Abstract
The purpose of this study was to identify new tumor suppressor microRNAs (miRs) in bladder cancer, carry out functional analysis of their suppressive role and identify their specific target genes. To explore tumor suppressor miRs in bladder cancer, miR microarray was performed using SV-HUC-1, T24, J82 and TCCSUP cells. Expression of miR-493 in bladder cancer (T24, J82 and TCCSUP) cells was down-regulated compared to normal SV-HUC-1 cells. Also the expression of miR-493 was significantly lower in bladder cancer tissues than in their corresponding non-cancerous tissues. Transfection of miR-493 into T24 or J82 cells decreased their cell growth and migration abilities. On the basis of this result, to identify potential miR-493 target genes, we used target scan algorithms to identify target oncogenes related to invasion and migration. MiR-493 decreased 3′UTR luciferase activity and protein expression of FZD4 and RhoC. MiR-493 also decreased binding of RhoC and Rock-1. MiR-493 is a new tumor suppressor microRNA in bladder cancer and inhibits cell motility through down-regulation of RhoC and FZD4.
Keywords: urothelial carcinoma, miR-493, RhoC, FZD4
INTRODUCTION
Bladder cancer is the 9th leading cause of death among men, accounting for 3 % of total cancer (1). The most common histological type of bladder cancer is urothelial carcinoma (UC), which was formerly known as transitional cell carcinoma (TCC) (2). Approximately 75 % of patients are “non-muscle invasive UC (pTa, pTis, pT1) and have a 5-year survival rate of between 88–98% (3). The common treatment for these patients is endoscopic resection (2, 4). Patients with muscle invasive UC are usually treated with radical cystectomy or chemoradiotherapy (2, 5). However half of muscle invasive UC patients develop subsequent metastatic disease after the first aggressive treatment (2, 6). Previous studies have identified several potential molecular biomarkers for bladder cancer (7, 8). Namely inactivation of tumor suppressor genes TP53 and Rb and Ras oncogene activation have been regarded as important key players in bladder cancer carcinogenesis (7).
MicroRNAs (miRs) are well known as examples of non-coding RNAs (9) and human miRNAs nownumber 1,100 based on microRNA.org (http://www.microrna.org/microrna/home.do). MiRNAs bind to the 3′UTR of target gene mRNA and repress translation or induce mRNA cleavage (10) thereby inhibiting translation from mRNA to protein. Aberrant expression of miRNAs occurs in bladder cancer. Decreased expression of tumor suppressor microRNAs result in increased expression of target oncogenes. In contrast, increased expression of oncogenic microRNAs leads to loss or decreased expression of target tumor suppressor genes. According to previous reports, a number of microRNA microarray studies have been done in bladder cancer patient samples and the expression level of several microRNAs (miR-17-5p, miR-23a, miR-23b, miR-26b, miR-103-1, miR-185, miR-203, miR-205, miR-221 and miR-223) were up-regulated in bladder cancer tissues (11). In contrast some miRNAs (miR-30-3p, miR-125, miR-133a, miR-145, miR-195 and miR-199a*) were down down-regulated in bladder cancer compared to normal bladder tissues (12). MicroRNA expression level in paired primary and metastatic bladder cancers was validated using real-time PCR. The data show that the expression levels of several microRNAs (miR-10b, miR-29a, miR-29b, miR-126, miR-142-5p, miR-146a, miR-146b-5p, miR-150, miR-155 and miR-342-3p) were up-regulated in metastatic bladder cancer tissues and some miRNAs (miR-143, miR-145 and miR-320) were down down-regulated (13). MiR-129 was found to be up-regulated in progressive bladder tumor and significantly associated with short survival (14). MiR-125b was down-regulated in bladder tumor tissues compared to normal bladder tissues and over-expression of miR-125b inhibited cell growth in bladder cancer cell lines (15). However there have been few reports regarding the detailed functional analysis of these miRNAs in bladder cancer.
