional activities in cardiomyocytes (Fig 1D, −2909 bp vs. −1807 bp reporters), suggesting this NKE is not functional. qChIP assays using a GATA4 antibody revealed that the most significant occupancy of GATA4 also occurred at −3 Kb and −200 bp of the TIS of Lrrc10 (Fig 2B), regions that contain GATA binding sites. GATA4 additionally occupies regions containing putative GATA binding sites at −5.5 Kb and −1.7 Kb, but to a much lesser extent (Fig 2B). These results indicate that high levels of occupancy by Nkx2-5 and GATA4 occur at conserved regions −3 Kb and −200 bp of the Lrrc10 locus in the heart, which contain cardiac regulatory elements (Fig 1D) and NKEs and GATA binding sites (Fig 1B).
Figure 2.
Lrrc10 is a target gene of Nkx2-5 and GATA4. (A, B) Nkx2-5 and GATA4 endogenously occupy specific regions of the Lrrc10 promoter. qChIP was performed on wildtype (WT) hearts at postnatal day 3 using (A) Nkx2-5 or (B) GATA4 specific antibodies and primers for conserved regions of the Lrrc10 promoter as indicated in Fig 1A (n=3). Preimmune serum (Pre IM) or nonspecific rabbit IgG was used as a control. (C–G) Lrrc10 expression is reduced in Nkx2-5−/ − hearts. (C) qRT-PCR shows markedly reduced expression of Lrrc10 in Nkx2-5−/ − embryos at E9.5 (n=3). Expression was normalized to 18S expression. (D) WT and (E) Nkx2-5−/ − embryos at E9.5 were subjected to whole-mount in situ hybridization using digoxigenin-labeled Lrrc10 antisense cRNA probes. Sense cRNA probes did not give any signal (data not shown). Transverse sections of labeled (F) WT and (G) Nkx2-5−/ − hearts reveal dramatically reduced expression of Lrrc10 in the Nkx2-5−/− heart. avc, atrioventricular canal; v, ventricle; lv, left ventricle; bc, bulbous cordis. *p<0.05, **p<0.01 compared to control.
To assess whether expression of Lrrc10 is regulated by Nkx2-5 in vivo, Lrrc10 expression was evaluated in Nkx2-5 knockout (Nkx2-5−/−) mice. Nkx2-5−/− mice die around embryonic day (E) 10 with arrested cardiogenesis after partial looping [9]. Evaluation of Lrrc10 transcript levels by qRT-PCR reveals a 57% reduction in Lrrc10 mRNA expression in Nkx2-5−/− hearts (Fig 2C), providing crucial evidence that Nkx2-5 is a key regulator of Lrrc10 expression in vivo. Lrrc10 mRNA level was normalized to a house keeping gene and other genes are expressed at normal levels in Nkx2-5−/− hearts despite severe morphological defects [9, 53], suggesting downregulation of Lrrc10 is specifically due to the absence of Nkx2-5. However, it cannot be ruled out that Lrrc10 expression is reduced in Nkx2-5−/− hearts as a consequence of gross morphogenetic abnormalities. Furthermore, whole mount in situ hybridization of E9.5 embryos demonstrates dramatically reduced expression of Lrrc10 in the Nkx2-5−/− heart as compared to the control heart (WT) (Fig 2D, E). Transverse sections of in situ labeled embryos show cardiomyocyte-specific expression of Lrrc10 in WT (Fig 2F) that is significantly reduced in Nkx2-5−/− hearts (Fig 2G). Notably, Lrrc10 is not expressed in the developing endocardium or epicardium (Fig 2F) [1]. Lrrc10 expression was not evaluated in Gata4 knockout embryos due to early embryonic lethality prior to the formation of the heart tube and the well-established genetic and functional redundancy of GATA factors such as cardiac expression of GATA5 and 6 [54, 55].
