Abstract
Secreted frizzled-related protein-5 (sFRP-5) has been identified as one of the secreted antagonists that bind Wnt protein. However, the functional significance of sFRP-5 in renal cell cancer (RCC) has not been reported. We hypothesized that sFRP-5 may be epigenetically downregulated through DNA methylation and histone modification and function as a tumor suppressor gene in RCC. Using tissue microarray and real-time RT-PCR, we found that sFRP-5 was significantly downregulated in kidney cancer tissues and cell lines, respectively. DNA bisulfite sequencing of the sFRP-5 promoter region in RCC cell lines showed it to be densely methylated whereas there was few promoter methylation in normal kidney. The sFRP-5 expression was restored and the acetylation of H3 and H4 histones associated with the sFRP-5 promoter region were significantly increased after treatment with demethylation agent (5-Aza-dc) and histone deacetylase inhibitor (TSA). When RCC cells were transfected with the sFRP-5 gene, significant inhibition of anchorage independent colony formation and cell invasion were observed compared to controls. The sFRP-5 transfection also significantly induced apoptosis in RCC cells. In conclusion, this is the first report documenting that the sFRP-5 is downregulated by promoter methylation and histone acetylation and functions as a tumor suppressor gene by inducing apoptosis in RCC cells.
Keywords: RCC, sFRP-5, methylation, histone modification, tumor suppressor
Introduction
The Wnt signaling pathway plays an important role during animal development.1,2 The main feature of the Wnt signaling pathway is the regulation of the transcription factor β-catenin, which together with members of the T cell factor lymphoid enhance factor (TCF/LEF) family, promotes the transcription of several genes that result in proliferation and reduced apoptosis.3,4 Recently, the Wnt/ β-catenin pathway has been shown to be involved in tumorigenesis involving several cell types. Aberrant expression of Wnt signaling has been reported in various human cancers, including breast cancer, colorectal cancer, non-small cell lung cancer (NSCLC) and bladder cancer.5-8
Secreted frizzled-related proteins (sFRPs), a family of five secreted glycoproteins, have been identified as possible negative modulators of the Wnt signal transduction pathway.9 sFRPs block Wnt signalling either by interacting with Wnt proteins to prevent them from binding to Fz (Frizzled) proteins those are receptors for Wnt and positive mediators of the Wnt signaling pathway, or by forming non-functional complexes with Fz proteins.10,11
Recently, it has become clear that aberrant epigenetic silencing of tumor suppressor genes by promoter DNA hypermethylation and histone deacetylation plays an important role in the pathogenesis of human cancer.12 Many candidate tumor suppressor genes silenced by DNA methylation and/or histone modification also have been reported in RCC.13-15 In addition, members of the sFRP family such as sFRP-1 and sFRP-2 have been reported to be epigenetically silenced in RCC.16-19
Secreted frizzled-related protein-5 (sFRP-5) is also a member of the sFRP family and downregulation by promoter hypermethylation has been reported recently in various human malignancies including gastric, cervical, hepatocellular, pancreatic, oral squamous cell, breast, colon and bladder cancer.5,20-28 With regard to RCC, a previous report from our laboratory showed that the methylation frequency of Wnt antagonists in clinical samples could serve as biomarkers for RCC,29 however, there have been no other previous reports investigating the function of sFRP-5 in RCC and the involvement of histone modifications in sFRP-5 repression. Therefore, in the present study, we hypothesized that sFRP-5 is epigenetically downregulated and functions as a tumor suppressor gene in RCC. To address this issue, we examined sFRP-5 mRNA and protein expression levels in RCC cell lines and human RCC tissues. We also looked at the methylation status of the sFRP-5 promoter region by bisulfite sequencing and the histone acetylation by ChIP assay. In addition, we overexpressed sFRP-5 by transfection in a RCC cell line (A498) and studied its effect on colony formation, invasion and apoptosis.
Materials and methods
Immunohistochemistry
A tissue microarray which consisted of twelve ccRCCs and matched adjacent tissue 1.5 cm away from the carcinoma was obtained from US Biomax, Inc. (KD481; Rockville, MD). Detailed information on the tumor samples can be found at http://www.biomax.us/index.php. Immunostaining was done on the tissue microarray using the UltraVision Detection System (Thermo Scientific, Seattle, WA) following the manufacture's protocol. Antigen retrieval was carried out by microwaving the slide in 10 mmol/L sodium citrate buffer. The slide was incubated overnight with a 1:100 dilution of anti-sFRP-5 antibody (cat#; LS-B307, LifeSpan Biosciences, Inc., Minneapolis, MN). A pathologist not involved in the present study evaluated the immunostaining under blind conditions. Immunohistochemical staining was graded on an arbitrary scale from 0 to 2; 0 representing negative expression (0–20% positive cells), 1 representing weakly positive expression (20–50% positive cells) and 2 representing strongly positive expression (50–100% positive cells). The renal tubular cells in normal tissues and tumorous cells (not stromal cells) in cancer tissues were scored and the scale was determined according to the average number of positive cells in 10 random fields of the slide.19
Cell culture and drug treatment
We used four human kidney cancer cell lines, Caki1, Caki2, ACHN and A498 obtained from the American Type Culture Collection (ATCC, Manassas, VA). The Caki1, Caki2 and A498 cell lines were incubated in RPMI 1640 medium supplemented with 10% fetal bovine serum. ACHN cells were maintained in Eagle's minimal essential medium supplemented with 10% fetal bovine serum. These cells were maintained in a humidified incubator (5% CO2) at 37 °C. For 5-aza-2’-deoxycytidine (5-Aza-dc; Sigma-Aldrich, St. Louis, MO) treatment, cells were treated with 5-Aza-dc (5μM) for 4 days. Culture medium and 5-Aza-dc were replaced daily. For the combined treatment, 300 nM of trichostatin A (TSA; Millipore, Bedford, MA) was added during the last 24 hours. Total RNA and genomic DNA were isolated using the RNeasy Mini kit (Qiagen, Valencia, CA) and QIAamp DNA mini kit (Qiagen), respectively, following the manufacturer's directions.