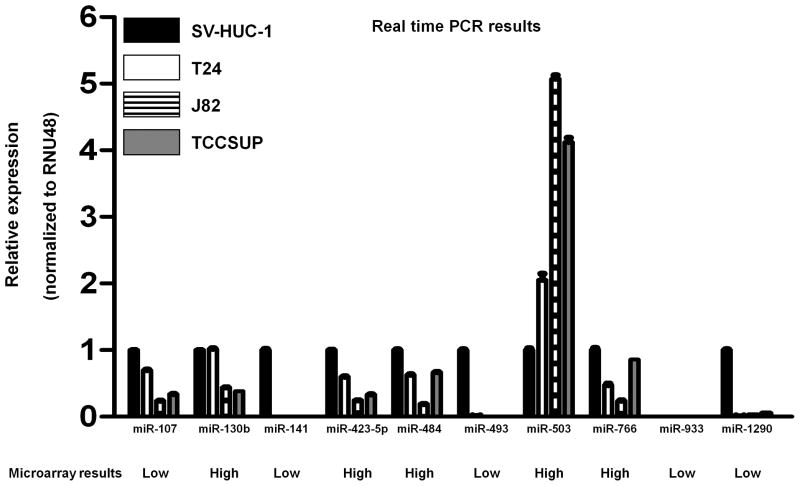
The aim of this study was to identify new tumor suppressor microRNAs that influence bladder cancer progression. Initially we performed microRNA microarray analysis to screen microRNAs related to bladder cancer using normal bladder cells (SV-HUC-1) and three bladder cancer cell lines (J82, T24, TCCSUP). We identified 10 microRNAs whose expression level in bladder cancer cell lines was significantly higher or significantly lower compared to normal bladder cell line, SV-HUC-1. Next we checked the microarray results by real-time PCR. Among 10 microRNAs, the expression of miR-493, miR-141 and miR-1290 was lower in bladder cancer cell lines and these results were consistent with the microarray data.
Based on microarray and real time PCR results, we hypothesized that miR-493 may be a potential tumor suppressive microRNA in bladder cancer and found that miR-493 expression was significantly lower in bladder cancer tissues. Thus we performed functional assays using miR-493. Transfection of miR-493 into bladder cancer cells decreased cell growth, invasion and migration. We also looked at potential target genes of microRNA-493 focusing on invasion and migration related ones. We initially used a target scan algorithm (microRNA.org) to identify genes RhoC and FZD4 as targets of miR-493 and validated the results with a second target scan algorithm (TargetScan). We also performed 3′UTR luciferase assays and Western analysis to look at target gene protein expression in miR-493 transfected bladder cancer cells. Finally, we knocked down FZD4 and RhoC mRNAs using a si-RNA technique to examine and confirm the mechanism of miR-493 tumor suppressive function.
MATERIALS AND METHODS
Cell lines and cell cultures
SV-HUC-1, T24, J82 and TCCSUP cells were purchased on February 2nd, 2010 from the ATCC (Manassas, VA, USA). No authentication was done by the authors. T24, J82 and TCCSUP cells originating from transitional cell carcinoma were selected as model transitional cell carcinoma cells. SV-HUC-1 cells derived from normal uroepithelium were used as control cells. SV-HUC-1 cells were cultured in F-12K Medium (ATCC) with 10% fetal bovine serum. T24 cells were cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum. J82 cells were cultured in MEM medium supplemented with 10% fetal bovine serum. TCCSUP cells were cultured in MEM medium supplemented with 10% fetal bovine serum, non-essential amino acids and 1 mM sodium pyruvate.
RNA extraction
MicroRNA and total RNA were extracted from cell lines using a miRNeasy Mini Kit and a RNeasy Mini Kit (Qiagen, Valencia, CA, USA). MicroRNAs from clinical samples were extracted using laser-capture-micro-dissection techniques with a miRNeasy FFPE Kit (Qiagen).
MicroRNA microarray
For microRNA microarray, total RNA was extracted from SV-HUC1, T24, J82 and TCCSUP cells using a miRNeasy Mini Kit. MicroRNA microarray analysis was carried out and analyzed by Phalanx Bio Inc. (Palo Alto, CA, USA). We selected 10 microRNAs whose expression level in bladder cancer cell lines was significantly higher (top 5 microRNAs; miR-130b, miR-423-5p, miR-484, miR-503, miR-766) or significantly lower (top5 microRNAs; miR-107, miR-141, miR-493, miR-933, miR-1290) compared to normal bladder SV-HUC-1 cells (Table S1).
Tissue array samples
A human bladder cancer tissue array was purchased from US Biomax (catalog#: BL801, US Biomax, Inc, Rockville, MD) to detect miR-493 localization and confirm miR-493 expression levels in bladder cancer tissues by in situ hybridization. Tissue array patient information is shown in Table S2.
Clinical Samples
Twenty-three normal and sixteen transitional cell carcinoma tissues in paraffin blocks were obtained from the Pathology Department of the Veterans Affairs Medical Center at San Francisco. Informed consent was obtained from patients from the Veterans Affairs Medical Center at San Francisco.