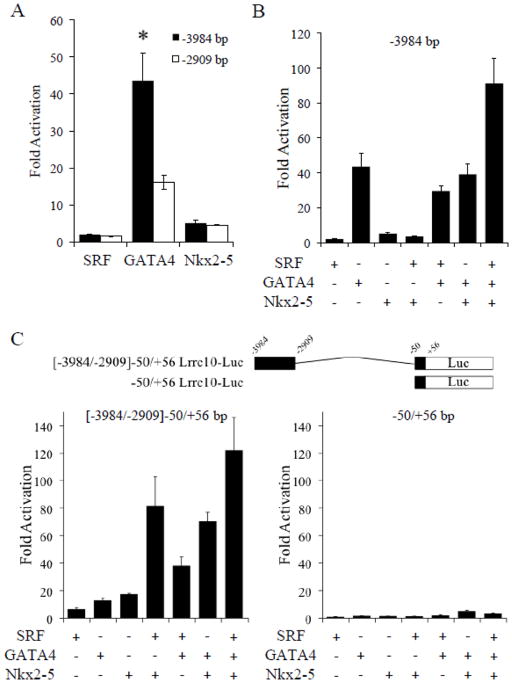
Because the Lrrc10 promoter is activated by SRF, Nkx2-5, and GATA4 (Fig 1F) and Nkx2-5 and GATA4 endogenously occupy the highly conserved DCE containing putative binding sites (Fig 2A, B), we tested whether these cardiac factors regulate the expression of Lrrc10 via this DCE. Cells were cotransfected with SRF, Nkx2-5, or GATA4 and a −3984 bp or a −2909 bp Lrrc10 reporter lacking the DCE harboring binding sites for these factors. A dramatic reduction in GATA4 mediated activation of the Lrrc10 promoter was observed when the conserved GATA site (−3044/−3041 bp) within the DCE was removed (Fig 3A). However, truncation of NKEs and SRF binding sites within this DCE failed to alter Nkx2-5- or SRF-mediated activation, respectively (Fig 3A). To investigate whether SRF, Nkx2-5, and GATA4 cooperatively regulate expression of Lrrc10, cells were cotransfected with the −3984 bp Lrrc10 reporter and SRF, Nkx2-5, and/or GATA4. Interestingly, although no one factor independently alters the transcriptional activity of any other factor, cotransfection of SRF, Nkx2-5, and GATA4 synergistically activates Lrrc10 expression (Fig 3B). Cotransfection of MEF2A did not affect the transcriptional activity of SRF, Nkx2-5, or GATA4 (data not shown), suggesting that MEF2 is not involved in regulation of Lrrc10 expression.
Figure 3.
The distal cardiac regulatory element (DCE) of Lrrc10 is regulated by SRF, GATA4, and Nkx2-5. (A) There is a conserved functional GATA4 binding site located in the DCE. 10T1/2 cells were cotransfectd with SRF, GATA4, or Nkx2-5 and a −3984 bp Lrrc10 reporter or a −2909 bp Lrrc10 reporter lacking the DCE containing three NKEs and conserved SRF and GATA factor binding sites. Deletion of the DCE significantly reduces GATA4-mediated activation of the Lrrc10 promoter while Nkx2-5 and SRF-mediated activation is unaffected. Nkx2-5, GATA4, and SRF regulate the DCE of Lrrc10. (B) 10T1/2 cells were cotransfected with the −3984 bp reporter and Nkx2-5, GATA4, and/or SRF. GATA4, Nkx2-5, and SRF together synergistically activate the Lrrc10 promoter. C) A reporter gene containing the DCE (−3984/−2909 bp) and the Lrrc10 promoter (−50/+56) or a reporter gene only containing the promoter was cotransfected with Nkx2-5, GATA4, and/or SRF. Nkx2-5, GATA4, and SRF cooperatively regulate the DCE of Lrrc10 (left). Negligible activation is observed on the Lrrc10 promoter alone (right). Activation is expressed relative to pcDNA3.1 vector control. *p<0.05 compared to −2909 bp reporter.
To determine if SRF, GATA4, or Nkx2-5 alone or in combination regulates Lrrc10 expression via the DCE (−3984/−2909 bp), a reporter gene containing only the DCE with the Lrrc10 promoter (−50/+56 bp) was constructed (Fig 3C, top). SRF, GATA4, or Nkx2-5 alone activates this DCE-containing reporter independent of the PCE (Fig 3C, left). Cotransfection of any two factors results in synergistic activation and cotransfection of SRF, GATA4, and Nkx2-5 results in robust activation (greater than 120-fold) of the DCE reporter (Fig 3C, left). In contrast, negligible activation is observed by the Lrrc10 promoter (−50/+56 bp) alone in response to SRF, GATA4, and/or Nkx2-5 (Fig 3C, right).