Bisulfite DNA sequencing
Bisulfite modification of genomic DNA was done using the EpiTect Bisulfite kit (Qiagen) following the manufacturer's protocol. The Genomic DNA from adult human normal kidney tissue (cat#; D1234142-50, BioChain, Hayward, CA) was also modified and used as a control. Primers for bisulfite genomic sequencing PCR were designed by using the online program MethPrimer30 or by referring to a previous report.24 The primer sequences and PCR conditions are shown in Table 1. The fragment amplified corresponded to the sFRP-5 promoter region −404 to −128 (the ATG start codon of sFRP-5 was defined as +1). All reactions were done as described previously from our laboratory.19 The amplification products were confirmed by electrophoresis on a 2% agarose gel and sequenced directly by an outside vendor (McLab, South San Francisco, CA).
Table 1.
Primer sequences and PCR conditions
| Primer sequence (5′-3′) | Annealing temperature (°C), PCR cycles | Product size (bp) | Reference | |
|---|---|---|---|---|
| Primers for RT-PCR | ||||
| SFRP5-S | GGCTTGAGCAGCTTCTTCTT | 62, 30 | 395 | 24 |
| SFRP5-AS | CACTCGGATACGCAGGTCTT | |||
| GAPDH-S | CAATGACCCCTTCATTGACC | 58, 30 | 135 | 8 |
| GAPDH-AS | TGGAAGATGGTGATGGGATT | |||
| Primers for sFRP5 bisulfite sequencing | ||||
| PAN-S | GTTAAGGTAGAGAGATAAGTAAGTG | 50, 40 | 752 | We designed |
| PAN-AS | ACTCCAACAAATTAAACAACC | |||
| BSP-S | GGGAGGTAGGGAGTTTTGGGGAGAA | 55, 40 | 277 | 24 |
| BSP-AS | CCCAAATAAATAACAACCTACRCTAC | |||
| Primers for ChIP PCR | ||||
| SFRP5-S | GAGGCGCCAGGATCAGTC | 60, 35 | 100 | We designed |
| SFRP5-AS | CCGGCCCTGACTCTACCC | |||
| GAPDH-S | TACTAGCGGTTTTACGGGCG | 60, 35 | 166 | 19 |
| GAPDH-AS | TCGAACAGGAGGAGCAGAGAGCGA |
Semi-quantitative and real time RT-PCR
Reverse transcription reactions were carried out with 2 μg of total RNA using a Reverse Transcription System Kit (Promega, Madison, WI). Semi-quantitative RT-PCR was done using REDTaq (Sigma-Aldrich). The primer sequences and PCR conditions are shown in Table 1. Quantitative real-time RT-PCR analysis was performed in triplicate with an Applied Biosystems Prism 7500 Fast Sequence Detection System using TaqMan universal PCR master mix according to the manufacturer's specifications (Applied Biosystems Inc., Foster City, CA). The TaqMan primer IDs purchased from Applied Biosystems are as follows, sFRP-5; Hs00169366_ml and GAPDH; Hs99999905_m1. The total RNA from adult human normal kidney tissue (cat#; R1234142-50, BioChain) was used as a control. The thermal cycler conditions were as follows: hold for 20 sec at 95 °C, followed by two-step PCR for 40 cycles of 95 °C for 3 sec, followed by 60°C for 30 sec. Levels of RNA expression were determined using the 7500 Fast System SDS software version 1.3.1 (Applied Biosystems).