Transfection
Pre-miR™ miRNA precursor (Negative Control/hsa-miR-493, Ambion), siRNA (control/RhoC/FZD4, Applied Biosystems, Invitorgen), Mock (without oligo-nucleotide) and co-transfection of Pre-miR™ miRNA Precursor/ pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) were transiently transfected into cells by Lipofectamine 2000 (Invitorgen). Anti-miR miRNA Inhibitors (negative control #1/has-miR-493, Applied Biosystems) were transiently transfected into cells by siPORT NeoFX Transfection Agent (Ambion).
Cell viability assay
Viability of T24 and J82 cells was measured by the MTS (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega, Madison, WI, USA) assay 4 days after transfection of Pre-miR™ miRNA Precursor. Cell viability was determined by absorbance measurements at 490 nm using SpectraMAX 190 (Molecular Devices, Sunnyvale, CA, USA). Data are presented as the mean value ± SD for triplicate experiments and compared to the level of miR-493/siRNA obtained in Mock or Control microRNA/siRNA transfected cells that is normalized to 100%.
Migration assay
Transwell membrane filter inserts (8.0 mm pore size; BD Biosciences) were set in 24-well plate. T24 and J82 cells transfected with Pre-miR™ miRNA Precursor or siRNA were harvested 72 hr after transfection and re-suspended in serum-free MEM medium. Aliquots (5×104 cells / 100 μl) of the prepared cell suspension were added into the upper chamber and the lower chamber was filled with 1 ml of media containing 10% FBS. Cells were incubated for 4 hr at 37°C in a 5% CO2 tissue culture incubator. After 4 hr, no migrated cells were removed from transwell membrane filter inserts using cotton-tipped swab, migrated cells were stained with Hema 3 STAIN SET (Fisher Scientific, Pittsburgh, PA, USA). Cells per three random fields of each membranes were counted with Nikon ECLIPSE TS100 (Nikon, Tokyo, Japan) at 100x magnification. Data are presented as the mean value ± SD for triplicate experiments and compared to the level of miR-493/siRNA obtained in Mock or Control microRNA/siRNA transfected cells that is normalized to 100%.
Wound healing assay
T24 and J82 cells were seeded to 6-well plates and transfected with Pre-miR™ miRNA Precursor or control. At 24 hr after tranfection, cells were transferred from 6-well plates to 12-well plates. After 48 hr, a wound was formed by scraping the cells with a 200 μl pipette tip and washed twice with medium. We observed cells at 0, 24 and 72 hr after scraping and photographed the cells with a microscope (Nikon, Tokyo, Japan). Data are presented as the mean value ± SD for experiments and compared to the level of miR-493/siRNA obtained in Mock or Control microRNA/siRNA transfected cells that is normalized to 100%.
Luciferase Reporter Assay
A pmirGLO Dual-Luciferase miRNA Target Expression Vector was used for 3′UTR luciferase assays (Promega, Madison, WI, USA). The target oncogenes of tumor suppressor miRNA-493 were selected based on target scan algorithms [microRNA.org (http://www.microrna.org/microrna/home.do) and TargetScan (http://www.targetscan.org/)]. The primer sequences used were as follows: RhoC forward primer, 5′ AAACTAGCGGCCGCTAGTtcTTGCCCCCTTTGACCTTCcT 3′; RhoC reverse primer, 5′ CTAGAgGAAGGTCAAAGGGGGCAAgaACTAGCGGCCGCTAGTTT 3′; FZD4 forward primer, 5′ AAACTAGCGGCCGCTAGTtcaGTTACCAGTGACCTTCaT 3′; FZD4 reverse primer, 5′ CTAGAtGAAGGTCACTGGTAACtgaACTAGCGGCCGCTAGTTT 3′; mutant RhoC forward primer, 5′ AAACTAGCGGCCGCTAGTtcCTTGGCGCTCTTCGGCCGcT 3′; mutant RhoC reverse primer, 5 ′ CTAGAgCGGCCGAAGAGCGCCAAGgaACTAGCGGCCGCTAGTTT 3′; mutant FZD4 forward primer, 5′ AAACTAGCGGCCGCTAGTtcaTTTCGGCTCTCGGCCGaT 3′; mutant FZD4 reverse primer, 5 ′ CTAGAtCGGCCGAGAGCCGAAAtgaACTAGCGGCCGCTAGTTT 3′. Bold shows PmeI (AAAC/GTTT) and XbaI (T/CTAGA) sites. Underline shows NotI internal site. Italic shows target sequence. Plasmids for 3′UTR luciferase assays were made as described previously (16). For 3′UTR lucifease assay, T24 cells were co-transfected with hsa-miR-493 and pmirGLO Dual-Luciferase miRNA Target Expression Vectors with wild type or mutant target sequence using Lipofectamine 2000. Luciferase assay was performed using the Dual-Luciferase® Reporter Assay System (Promega) at 48 hr after transfection. Data are presented as the mean value ± SD for triplicate experiments and compared to the level of wild type sequence vectors obtained in mutant type sequence vectors transfected cells that is normalized to 100%.