To investigate whether Nkx2-5 and GATA4 cooperatively regulate Lrrc10 transcription via the PCE, the −178 bp Lrrc10 reporter was cotransfected with Nkx2-5 and GATA4 alone or together. Results indicate that Nkx2-5 or GATA4 alone induce Lrrc10 reporter activity 18- and 54-fold, respectively, while together Nkx2-5 and GATA4 synergistically activate Lrrc10 reporter activity 184-fold (Fig 4A). Interestingly, there are two NKEs that are each adjacent to GATA binding sites (Fig 1B, 4B) within the PCE. To identify the functional binding sites that are responsible for the Nkx2-5- and GATA4-mediated activity of the PCE, we mutated the NKEs and GATA sites within this region (Fig 4B). A −178 bp Lrrc10 reporter or a mutant construct containing mutations in one or both of the NKEs or GATA sites was cotransfected with Nkx2-5 or GATA4 (Fig 4C, D). Mutation of either one of the NKEs in the PCE (pmN/G or dmN) diminished Nkx2-5-mediated activation. When both NKEs were mutated (pmN/G/dmN), Nkx2-5-mediated reporter activity was reduced further (Fig 4C). Mutation of either GATA site (pmN/G or dmG) drastically reduced GATA4-mediated activation and mutation of both GATA binding sites (pmN/G/dmG) almost abrogated activation by GATA4 (Fig 4D). Taken together, reporter assay data indicate that NKEs in the PCE, but not in the DCE, play important roles in activating Lrrc10 transcription, whereas GATA sites in both the DCE and PCE are crucial.
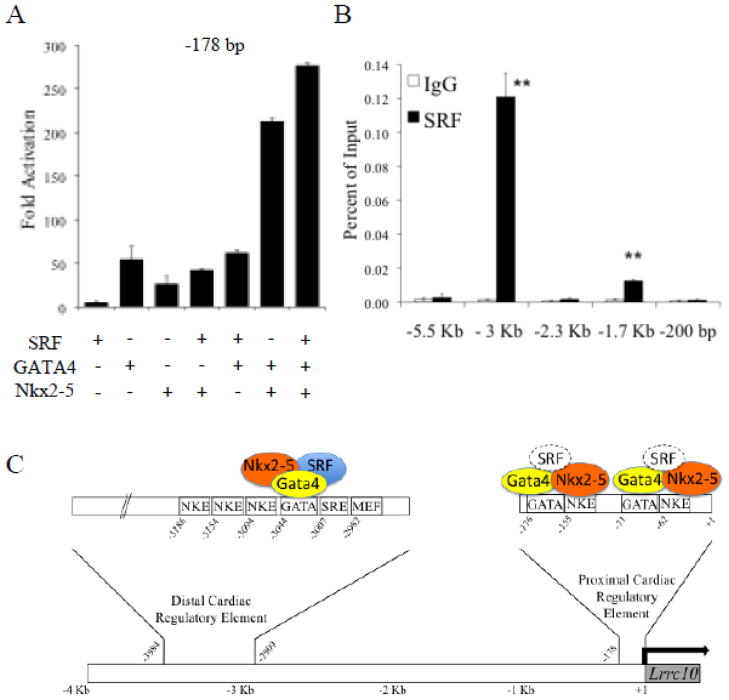
To investigate whether SRF contributes to activation of the PCE, the −178 bp Lrrc10 reporter was cotransfected with Nkx2-5, GATA4, and/or SRF. Nkx2-5 and GATA4 synergistically activate the reporter but SRF alone does not activate the PCE. When cotransfected, SRF augments synergistic activation of the −178 bp reporter by Nkx2-5 and GATA4 (Fig 5A). To determine if SRF endogenously occupies regulatory regions of Lrrc10 in the heart, qChIP assays were performed with an SRF antibody or IgG control (Fig 5B). There is robust accumulation of SRF at the conserved region at −3 Kb of the Lrrc10 TIS (Fig 5B), supporting cooperative regulation of the DCE by Nkx2-5, GATA4, and SRF (Fig 3B, C). Although a low level of accumulation of SRF is observed at the −1.7 Kb region, there are no cardiac regulatory elements (Fig 1D, E, −1807 bp vs. −999 bp reporters) or SRF binding sites (SREs) located within this region. In contrast, no accumulation of SRF was observed at the PCE. Based on our findings, our working model of transcriptional regulation of Lrrc10 is depicted in Fig 5C (See Discussion).
Figure 5.
Roles of SRF in regulation of Lrrc10 expression and working model. (A) A −178 bp Lrrc10 reporter was cotransfected with Nkx2-5, GATA4, and/or SRF. (B) SRF endogenously occupies the DCE of Lrrc10. qChIP was performed on WT hearts at postnatal day 3 using an SRF antibody or IgG as a control (n=3). (C) Working model of Lrrc10 transcriptional regulation. The DCE located at around −3Kb is endogenously occupied in the heart and cooperatively activated by SRF, Nkx2-5, and GATA4, and contains a highly conserved functional GATA4 binding site. The PCE of Lrrc10 located within 200 bp of the transcription initiation site (TIS) is endogenously occupied and synergistically activated by Nkx2-5 and GATA4 via their respective binding sites and SRF may augment activation by Nkx2-5 and GATA4. Dotted outline of SRF denotes lack of endogenous accumulation in the heart as evidenced by qChIP as shown in (B). **p<0.01 compared to IgG.
