Expansions of simple repetitive sequences in genomic DNA are responsible for many human hereditary disorders, including such debilitating diseases as fragile X syndrome, Huntington's disease, myotonic dystrophy and the familial form of Lou Gehrig's disease. Simple DNA repeats are usually variable in length, but there is a “healthy” range of lengths for each repeat. Occasionally, however, a particular repeat undergoes expansion beyond the threshold of the healthy range. Once this occurs, such a repeat will undergo further expansions as it is transmitted through generations, which ultimately changes the pattern of gene expression, resulting in various pathologies, including the diseases mentioned above. Importantly, only one repeat becomes unstable in a given pedigree, suggesting that there might be a cis-acting triggering event for the primary expansion, i.e. some change in the genome structure around the repeat. Several years ago, based on the findings of Cleary et al. (2002) and others, we proposed several models to explain repeat expansion, one of which we termed the “ori-switch” (Mirkin and Smirnova, 2002). The ori-switch model postulated that this triggering event could be a switch in the direction of replication through the repeat owing to the utilization of a different replication origin (Fig. 1). The paper by Gerhardt et al in the current issue of Molecular Cell (Gerhardt et al., 2014) provides beautiful experimental evidence in support of the ori-switch model for repeat expansion.
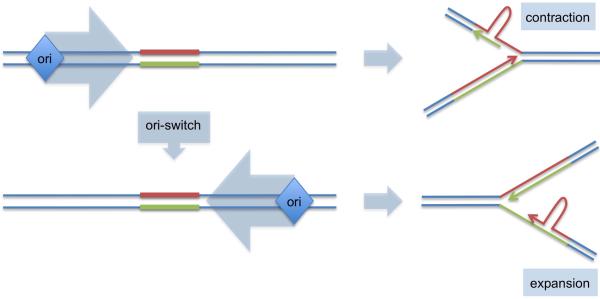
Fig. 1.
The ori-switch model of repeat expansions
Red - structure-prone strand; green – complementary strand. See text for details.
DNA replication likely plays important role in the repeat expansion, although DNA repair and recombination were also implicated in this process. In primate cells, expandable DNA repeats stall replication forks (Voineagu et al., 2009) becoming unstable in the course of DNA replication (Cleary et al., 2002). The fact that direction of replication through expandable repeats affects their stability is well documented in bacteria and yeast (Kim and Mirkin, 2013); recently it was also confirmed for a human chromosomal locus (Liu et al., 2010). This effect could be grounded in the fact that complementary DNA strands of these repeats differ in their ability to form transient secondary structures that ultimately lead to expansions or contractions. During replication, a repetitive sequence that appears in the leading strand template and in the nascent lagging strand is complementary to the repetitive sequence in the lagging strand template and nascent leading strand (Fig. 1). Positioning of a repetitive run within the replication fork combined with its structure-forming potential can influence the pattern of repeat instability. Simply put, the “ori-switch” model suggests that a change in the replication direction through the repeat provokes formation of secondary structures in nascent DNA strands, thus, causing expansions. Which of the nascent DNA strand, leading or lagging is better suited for the expansions remains unclear and is a subject of intense discussions (Kim and Mirkin, 2013).
While studies conducted in various model systems have been generally in-line with the ori-switch model, analysis of replication direction in cells isolated from patients with various repeat expansion diseases has yielded conflicting results. One study suggested that in myotonic dystrophy patients, the mutated allele is replicated from the opposite side compared to the normal allele (Cleary et al., 2010). However, no such difference has been detected for patients with Huntington disease (Nenguke et al., 2003). Progress in this area has been complicated by two problems: (1) the nascent strand abundance method used in mapping of mammalian replication origins works at the level of cell population average complicating data analysis, particularly in heterozygous cell cultures; (2) origins were mapped in differentiated cell cultures that are not particularly prone to expansions.
Gerhardt et al (2014)managed to successfully overcome these problems in their study of replication of expanded (CGG)n repeats in the FMR1 gene responsible for fragile X syndrome. First, replication origins were mapped using an approach called single molecule analysis of replicated DNA (SMARD) developed in Carl Schildkraut's lab. In brief, it combines stretching DNA on a microscope slide followed by in situ hybridization and immunofluorescence, thus allowing one to detect replication direction in any given chromosomal segment at a single-molecule resolution. Second, they looked at the replication of expanded (CGG)n repeats in two lines of fragile X human embryonic stem cells (hESCs). This focus was crucial, as genetic data implied that (CGG)n repeats expand during either oogenesis, or early embryogenesis.
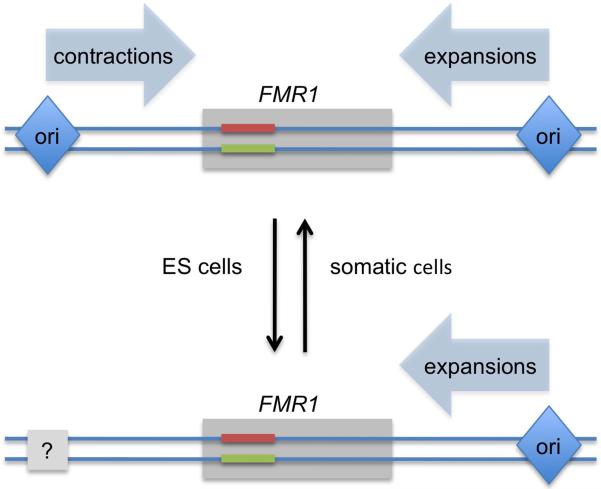
Using these advances, they found that the direction of replication was indeed altered in fragile X hESCs compared to the control hESC lines: while in control cells, FMR1 locus was replicated with equal probability by replication forks moving from the left and from the right, in fragile X cells, it was replicated exclusively by the forks approaching from the right. These data suggest that the origin(s) of replication located on the left side of the FMR1 gene, was not firing properly in the X chromosome carrying expanded (CGG)n repeat (Fig. 2). A chicken-and-egg problem still persists in these data: is the observed ori-switch a cause or a consequence of repeat expansions? In somatic cells, expansions of (CGG)n repeats beyond 200 copies induce heterochromatization of the FMR1 gene and surrounding chromosomal segment, causing its very late replication and chromosomal fragility (Hansen et al., 1993). Could heterochromatinization also cause ori-switch? Remarkably, expanded (CGG)n repeats didn't cause significant heterochromatinization in fragile X hESCs studied by Gerhardt et al (2014) as the FMR1 gene remained transcriptionally active. Thus, a change in the direction of replication is unlikely to be a consequence of heterochromatinization, arguing that it is, in fact, the cause of repeat expansion.