Chromatin Immunoprecipitation (ChIP) assay
ChIP assays were performed on cell line DNA using the EZ-ChIP Kit (Millipore, Billerica, MA) and followed the manufacturer's protocols. Chromatin was prepared from confluent cells and sonicated by using a Bioruptor (Diagenode, Sparta, NJ), then immunoprecipitated with each antibody. Antibodies for acetylated histone H3 (AcH3, 06-599, Millipore) and acetylated histone H4 (AcH4, 06-866, Millipore) were used in the immunoprecipitations. The immunoprecipitated DNA was eluted in a total volume of 200μl, and 5 μl of each DNA sample were analyzed by semi-quantitative PCR and quantitative real time PCR using SYBR® Green PCR Master Mix (Applied Biosystems). Semi-quantitative PCR was performed as described previously.19 The thermal cycler conditions for quantitative real time PCR were as follows: hold for 10 min at 95 °C, followed by three-step PCR for 40 cycles of 95 °C for 30 sec, 60°C for 30 sec and 72°C for 30 sec. Cycle threshold (CT) value was used for relative quantification of sFRP-5 expression. Fold changes relative to input ΔCT were calculated as 2−ΔΔCT.31 PCR primers were designed to amplify the corresponding promoter region of sFRP-5 (−180 to −81; the ATG start codon of sFRP-5 was defined as +1). The primer pairs used for ChIP assays and PCR conditions are shown in Table 1.
sFRP-5 transfection
Plasmids containing the human full-length cDNA fragment of sFRP-5 (sFRP-5; cat#RC208391) and empty vector (EV; cat#PS100001) were purchased from Origene (Rockville, MD). A498 cells (1 × 105 cells per well of 6-well plate) were transfected with either sFRP-5 or EV using FuGENE HD (Roche Applied science, Indianapolis, IN) according to the manufacturer's protocol. The transfected A498 cells were selected with 400μg/ml of G418 (Invitrogen, Carlsbad, CA). Cell lines stably expressing sFRP-5 (A498-sFRP-5) and transfected with empty vector (A498-EV) were maintained with 200μg/ml of G418.
Soft agar colony formation and invasion assay
Soft agar colony formation was done with EV-transfected stable A498 cells (A498-EV) and sFRP-5-transfected stable A498 cells (A498-sFRP-5) using a CytoSelect Cell Transformation Assay kit (Cell Biolabs, San Diego, CA) according to the manufacturer's protocol. Cells were incubated 7 days in semisolid agar media before being solubilized and detected by using the provided MTT solution in a microplate reader (Spectra MAX 190; Molecular Devices Co., Sunnyvale, CA) at OD570nm. Data are the mean ± S.D. of 6 independent experiments. Invasion assay was conducted with A498-EV and A498-sFRP-5 using the BioCoat Matrigel Invasion Chambers (BD Biosciences, Bedford, MA). Cells were incubated for 22 h at 37°C in a 5% CO2 atmosphere. Those cells that migrated through the membrane to the lower surface were stained with Hema-3 (Fisher Scientific, Middletown, VA) and counted with a microscope. Four random fields were chosen for each membrane, and the results were shown as migrated cells per field.
Apoptosis analysis
A498 cells transiently transfected with EV or sFRP-5 were harvested 72 hours after transfection by trypsinization and stained using an ANNEXIN V-FITC /7-AAD KIT (Beckman Caulter, Inc. Fullerton, CA) according to the manufacturer's protocol and immediately analyzed by flow cytometry (Cell Lab Quanta SC; Beckman Coulter, Inc.). Experiments were done in triplicate.
Statistical analysis
Data are shown as mean values ± standard deviation (SD). The Student's t-test was used to compare the two different groups. P values of less than 0.05 were regarded as statistically significant. All statistical analyses were performed using StatView version 5.0 for Windows.
Results
Immunostaining of sFRP-5 in normal human kidney and kidney cancer specimens
To examine the expression levels of sFRP-5 in malignant and normal kidney tissues, immunostaining was performed on a tissue microarray using antibody to sFRP-5. The macroscopic appearance of the tissue microarray and typical immunostaining of sFRP-5 in normal and ccRCC specimens (x40 and x200) are shown in Fig.1A. The staining was much stronger in normal specimens than in ccRCC. The mean expression score (1.864±0.351) in normal specimens (N=22) was significantly higher compared to that (1.000±0.590) in ccRCC specimens (N=24) (p<0.0001; Fig.1b). This result suggests that sFRP-5 protein expression is downregulated in human ccRCC.
FIGURE 1.
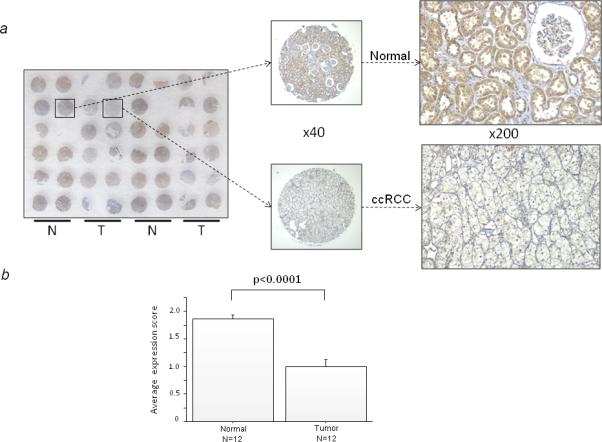
sFRP-5 immunohistochemistry in normal human kidney and ccRCC tissues. (a) Macroscopic appearance of the sFRP-5-stained tissue microarray is shown with typical immunostaining of sFRP-5 in normal kidney tissue and in ccRCC tissue (x40 and x200). N and T indicate normal and tumor, respectively. (b) Mean ± SD in immunostaining score between normal tissue and ccRCC tissue. Expression score was defined as described in Materials and Methods.