4. Discussion
Cardiac-specific expression of Lrrc10 begins very early in the precardiac region [1]. Lrrc10 expression at the mRNA [56] and protein levels is significantly increased in the heart soon after birth, suggesting a transcriptional regulatory mechanism mediates the cardiac-specific upregulation of Lrrc10. Since Lrrc10 expression is remarkably cardiac-specific [4, 56] and is dynamically regulated during development, it is critical to determine the molecular mechanisms that regulate expression of Lrrc10. Here, we provide in vivo evidence that Lrrc10 is a novel target gene of the cardiac transcription factors Nkx2-5 and GATA4 by showing that these factors occupy the cardiac regulatory regions of Lrrc10 in the heart and that deletion of Nkx2-5 dramatically reduces Lrrc10 expression. We identify two cardiac regulatory elements located around −3 Kb (DCE) and −200 bp (PCE) of the Lrrc10 TIS, which confer Lrrc10 reporter activity specifically in cardiomyocytes but are inactive in fibroblasts.
The DCE at −3 Kb from the TIS of Lrrc10 is endogenously occupied and cooperatively activated by SRF, Nkx2-5, and GATA4. Deletion of the DCE markedly reduces GATA4-mediated, but not SRF- or Nkx2-5-mediated activation of the Lrrc10 promoter, suggesting that the conserved GATA site (−3044/−3041) in the DCE plays a major functional role (Fig 5C). Importantly, GATA4, SRF, and Nkx2-5 cooperatively activate transcription via this DCE independent of the PCE. GATA4 may be involved in recruiting SRF and/or Nkx2-5 or perhaps other yet unidentified cardiac transcription factors to the DCE to regulate Lrrc10 expression. Although there are other GATA sites within the DCE, these sites are not conserved in human, suggesting that the conserved GATA site at −3044/−3041 bp, which is in closest proximity to the SRE and NKEs in this region, is likely the GATA4-responsive DNA sequence.
Indeed, formation of higher order transcriptional complexes among cardiac transcription factors including SRF, Nkx2-5, and GATA4 is often needed for cooperative regulation of transcription. For example, Nkx2-5 autoregulates its own transcription in the second heart field via conserved enhancers by combinatorial but not individual expression of Nkx2-5, SRF, GATA4, GATA6, myocardin, and p300 [57]. The Xenopus MLC2 gene contains GATA4 binding sites that flank a YY1/CarG-like site that mediate its myocardial expression [58] and the zebrafish ventricular myosin heavy chain (vmhc) promoter contains a proximal cardiac enhancer element with multiple NKEs and GATA factor binding sites upstream of an SRF binding site [59], both similar to the configuration observed in the DCE of Lrrc10. In the case of Lrrc10, the SRF binding site within the DCE is not critical for SRF-mediated activation, but SRF cooperates with Nkx2-5 and GATA4 (Fig 5B).
We also identify a PCE within the −178 bp region of the Lrrc10 promoter that is sufficient to confer cardiac-specific expression and is synergistically activated by Nkx2-5 and GATA4. The PCE is endogenously occupied by Nkx2-5 and GATA4. The PCE contains two NKEs (−155/−149 and −62/−56 bp) with adjacent upstream GATA binding motifs that are critical for Nkx2-5- and GATA4-mediated activation, respectively. Although SRF augments synergistic activation of the PCE by Nkx2-5 and GATA4, SRF does not significantly accumulate at the PCE in the heart when assayed by qChIP. Cooperation of SRF with Nkx2-5 and GATA4 at the PCE may be observed as a consequence of overexpression in transfection assays. Alternatively, accumulation of SRF at the PCE may not be detectable under the experimental ChIP conditions employed. Therefore, SRF may regulate Lrrc10 expression primarily via the DCE in vivo.
Although a previous study attempted to characterize the cis elements mediating cardiac-specific expression of Lrrc10, experiments were performed only in vitro using NIH3T3 fibroblast cells [6], which are not suitable cells for identifying regulatory elements that confer cardiac-specific expression. Moreover, the functional significance of the cis elements was not investigated in cardiomyocytes nor was regulation of Lrrc10 expression evaluated in vivo. In contrast, we clearly demonstrate that fibroblasts do not activate reporter genes containing the Lrrc10 promoter regions (Fig 1E). Nonetheless, Fan et al. reported that a different GATA binding site (−2894/−2889 bp) downstream of the DCE is dispensable while the GATA site at −71/−68 bp is important for GATA4-mediated activation [6], supporting our finding that the GATA sites within the DCE (−3044/−3041 bp) and PCE (−171/−168 and −71/−68 bp) are the GATA4 responsive DNA sequences. In addition, the MEF2 site in the DCE was not functional, consistent with the previous study [6].