Quantitative RT-PCR
Extracted total RNA was reverse-transcribed into single stranded cDNA using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA) and a TaqMan microRNA reverse transcription kit (Applied Biosystems). Real-Time PCR was performed using first-strand cDNA with TaqMan Fast Universal PCR Master Mix (Applied Biosystems). The assay numbers for the mRNA endogenous control (beta-actin), target gene, microRNA endogenous control (RNU48) and target microRNAs were as follows: beta-actin (Hs99999903_m1), RhoC (Hs00747110_s1), FZD4 (Hs00201853_m1), RNU48 (001006), miR-493 (002364), miR-107 (000443), miR-130b (000456), miR-141 (000463, miR-423-5p (002340), miR-484 (001821), miR-503 (001048), miR-766 (001986), miR-933 (002176) and miR-1290 (002863). Quantitative PCR was performed with an Applied Biosystems Prism 7500 Fast Sequence Detection System (Applied Biosystems). Quantitative PCR parameters for cycling were as follows: 95°C for 20 sec 40 cycles of PCR at 95°C for 3 sec, and 60°C for 30 sec. All reactions were done in a 10-μl reaction volume in triplicate. The mRNA and miR expression level were determined using the 2−deltaCT method.
Western Analysis
At 72 hr after transfection, cells were washed in ice-cold PBS and added to RIPA lysis and extraction buffer (Fisher Scientific, Pittsburgh, PA, USA) containing Protease Inhibitor Cocktail I (Millipore, Billerica, MA, USA). Dishes were incubated for 5 min on ice and cells were collected with a cell lifter and rotated for 30 min at 4°C followed by centrifugation at 12,000 g for 20 min at 4°C. Total protein was analyzed by Western blotting using primary antibodies, followed by anti-mouse and -rabbit IgG HRP-conjugated secondary antibodies (#7076, #7074, Cell Signaling Technology, Beverly, MA, USA) and anti-rat IgG-B (sc-2041, Santa Cruz Biotechnology, Inc) / Streptavidin – HRP (Invitrogen) and were visualized with LumiGLO Reagent and peroxidet reagent (Cell Signaling Technology). The primary antibodies used were anti-RhoC Antibody (#3430, Cell Signaling Technology), anti-RhoA Antibody (#2117, Cell Signaling Technology), FZD4 Antibody (#LS-C6904, LifeSpan BioSciences, Inc.), and anti-beta-actin (#3700, Cell Signaling Technology) antibodies. Western analysis was repeated independently two times.
Immunoprecipitation
At 72 hr after transfection, cells were washed in ice-cold PBS and added to cell lysis buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% Triton X-100) containing Protease Inhibitor Cocktail I (Millipore, Billerica, MA, USA). Dishes were incubated for 5 min on ice and cells were collected with a cell lifter and transferred to a 1.5 ml tube. Samples were sonicated on ice three times for 5 sec followed by centrifugation at 14,000 × g for 10 min at 4°C. The supernatants were transferred to new 1.5 ml tubes. For cell lysate pre-clearing, protein G agarose was added to sample tubes. Sample tubes were incubated at 4°C for 1 hr with gentle rocking followed by centrifugation at 14,000 × g for 10 min at 4°C. The supernatants were transferred to new 1.5 ml tubes. Anti-Rock-1 antibody (#ab45171, Abcam, Cambridge, MA, USA) or anti-control antibody (#2729, Cell Signaling Technology) was added to sample tubes and their tubes were incubated with gentle rocking overnight at 4°C. Protein G was added to sample tubes and their sample tubes were incubated with gentile rocking for 2 hr 4°C followed by centrifugation at 14,000 × g for 30 sec at 4°C. Samples were washed five times with cell lysis buffer. 2x sample buffer was added to sample tubes and vortexed followed by centrifugation at 14,000 × g for 30 sec. Samples were incubated for 5 min at 95°C followed by centrifugation at 14,000 × g for 1 min. The supernatants were analyzed by Western blotting. Immunoprecipitation was repeated independently two times.