Fig. 2.
A developmentally-regulated replication switch, which disrupts homeostasis of a repeat's length in hESCs See text for details.
Gerhardt et al (2014) also looked at the replication pattern of (CGG)n-containing FMR1 locus after the fragile X hESCs underwent differentiation into embryoid bodies. Amazingly, the replication pattern they discovered at that stage was indistinguishable from control cells, suggesting that the inactivation of the left replication origin was a developmentally regulated event, only evident at the earliest embryonic stages. This notion provides an explanation for the long-known puzzling fact that while expanded repeats tend to be relatively stable in somatic cells, they undergo frequent expansions during intergenerational transmissions, thus increasing the severity of the disease in the next generation. Data of Gerhardt et al argue that early embryonic development is the likely stage for repeat expansions in fragile X syndrome.
Based on their findings, Gerhardt et al (2014) suggest a model for repeat expansion in fragile X patients (Fig. 2). In normal individuals, the (CGG)n-containing FMR1 locus is replicated with equal probability from left and right directions. When it is replicated by the fork that approaches from left, such that the structure-prone CGG-strand is in the lagging strand template, it occasionally undergoes contractions. When it is replicated by the fork that approaches from right, such that the CGG-strand is in the nascent lagging strand, it occasionally undergoes expansions (Fig. 1). Thus, in normal cells, expansions and contractions balance each other out, leading to homeostasis of the (CGG)n repeat's length. The triggering event in fragile X pedigrees is developmentally-regulated inactivation of the replication origin located on the left side of FMR1, such that during early embryonic development, the (CGG)n repeat is predominantly replicated by the replication fork moving from the right, thus predisposing it to expansions. The beauty of this model is that it addresses two mysteries of repeat expansion diseases: why only one particular repeat keeps expanding in a given pedigree, and why it predominantly expands during intergenerational transmissions, but not in somatic tissues.
That said, a lot more work must still be done to substantiate this model and explore its generality. For example, it is yet to be demonstrated that expansions in their fragile X hESCs as expected for (CGG)n repeats replicated exclusively from the origin on the right. While Gerhardt et al. (2014) did observe replication fork stalling at (CGG)n repeats, consistent with previous studies, it is unclear why the stalling observed for ~450 (CGG)n repeats in fragile X cells was of the same magnitude as that for ~30 (CGG) repeats in normal cells. In a different study, replication stalling at (CGG)n repeats in mammalian cells was length-dependent (Voineagu et al., 2009). The replication analysis in hESCs from the fragile X pre-mutation carriers should also be performed to confirm the model. Further, the DNA mismatch repair protein MSH2 was found to be rate limiting for repeat expansions in a fragile X mouse model (Lokanga et al., 2013). Is there a role for MSH2 protein in the fragile X hESCs?
It would be of great interest to sequence the genomic area to the left of FMR1 from fragile X hESCs to find out why the left replication origin has turned off. One might expect to discover a genetic determinant of developmentally regulated origin activity since the propensity of (CGG)n repeat to expand persists in generations.
Finally, Gerhardt et al (2014) so far have only applied their SMARD technique to analyze replication of in fragile X hESCs. It would be great to apply this powerful methodology to other repeat expansion diseases and see if a developmentally regulated replication switch is their general tendency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- Cleary JD, Tome S, Lopez Castel A, Panigrahi GB, Foiry L, Hagerman KA, Sroka H, Chitayat D, Gourdon G, Pearson CE. Tissue- and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nat StructMolBiol. 2010;17:1079–1087. doi: 10.1038/nsmb.1876. [DOI] [PubMed] [Google Scholar]
- Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell. 2014 doi: 10.1016/j.molcel.2013.10.029. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Canfield TK, Lamb MM, Gartler SM, Laird CD. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- Kim JC, Mirkin SM. The balancing act of DNA repeat expansions. CurrOpin Genet Dev. 2013;23:280–288. doi: 10.1016/j.gde.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) × (CAG) repeat hairpins in human cells. Nat ChemBiol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokanga RA, Zhao XN, Usdin K. The Mismatch Repair Protein MSH2 is Rate Limiting for Repeat Expansion in a Fragile X Premutation Mouse Model. Hum Mutat. 2013 doi: 10.1002/humu.22464. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM, Smirnova EV. Positioned to expand. Nat Genet. 2002;31:5–6. doi: 10.1038/ng0502-5. [DOI] [PubMed] [Google Scholar]
- Nenguke T, Aladjem MI, Gusella JF, Wexler NS, Arnheim N. Candidate DNA replication initiation regions at human trinucleotide repeat disease loci. Hum Mol Genet. 2003;12:1021–1028. doi: 10.1093/hmg/ddg111. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat StructMolBiol. 2009;16:226–228. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]