Nkx2-5 and GATA4 physically interact via the homeobox domain of Nkx2-5 and a C-terminal zinc finger of GATA4 to synergistically activate cardiac target gene expression [16–19]. The promoters of essential cardiac genes such as α-cardiac actin [16], ANF [15, 18], CARP [60], Id2 [45] and the A1 adenosine receptor [61] are synergistically activated by Nkx2-5 and GATA4. Binding to adjacent NKE and GATA sites by Nkx2-5 and GATA4, respectively, is required to cooperatively activate the minimal (−150 bp) cardiac enhancer of connexin40 [22] and to synergistically activate the Id2 [45] and A1 adenosine receptor [61] promoters. In contrast, regulation of CARP requires GATA4, but not Nkx2-5, binding to DNA [60], and neither Nkx2-5 nor GATA4 binds DNA to activate cardiac α-actin expression but are instead recruited to its promoter by SRF [16, 17, 20, 21]. Conversely, in the case of Lrrc10, SRF is likely recruited to the DCE by GATA4 and to the PCE by GATA4 and/or Nkx2-5 to induce full activation of Lrrc10 transcription.
Most notably, the ANF enhancer element within −270 bp of the TIS contains two adjacent NKEs with upstream GATA binding sites in very close proximity (within 25 bp), an arrangement very similar to the PCE of Lrrc10, which contains two adjacent NKEs with GATA sites within 15 bp upstream. The transcriptional activity of GATA4 and Nkx2-5 and corresponding DNA binding sites are required for synergistic activation of ANF [15, 18]. Due to the close proximity of GATA binding sites and NKEs in the PCE of Lrrc10 and dramatic reduction in activation when either GATA site or either NKE is mutated, we speculate that GATA4 and Nkx2-5 bind their respective DNA binding sites and physically interact with each other to potentiate their ability to activate Lrrc10 expression. However, further investigation is required to determine the critical endogenous cardiac regulatory sequences and exact molecular mechanism of Nkx2-5 and GATA4-mediated activation of Lrrc10. Regardless of the mechanism, our data taken together demonstrate that Lrrc10 is a bona fide cardiac target gene of Nkx2-5 and GATA4.
The PCE of mouse Lrrc10 maps to the 5′UTR of the human LRRC10 genomic locus, suggesting that regulatory elements may reside within the 5′UTR of human LRRC10, which has been reported in other genes. The expression of rat carnitine palmitoyltransferase Iβ [62] and human 5-hydroxytryptamine receptor 4 [63] is regulated by an enhancer element in the 5′UTR that contains putative overlapping NKE and GATA binding sites or a putative NKE, respectively. Zebrafish ventricular myosin heavy chain (vmhc) contains overlapping NKEs in the second intron [59, 64] that are functionally important in the regulation of ventricular specific expression [64]. Additionally, splice variants 1A and 1C of the mouse cardiac sodium channel (Scn5a) contain cardiac enhancer elements within the 5′UTR and within the first exon, respectively [65]. Importantly, the human integrin β3 gene contains a functional regulatory element in the 5′UTR that maps to the 5′ flanking promoter region in avians [66]. Therefore, 5′UTRs in other genes have been shown to be critical in regulation of cardiac gene expression, as is likely the case for human LRRC10.
5. Conclusions
We identify the cardiac-specific factor Lrrc10 as a novel target gene of Nkx2-5 and GATA4. Distal (around −3 Kb) and proximal (within−200 bp) cardiac regulatory elements upstream of Lrrc10 are critical for mediating its expression and are occupied by Nkx2-5 and GATA4 in the heart. We characterize how Lrrc10 is transcriptionally regulated by Nkx2-5 and GATA4 in cooperation with SRF, providing fundamental insight into gene regulatory mechanisms necessary for proper cardiac development and function.
Supplementary Material
Highlights.
We characterize mechanisms mediating the cardiac-specific expression of Lrrc10.
Distal and proximal cardiac cis- regulatory elements mediate expression of Lrrc10.
Nkx2-5, GATA4, and SRF cooperatively regulate transcription of Lrrc10.