Methylation status of the sFRP-5 promoter region
The methylation status at 17 CpG sites of the sFRP-5 CpG island was characterized in four RCC cell lines and normal kidney DNA by bisulfite genomic sequencing (Fig. 2a, b and c). These analyses indicated that the CpG islands in four RCC cell lines showed essentially the same methylation status. The promoter regions checked in these RCC cell lines were densely methylated whereas the same region in normal kidney was mostly unmethylated.
FIGURE 2.
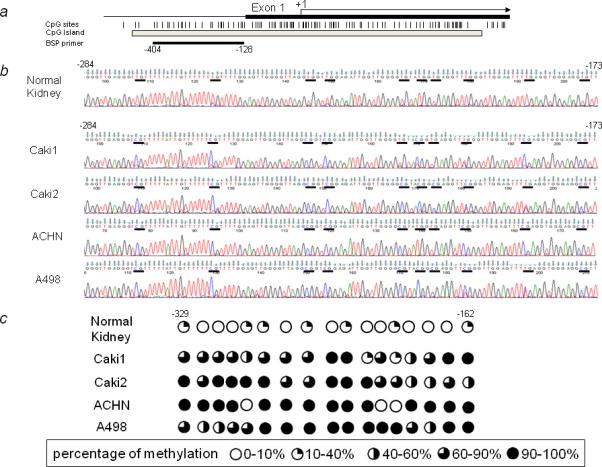
Methylation of the human sFRP-5 gene promoter. (a) Schematic representation of the promoter region of the human sFRP-5 gene and primer location. The vertical lines represent the location of CpG dinucleotides, and the gray box shows the CpG island. The black box indicates the first exon, and the arrow indicates the approximate position of the translation start site. The black horizontal line under the gray box indicates the region examined by bisulfite DNA sequencing. (b) Representative results of bisulfite sequencing for normal kidney and four RCC cell lines (from −284 to −173). The horizontal bars indicate CpG sites. (c) Methylation mapping of 17 CpG sites of the sFRP-5 promoter region obtained from bisulfite sequencing in normal kidney and RCC cell lines (from −329 to −162). Percentage of methylation was classified as indicated.
Downregulation of sFRP-5 mRNA expression and restoration by 5-Aza-dc and TSA treatment
We checked sFRP-5 mRNA expression in RCC cell lines and in human normal kidney RNA by real time RT-PCR. As shown in Figure 3a, sFRP-5 mRNA expression was significantly downregulated in RCC cell lines compared to human normal kidney RNA (p<0.01). We also examined the changes in sFRP-5 expression after treatment of cells with 5-Aza-dc, an inhibitor of DNA methyltransferases (DNMTs) and TSA, a histone deacetylase (HDAC) inhibitor. After the combined treatment with 5-Aza-dc and TSA, sFRP-5 expression was restored in all RCC cell lines tested (Fig. 3b). Treatment using only 5-Aza-dc also restored sFRP-5 mRNA expression indicating that downregulation of sFRP-5 expression in RCC cell lines correlates with CpG methylation of the sFRP-5 promoter.
FIGURE 3.
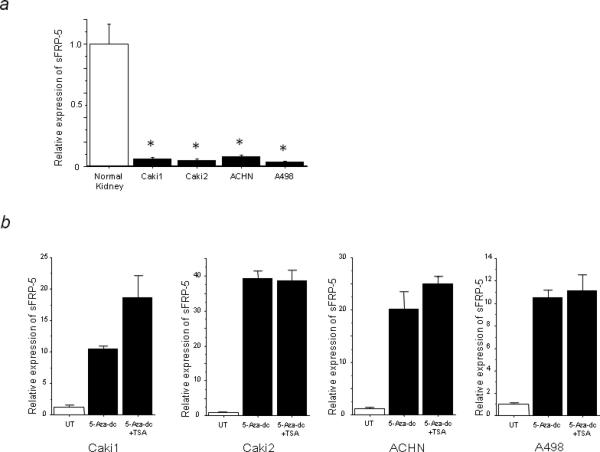
Downregulation of sFRP-5 mRNA expression and restoration by combined 5-Aza-dc and TSA treatment. (a) sFRP-5 mRNA expression in RCC cell lines is significantly downregulated compared to human normal kidney RNA (* p<0.01). (b) 5-Aza-dc and combined (5-Aza-dc and TSA) treatment restored sFRP-5 mRNA expression in RCC cell lines. UT indicates untreated control.
Histone acetylations associated with the sFRP-5 promoter region
To further investigate whether the transcriptional upregulation of sFRP-5 was associated with alternations in histone acetylation status at the promoter region, we examined local histone acetylation in the chromatin associated with the sFRP-5 promoter region using AcH3 and AcH4 antibodies by ChIP assay. We found that 5-Aza-dc and TSA treatment significantly increased the state of acetylation of H3 and H4 histones associated with an open, transcriptionally active regulatory region in the sFRP-5 gene in all cell lines (Fig.4a and b). There was no PCR amplification in control immunoprecipitations without antibody (data not shown).