Acknowledgments
The authors would like to acknowledge Dr. Aseem Z. Ansari (University of Wisconsin-Madison) for kindly providing the Nkx2-5 antibody and Dr. Jay Patrick (Washington University, St. Louis, MO) for providing reagents. This work was supported by grants from the National Institutes of Health (HL067050) and the American Heart Association (12GRNT12070021) to YL, predoctoral fellowship to MJB from the American Heart Association (11PRE5580012), and the Molecular and Environmental Toxicology Training Grant from the National Institute for Environmental Health Sciences (T32ES007015). KHL was funded by the American Heart Association (12BGIA12060120) and the South Carolina COBRE for Developmentally-based Cardiovascular Disease (NCRR P20RR016434).
Footnotes
Disclosure Statement
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim KH, Kim TG, Micales BK, Lyons GE, Lee Y. Dynamic expression patterns of leucine-rich repeat containing protein 10 in the heart. Dev Dyn. 2007;236:2225–34. doi: 10.1002/dvdy.21225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakane T, Satoh T, Inada Y, Nakayama J, Itoh F, Chiba S. Molecular cloning and expression of HRLRRP, a novel heart-restricted leucine-rich repeat protein. Biochem Biophys Res Commun. 2004;314:1086–92. doi: 10.1016/j.bbrc.2003.12.202. [DOI] [PubMed] [Google Scholar]
- 3.Kim KH, Antkiewicz DS, Yan L, Eliceiri KW, Heideman W, Peterson RE, et al. Lrrc10 is required for early heart development and function in zebrafish. Dev Biol. 2007;308:494–506. doi: 10.1016/j.ydbio.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adameyko, Mudry RE, Houston-Cummings NR, Veselov AP, Gregorio CC, Tevosian SG. Expression and regulation of mouse SERDIN1, a highly conserved cardiac-specific leucine-rich repeat protein. Dev Dyn. 2005;233:540–52. doi: 10.1002/dvdy.20368. [DOI] [PubMed] [Google Scholar]
- 5.Brody MJ, Hacker TA, Patel JR, Feng L, Sadoshima J, Tevosian SG, et al. Ablation of the cardiac-specific gene leucine-rich repeat containing 10 (lrrc10) results in dilated cardiomyopathy. PLoS One. 2012;7:e51621. doi: 10.1371/journal.pone.0051621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan X, Yang Q, Wang Y, Zhang Y, Wang J, Yuan J, et al. Cloning and characterization of the cardiac-specific Lrrc10 promoter. BMB Rep. 2011;44:123–8. doi: 10.5483/BMBRep.2011.44.2.123. [DOI] [PubMed] [Google Scholar]
- 7.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, et al. Nkx2–5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–86. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 8.Terada R, Warren S, Lu JT, Chien KR, Wessels A, Kasahara H. Ablation of Nkx2–5 at mid-embryonic stage results in premature lethality and cardiac malformation. Cardiovasc Res. 2011 doi: 10.1093/cvr/cvr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–80. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 10.Briggs LE, Takeda M, Cuadra AE, Wakimoto H, Marks MH, Walker AJ, et al. Perinatal loss of Nkx2-5 results in rapid conduction and contraction defects. Circ Res. 2008;103:580–90. doi: 10.1161/CIRCRESAHA.108.171835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda M, Briggs LE, Wakimoto H, Marks MH, Warren SA, Lu JT, et al. Slow progressive conduction and contraction defects in loss of Nkx2-5 mice after cardiomyocyte terminal differentiation. Lab Invest. 2009;89:983–93. doi: 10.1038/labinvest.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–76. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 13.Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–7. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–11. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, Shioi T, Kasahara H, Jobe SM, Wiese RJ, Markham BE, et al. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–9. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sepulveda JL, Belaguli N, Nigam V, Chen CY, Nemer M, Schwartz RJ. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–15. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sepulveda JL, Vlahopoulos S, Iyer D, Belaguli N, Schwartz RJ. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J Biol Chem. 2002;277:25775–82. doi: 10.1074/jbc.M203122200. [DOI] [PubMed] [Google Scholar]
- 18.Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–96. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiojima I, Komuro I, Oka T, Hiroi Y, Mizuno T, Takimoto E, et al. Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4. J Biol Chem. 1999;274:8231–9. doi: 10.1074/jbc.274.12.8231. [DOI] [PubMed] [Google Scholar]
- 20.Chen CY, Croissant J, Majesky M, Topouzis S, McQuinn T, Frankovsky MJ, et al. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet. 1996;19:119–30. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 21.Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol Cell Biol. 1996;16:6372–84. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linhares VL, Almeida NA, Menezes DC, Elliott DA, Lai D, Beyer EC, et al. Transcriptional regulation of the murine Connexin40 promoter by cardiac factors Nkx2-5, GATA4 and Tbx5. Cardiovasc Res. 2004;64:402–11. doi: 10.1016/j.cardiores.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim TG, Chen J, Sadoshima J, Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol Cell Biol. 2004;24:10151–60. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Berlo JH, Elrod JW, Aronow BJ, Pu WT, Molkentin JD. Serine 105 phosphorylation of transcription factor GATA4 is necessary for stress-induced cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2011;108:12331–6. doi: 10.1073/pnas.1104499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–7. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 26.Haworth KE, Kotecha S, Mohun TJ, Latinkic BV. GATA4 and GATA5 are essential for heart and liver development in Xenopus embryos. BMC Dev Biol. 2008;8:74. doi: 10.1186/1471-213X-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–18. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–28. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, et al. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2009;326:368–77. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belaguli NS, Sepulveda JL, Nigam V, Charron F, Nemer M, Schwartz RJ. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol Cell Biol. 2000;20:7550–8. doi: 10.1128/mcb.20.20.7550-7558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore ML, Wang GL, Belaguli NS, Schwartz RJ, McMillin JB. GATA-4 and serum response factor regulate transcription of the muscle-specific carnitine palmitoyltransferase I beta in rat heart. J Biol Chem. 2001;276:1026–33. doi: 10.1074/jbc.M009352200. [DOI] [PubMed] [Google Scholar]
- 32.Brown CO, 3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–69. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 33.Oka T, Maillet M, Watt AJ, Schwartz RJ, Aronow BJ, Duncan SA, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98:837–45. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 34.Funke-Kaiser H, Lemmer J, Langsdorff CV, Thomas A, Kovacevic SD, Strasdat M, et al. Endothelin-converting enzyme-1 (ECE-1) is a downstream target of the homeobox transcription factor Nkx2-5. FASEB J. 2003;17:1487–9. doi: 10.1096/fj.02-0700fje. [DOI] [PubMed] [Google Scholar]
- 35.Barth JL, Clark CD, Fresco VM, Knoll EP, Lee B, Argraves WS, et al. Jarid2 is among a set of genes differentially regulated by Nkx2.5 during outflow tract morphogenesis. Dev Dyn. 2010;239:2024–33. doi: 10.1002/dvdy.22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riazi AM, Takeuchi JK, Hornberger LK, Zaidi SH, Amini F, Coles J, et al. NKX2-5 regulates the expression of beta-catenin and GATA4 in ventricular myocytes. PLoS One. 2009;4:e5698. doi: 10.1371/journal.pone.0005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy AM, Thompson WR, Peng LF, Jones L., 2nd Regulation of the rat cardiac troponin I gene by the transcription factor GATA-4. Biochem J. 1997;322 (Pt 2):393–401. doi: 10.1042/bj3220393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ip HS, Wilson DB, Heikinheimo M, Tang Z, Ting CN, Simon MC, et al. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol Cell Biol. 1994;14:7517–26. doi: 10.1128/mcb.14.11.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhalla SS, Robitaille L, Nemer M. Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J Biol Chem. 2001;276:11439–45. doi: 10.1074/jbc.M100208200. [DOI] [PubMed] [Google Scholar]
- 40.Pikkarainen S, Tokola H, Majalahti-Palviainen T, Kerkela R, Hautala N, Bhalla SS, et al. GATA-4 is a nuclear mediator of mechanical stretch-activated hypertrophic program. J Biol Chem. 2003;278:23807–16. doi: 10.1074/jbc.M302719200. [DOI] [PubMed] [Google Scholar]
- 41.Thuerauf DJ, Hanford DS, Glembotski CC. Regulation of rat brain natriuretic peptide transcription. A potential role for GATA-related transcription factors in myocardial cell gene expression. J Biol Chem. 1994;269:17772–5. [PubMed] [Google Scholar]
- 42.Molkentin JD, Kalvakolanu DV, Markham BE. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the alpha-myosin heavy-chain gene. Mol Cell Biol. 1994;14:4947–57. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durocher D, Chen CY, Ardati A, Schwartz RJ, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–55. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Morishima M, Zheng M, Uchino T, Mannen K, Takahashi A, et al. Transcription factors Csx/Nkx2.5 and GATA4 distinctly regulate expression of Ca2+ channels in neonatal rat heart. J Mol Cell Cardiol. 2007;42:1045–53. doi: 10.1016/j.yjmcc.2007.03.905. [DOI] [PubMed] [Google Scholar]