FIGURE 4.
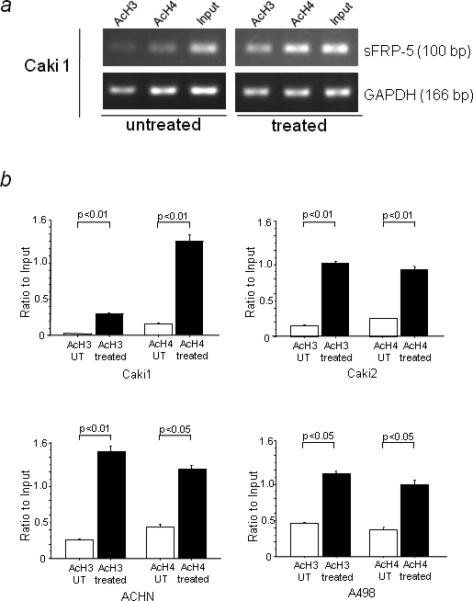
Induction of histone acetylation at the sFRP-5 promoter by 5-Aza-dc and TSA. (a) Representative ChIP-PCR results with Caki1 cells. DNA fragments corresponding to the sFRP-5 promoter region were amplified by semi-quantitative PCR. (b) Quantitative real time PCR results. Fold changes relative to input ΔCT were calculated as 2−ΔΔCT. UT indicates untreated control.
Suppression of anchorage-independent colony formation and cell invasion by sFRP-5 transfection
Next, we examined the effect of sFRP-5 overexpression in A498 cells using soft agar colony formation and cell invasion assays. As shown in Figure 5a(1) and (2) left, sFRP-5 transcription was significantly increased at 72 hrs after transient transfection with sFRP-5 but not in untreated (UT) cells or those treated with empty vector (EV) (p<0.01). Stable cell line A498-sFRP5 also had significantly higher levels of sFRP-5 transcription compared to A498-EV (p<0.01, Fig. 5a (2) right) and we used these stable cell lines for soft agar colony formation and invasion assays. In soft agar colony formation assay, we found a significant reduction of anchorage independent colony formation in A498-sFRP-5 compared to A498-EV (p<0.05, Fig. 5b). As shown in Figure 5c, the number of invasive cells of A498-sFRP-5 (89.67±4.33) was also significantly decreased compared to that of A498-EV (62.33±3.06; p<0.05). These results suggest that sFRP-5 expression reduced anchorage independent colony formation efficacy and the invasive ability of A498 cells.
FIGURE 5.
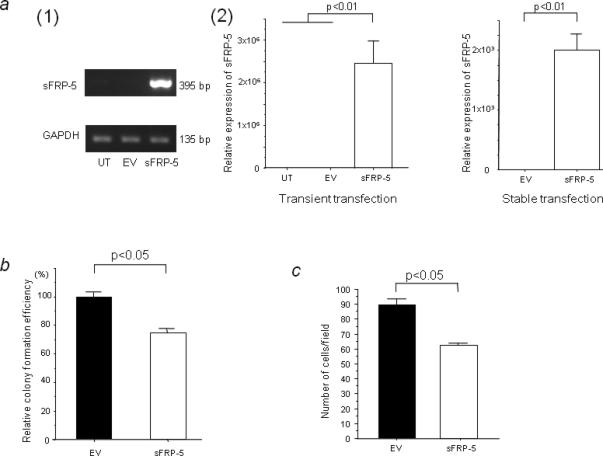
The effect of sFRP-5 overexpression on A498 cells. (a) (1) Semi-quantitative RT-PCR for sFRP-5. A498 cells were transiently transfected with empty vector (EV) or with sFRP-5. GAPDH was used as a control for RNA quality and loading. (2) Real-time RT-PCR for sFRP-5 transient (left) and stable transfection (right). Levels of sFRP-5 mRNA expression were quantified by real-time RT-PCR. (b) Effect of sFRP-5 transfection on A498 colony formation efficiency in soft agar colony formation assay using stably transfected cells (A498-EV and A498-sFRP-5). The colony formation efficiency of A498-sFRP-5 was calculated assuming that the colony formation ability of A498-EV was 100% using the absorbance obtained from MTT assay. (c) Invasion assay using the same cells in (b). The data expressed in the graph is the mean values ± SD of three independent experiments.
sFRP-5 transfection induces apoptosis
We also evaluated apoptosis by using flow cytometry with A498 cells which were transiently transfected with EV or sFRP-5. As shown in Figure 6a, the apoptotic cell fraction was 6.17% in EV-transfected A498 cells and 16.45% in those transfected with sFRP-5 at 72 hours after transfection. Thus sFRP-5 transient transfection significantly induced apoptosis in A498 cells (p<0.05; Fig. 6b).
FIGURE 6.