- 45.Lim JY, Kim WH, Kim J, Park SI. Induction of Id2 expression by cardiac transcription factors GATA4 and Nkx2.5. J Cell Biochem. 2008;103:182–94. doi: 10.1002/jcb.21396. [DOI] [PubMed] [Google Scholar]
- 46.Kim TG, Kraus JC, Chen J, Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem. 2003;278:42247–55. doi: 10.1074/jbc.M307386200. [DOI] [PubMed] [Google Scholar]
- 47.Kim TG, Jung J, Mysliwiec MR, Kang S, Lee Y. Jumonji represses alpha-cardiac myosin heavy chain expression via inhibiting MEF2 activity. Biochem Biophys Res Commun. 2005;329:544–53. doi: 10.1016/j.bbrc.2005.01.154. [DOI] [PubMed] [Google Scholar]
- 48.Johansen FE, Prywes R. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol Cell Biol. 1993;13:4640–7. doi: 10.1128/mcb.13.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mysliwiec MR, Carlson CD, Tietjen J, Hung H, Ansari AZ, Lee Y. Jarid2 (Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J Biol Chem. 2012;287:1235–41. doi: 10.1074/jbc.M111.315945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mysliwiec MR, Bresnick EH, Lee Y. Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression. J Biol Chem. 2011 doi: 10.1074/jbc.M110.205146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–7. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–9. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–66. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 54.Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317:614–9. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xin M, Davis CA, Molkentin JD, Lien CL, Duncan SA, Richardson JA, et al. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc Natl Acad Sci U S A. 2006;103:11189–94. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mysliwiec MR, Kim TG, Lee Y. Characterization of zinc finger protein 496 that interacts with Jumonji/Jarid2. FEBS Lett. 2007;581:2633–40. doi: 10.1016/j.febslet.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark CD, Zhang B, Lee B, Evans SI, Lassar AB, Lee KH. Evolutionary conservation of Nkx2.5 autoregulation in the second heart field. Dev Biol. 2013;374:198–209. doi: 10.1016/j.ydbio.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Latinkic BV, Cooper B, Smith S, Kotecha S, Towers N, Sparrow D, et al. Transcriptional regulation of the cardiac-specific MLC2 gene during Xenopus embryonic development. Development. 2004;131:669–79. doi: 10.1242/dev.00953. [DOI] [PubMed] [Google Scholar]
- 59.Park JS, Kim HS, Kim JD, Seo J, Chung KS, Lee HS, et al. Isolation of a ventricle-specific promoter for the zebrafish ventricular myosin heavy chain (vmhc) gene and its regulation by GATA factors during embryonic heart development. Dev Dyn. 2009;238:1574–81. doi: 10.1002/dvdy.21964. [DOI] [PubMed] [Google Scholar]
- 60.Kuo H, Chen J, Ruiz-Lozano P, Zou Y, Nemer M, Chien KR. Control of segmental expression of the cardiac-restricted ankyrin repeat protein gene by distinct regulatory pathways in murine cardiogenesis. Development. 1999;126:4223–34. doi: 10.1242/dev.126.19.4223. [DOI] [PubMed] [Google Scholar]
- 61.Rivkees SA, Chen M, Kulkarni J, Browne J, Zhao Z. Characterization of the murine A1 adenosine receptor promoter, potent regulation by GATA-4 and Nkx2.5. J Biol Chem. 1999;274:14204–9. doi: 10.1074/jbc.274.20.14204. [DOI] [PubMed] [Google Scholar]
- 62.Wang GL, Moore ML, McMillin JB. A region in the first exon/intron of rat carnitine palmitoyltransferase Ibeta is involved in enhancement of basal transcription. Biochem J. 2002;362:609–18. doi: 10.1042/0264-6021:3620609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maillet M, Gastineau M, Bochet P, Asselin-Labat ML, Morel E, Laverriere JN, et al. Functional studies of the 5′-untranslated region of human 5-HT4 receptor mRNA. Biochem J. 2005;387:463–71. doi: 10.1042/BJ20040744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang R, Xu X. Transient and transgenic analysis of the zebrafish ventricular myosin heavy chain (vmhc) promoter: an inhibitory mechanism of ventricle-specific gene expression. Dev Dyn. 2009;238:1564–73. doi: 10.1002/dvdy.21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shang LL, Dudley SC., Jr Tandem promoters and developmentally regulated 5′- and 3′-mRNA untranslated regions of the mouse Scn5a cardiac sodium channel. J Biol Chem. 2005;280:933–40. doi: 10.1074/jbc.M409977200. [DOI] [PubMed] [Google Scholar]
- 66.Wilhide CC, Jin Y, Guo Q, Li L, Li SX, Rubin E, et al. The human integrin beta3 gene is 63 kb and contains a 5′-UTR sequence regulating expression. Blood. 1997;90:3951–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.