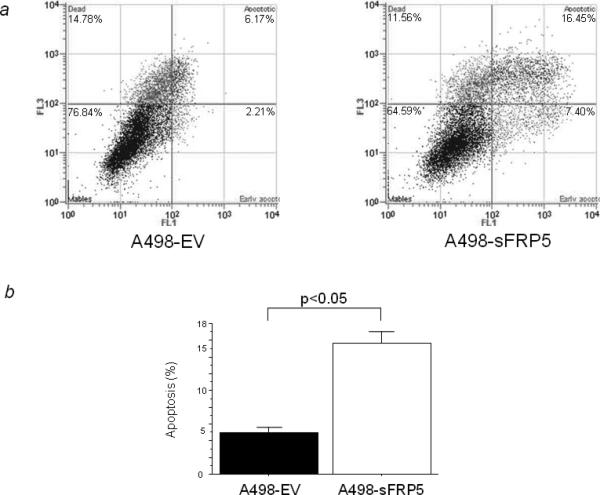
Promotion of apoptosis by sFRP-5 transfection. (a) Effect of sFRP-5 transfection on apoptosis in A498 cells. AnnexinV-FITC/7-AAD staining discriminates between cells in early (lower right quadrant) and advanced apoptotic states (upper right quadrant). Viable cells are double negative (lower left quadrant). The representative apoptotic cell fractions in EV or sFRP-5-transfected A498 cells are shown. (b) Proportion of apoptotic cells. The data expressed is the percentage of apoptotic cells out of the total measured cell population of A498 cells.
Discussion
RCC is the third most common genitourinary cancer, with 54,390 predominantly male cases and 13,010 deaths in the United States in 2008.32 Recently, the Wnt signaling pathway has been shown to be involved in tumorigenesis involving several cell types.5-8 The sFRPs have been identified as possible antagosists of the Wnt signaling pathway9 and sFRP-5, a member of the sFRP family, has been reported to be downregulated by promoter hypermethylation in various human cancers.5, 20, 22-27
In this study we found that sFRP-5 was significantly downregulated at the protein level in human ccRCC tissue microarrays and at the mRNA level in human RCC cell lines. Bisulfite sequencing of the sFRP-5 promoter region showed it to be densely methylated in RCC cell lines whereas there was few methylation in normal human kidney DNA. In addition, treatment with demethylation agent (5-Aza-dc) and histone deacetylase inhibitor (TSA) restored sFRP-5 mRNA expression in RCC cell lines. Our results indicate that downregulation of sFRP-5 expression in RCC cell lines and human cancer tissues correlate with CpG methylation of the sFRP-5 promoter. These results are consistent with studies in other types of cancer mentioned above. However, these previous reports did not examined histone modifications in the sFRP-5 gene. Several tumor suppressor genes are known to be repressed even though they have hypomethylated promoters.33, 34 This indicates that although DNA methylation is a major epigenetic mechanism for gene silencing, there are other epigenetic silencing pathways independent of DNA methylation.35 It is generally known that aberrant DNA methylation and histone modifications work together to silence many tumor suppressor genes in human cancer.36,37
Therefore, we examined the effect of a DNA methyltransferase inhibitor (5-Aza-dc) and a histone deacetylase inhibitor (TSA) on histone acetylation (AcH3, AcH4) associated with the sFRP-5 promoter region. We found that 5-Aza-dc and TSA treatment altered the state of chromatin modification by increasing the acetylation of histones associated with sFRP-5 genes. These data suggest that the induction of sFRP-5 gene is at least partially mediated by changes in histone acetylation. Taken together, the sFRP-5 gene may be downregulated by epigenetic events including DNA methylation and histone modification.
Using A498 cells transfected with a sFRP-5 expression plasmid, we also demonstrated that re-expression of sFRP-5 significantly inhibited anchorage independent colony formation and cell invasion. These results indicate that sFRP-5 functions as a tumor suppressor in RCC cells. Recent studies have revealed that sFRPs are tumor suppressor candidates.5,14,17,38 The expression of sFRPs is frequently silenced by promoter hypermethylation in a variety of cancers. Restored expression of sFRPs has been shown to inhibit cell growth in vitro and tumor growth in vivo.38 For instance, Gumz et al. reported that stable reexpression of sFRP-1 in RCC cells resulted in decreased expression of Wnt target genes, decreased growth in cell culture, inhibition of anchorage-independent growth, and decreased tumor growth in athymic nude mice.17 As for sFRP-5, Zhao et al. have shown that the expression of sFRP-5 was inversely correlated with the expression of MMP-7 and MT1-MMP, well known as positive regulators of invasion and metastasis, in gastric cancer cells.20 Also, Lin et al. have found that the restoration of sFRP-5 suppressed colony formation, invasive ability and inhibited expression of Wnt/ β-catenin downstream genes such as c-myc and cyclin D1 in cervical cancer cells.21 Thus our current studies are in agreement with these previous reports.
In the present study we have also shown that restoration of sFRP-5 expression significantly induces apoptosis in RCC cells. Targeted inhibition of the Wnt signaling pathway by sFRP-5 transfection has been shown in other cancers.23,27 Positive Wnt signaling has been reported to have anti-apoptotic activity in several cell types39,40 and our results and others suggest that sFRP-5 restoration leads to increased apoptosis in several types of cancer cells.
Recently, there is increasing evidence showing that some sFRPs have oncogenic functions. Under certain circumstances, some sFRPs are overexpressed in cancer41 and have growth promoting42 or anti-apoptotic effects.43 The apparently contradictory roles of sFRPs in these studies may be due to the different Wnt ligands present in different cells, biphasic responses to different concentrations of sFRPs, and the binding affinities and specificities of different sFRPs for Wnts.10 Further work will be needed to have a better understanding of the specific relationship between sFRPs including sFRP-5 and Wnt signaling.
In conclusion, we have shown for the first time that sFRP-5 is frequently downregulated in RCC cell lines and clinical ccRCC tissues. Our results suggest that the downregulation is caused by a combination of DNA methylation and histone modification. In addition, sFRP-5 overexpression inhibited colony formation and cell invasion and induced apoptosis in RCC cells. These results suggest that sFRP-5 functions as a tumor suppressor gene in RCC and that restoration of sFRP-5 may offer a new therapeutic approach for the treatment of RCC patients.
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript. This study was supported by Grants RO1CA130860, and T32DK007790 from the NIH, VA REAP award, and Merit Review grants (PI: R.D.).
References
- 1.Guillén-Ahlers H. Wnt signaling in renal cancer. Curr Drug Targets. 2008;9:591–600. doi: 10.2174/138945008784911813. [DOI] [PubMed] [Google Scholar]
- 2.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 3.Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–42. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Guttridge DC, You Z, Zhang Z, Fribley A, Mayo MW, Kitajewski J, Wang CY. Wnt-1 signaling inhibits apoptosis by activating beta-catenin/T cell factor-mediated transcription. J Cell Biol. 2001;152:87–96. doi: 10.1083/jcb.152.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, Knüchel R, Dahl E. Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29:991–8. doi: 10.1093/carcin/bgn076. [DOI] [PubMed] [Google Scholar]
- 6.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 7.Uematsu K, He B, You L, Xu Z, McCormick F, Jablons DM. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene. 2003;22:7218–21. doi: 10.1038/sj.onc.1206817. [DOI] [PubMed] [Google Scholar]
- 8.Urakami S, Shiina H, Enokida H, Kawakami T, Tokizane T, Ogishima T, Tanaka Y, Li LC, Ribeiro-Filho LA, Terashima M, Kikuno N, Adachi H, et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin Cancer Res. 2006;12:383–91. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- 9.Heller RS, Dichmann DS, Jensen J, Miller C, Wong G, Madsen OD, Serup P. Expression patterns of Wnts, Frizzleds, sFRPs, and misexpression in transgenic mice suggesting a role for Wnts in pancreas and foregut pattern formation. Dev Dyn. 2002;225:260–70. doi: 10.1002/dvdy.10157. [DOI] [PubMed] [Google Scholar]
- 10.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–34. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 11.Huang HC, Klein PS. The Frizzled family: receptors for multiple signal transduction pathways. Genome Biol. 2004;5:234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellebrekers DM, Griffioen AW, van Engeland M. Dual targeting of epigenetic therapy in cancer. Biochim Biophys Acta. 2007;1775:76–91. doi: 10.1016/j.bbcan.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, Saini S, Tanaka Y, Dahiya AV, Khatri G, Dahiya R. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–70. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris MR, Gentle D, Abdulrahman M, et al. Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. Br J Cancer. 2008;98:496–501. doi: 10.1038/sj.bjc.6604180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reu FJ, Leaman DW, Maitra RR, Bae SI, Cherkassky L, Fox MW, Rempinski DR, Beaulieu N, MacLeod AR, Borden EC. Expression of RASSF1A, an epigenetically silenced tumor suppressor, overcomes resistance to apoptosis induction by interferons. Cancer Res. 2006;66:2785–93. doi: 10.1158/0008-5472.CAN-05-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awakura Y, Nakamura E, Ito N, Kamoto T, Ogawa O. Methylation-associated silencing of SFRP1 in renal cell carcinoma. Oncol Rep. 2008;20:1257–63. [PubMed] [Google Scholar]
- 17.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, Sun LZ, Ahlquist DA, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–9. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 18.Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, Knüchel R, Klopocki E, Sauter G, Simon R, Wieland WF, Walter B, et al. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26:5680–91. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- 19.Kawamoto K, Hirata H, Kikuno N, Tanaka Y, Nakagawa M, Dahiya R. DNA methylation and histone modifications cause silencing of Wnt antagonist gene in human renal cell carcinoma cell lines. Int J Cancer. 2008;123:535–42. doi: 10.1002/ijc.23514. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C, Bu X, Zhang N, Wang W. Downregulation of SFRP5 expression and its inverse correlation with those of MMP-7 and MT1-MMP in gastric cancer. BMC Cancer. 2009;9:224. doi: 10.1186/1471-2407-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YW, Chung MT, Lai HC, De Yan M, Shih YL, Chang CC, Yu MH. Methylation analysis of SFRP genes family in cervical adenocarcinoma. J Cancer Res Clin Oncol. 2009;135:1665–74. doi: 10.1007/s00432-009-0613-5. [DOI] [PubMed] [Google Scholar]
- 22.Chung MT, Sytwu HK, Yan MD, Shih YL, Chang CC, Yu MH, Chu TY, Lai HC, Lin YW. Promoter methylation of SFRPs gene family in cervical cancer. Gynecol Oncol. 2009;112:301–6. doi: 10.1016/j.ygyno.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, Shinomura Y. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–89. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- 24.Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, Burgart LJ, Ahlquist DA. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett's esophagus. Int J Cancer. 2005;116:584–91. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- 25.Sogabe Y, Suzuki H, Toyota M, Ogi K, Imai T, Nojima M, Sasaki Y, Hiratsuka H, Tokino T. Epigenetic inactivation of SFRP genes in oral squamous cell carcinoma. Int J Oncol. 2008;32:1253–61. doi: 10.3892/ijo_32_6_1253. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, Mori M, Hirata K, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–56. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 28.Urakami S, Shiina H, Enokida H, Kawakami T, Kawamoto K, Hirata H, Tanaka Y, Kikuno N, Nakagawa M, Igawa M, Dahiya R. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12:2109–16. doi: 10.1158/1078-0432.CCR-05-2468. [DOI] [PubMed] [Google Scholar]
- 29.Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, Igawa M, Dahiya R. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–97. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- 30.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 31.Gavin DP, Kartan S, Chase K, Jayaraman S, Sharma RP. Histone deacetylase inhibitors and candidate gene expression: An in vivo and in vitro approach to studying chromatin remodeling in a clinical population. J Psychiatr Res. 2009;43:870–6. doi: 10.1016/j.jpsychires.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Garcia JA, Cowey CL, Godley PA. Renal cell carcinoma. Curr Opin Oncol. 2009;21:266–71. doi: 10.1097/CCO.0b013e32832a05c8. [DOI] [PubMed] [Google Scholar]
- 33.Zhu WG, Dai Z, Ding H, Srinivasan K, Hall J, Duan W, Villalona-Calero MA, Plass C, Otterson GA. Increased expression of unmethylated CDKN2D by 5-aza-2'-deoxycytidine in human lung cancer cells. Oncogene. 2001;20:7787–96. doi: 10.1038/sj.onc.1204970. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawachi T, Soejima H, Urano T, Zhao W, Higashimoto K, Satoh Y, Matsukura S, Kudo S, Kitajima Y, Harada H, Furukawa K, Matsuzaki H, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22:8835–44. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Joh K, Yatsuki H, Zhao W, Soejima H, Higashimoto K, Noguchi M, Yokoyama M, Iwasaka T, Mukai T. Retinoic acid receptor beta2 is epigenetically silenced either by DNA methylation or repressive histone modifications at the promoter in cervical cancer cells. Cancer Lett. 2007;247:318–27. doi: 10.1016/j.canlet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Baylin SB, Höppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986;46:2917–22. [PubMed] [Google Scholar]
- 37.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–15. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, He B, You L, Jablons DM. Roles of secreted frizzled-related proteins in cancer. Acta Pharmacol Sin. 2007;28:1499–504. doi: 10.1111/j.1745-7254.2007.00692.x. [DOI] [PubMed] [Google Scholar]
- 39.Dehner M, Hadjihannas M, Weiske J, Huber O, Behrens J. Wnt signaling inhibits Forkhead box O3a-induced transcription and apoptosis through up-regulation of serum- and glucocorticoid-inducible kinase 1. J Biol Chem. 2008;283:19201–10. doi: 10.1074/jbc.M710366200. [DOI] [PubMed] [Google Scholar]
- 40.Tapia JC, Torres VA, Rodriguez DA, Leyton L, Quest AF. Casein kinase 2 (CK2) increases survivin expression via enhanced beta-catenin-T cell factor/lymphoid enhancer binding factor-dependent transcription. Proc Natl Acad Sci U S A. 2006;103:15079–84. doi: 10.1073/pnas.0606845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Han Q, Zhao W, Bentel J, Shearwood AM, Zeps N, Joseph D, Iacopetta B, Dharmarajan A. Expression of sFRP-4 and beta-catenin in human colorectal carcinoma. Cancer Lett. 2006;231:129–37. doi: 10.1016/j.canlet.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 42.Roth W, Wild-Bode C, Platten M, Grimmel C, Melkonyan HS, Dichgans J, Weller M. Secreted Frizzled-related proteins inhibit motility and promote growth of human malignant glioma cells. Oncogene. 2000;19:4210–20. doi: 10.1038/sj.onc.1203783. [DOI] [PubMed] [Google Scholar]
- 43.Han X, Amar S. Secreted frizzled-related protein 1 (SFRP1) protects fibroblasts from ceramide-induced apoptosis. J Biol Chem. 2004;279:2832–40. doi: 10.1074/jbc.M308102200. [DOI] [PubMed] [Google Scholar]