In situ hybridization
Tissue arrays were purchased from US Biomax, inc. (Rockville, MD, USA). For in situ hybridization, IsHyb In Situ Hybridization (ISH) Kit was used according to the manufacturer’s instructions (BioChain, Hayward, CA, USA). A tissue array slide was incubated overnight with a 1:500 dilution of miRCURY LNA Detection probe 5′-DIG labeled for miR-493 (EXIQON, Woburn, MA, USA) and overnight at 4°C with a 1:200 dilution of AP-conjugated anti-digoxingenin antibody. BM Purple; AP substrate precipitating, (NBT/BCIP ready-to-use solution) (Roche, Indianapolis, IN, USA) was used as detector. Staining intensity of in situ hybridization was divided into 0, no staining; 1, positive staining; 2, strong positive staining.
siRNA Knockdown of RhoC and FZD4 mRNA
T24 and J82 cells were transfected with RhoC and FZD4 siRNA (RhoC Silencer® Select Validated siRNA, siRNA ID s97 and FZD4 Silencer® Select Validated siRNA, siRNA ID s15840, Applied Biosystems), RhoC and FZD4 siRNA (RHOC Stealth RNAi siRNA HSS100662 and FZD4 Stealth RNAi siRNA HSS188730, Invitrogen) or negative control siRNA using Lipofectamine 2000 according to the manufacturer’s instructions.
Statistical Analysis
All statistical analyses were performed using GraphPad prism 5 software (GraphPad Software, San Diego, CA, USA). A p-value of < 0.05 was regarded as statistically significant.
RESULTS
Expression level of miR-493 in cell lines and primary tissues
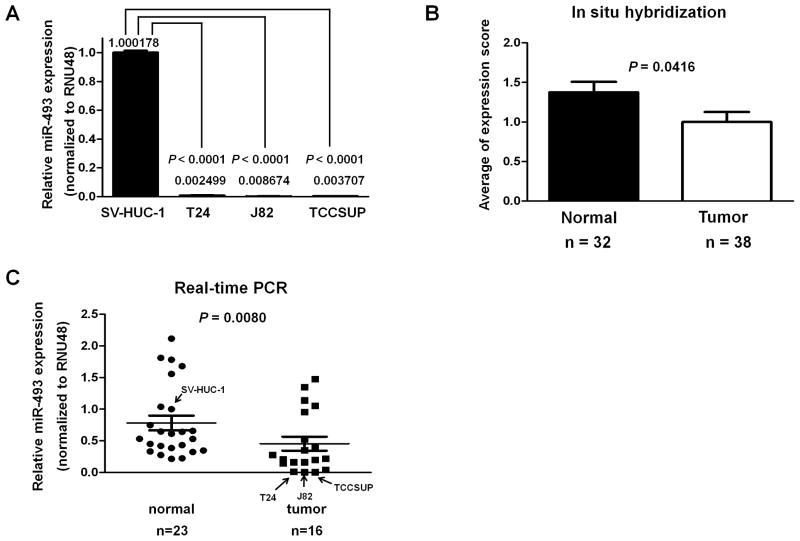
To identify tumor suppressor microRNAs in bladder cancer, we performed a miR microarray using SV-HUC-1 as a normal bladder cell line and three bladder cancer cell lines, T24, J82 and TCCSUP, and selected 10 microRNAs for our study (Table S1). Namely the expression of 5 microRNAs were significantly higher (top 5) and that of 5 microRNAs were significantly lower (top 5) in bladder cancer cell lines compared to normal bladder cells. To confirm the expression of these 10 microRNAs, we performed real-time PCR and found that miR-107, miR-141 miR-493 and miR-1290 expression in bladder cancer cells was significantly lower than that in SV-HUC-1 (P < 0.0001, Figure 1, Figure 2A). MiR-933 expression was not detected in bladder cell lines using real-time PCR. The real-time PCR data showed similar results as the microarray data for miR-107, miR-141, miR-493, miR-503 and miR-1290 while some miRs showed opposite results (miR-130b, miR-423-5p, miR-484 and miR-766) was higher compared to SV-HUC-1.
Figure 1. Expression level of ten microRNAs in bladder normal and cancer cell lines.
Ten microRNAs were selected based on microRNA-microarray data (Phalanx Bio Inc). Low: MicroRNA expression is significantly lower in bladder cancer cell lines compared to normal cell line (SV-HUC-1) based on microRNA-microarray data. High: MicroRNA expression is significantly higher in bladder cancer cell lines compared to normal cell line (SV-HUC-1) based on microRNA-microarray data. The ten microRNA expression levels in SV-HUC-1 and bladder cancer cell lines were also measured by real-time PCR and normalized to RNU48. Data are presented as mean value ± SD for three independent experiments and compared to the level in SV-HUC-1 cells normalized as 1.
Figure 2. Expression of miR-493 in bladder cell lines and human bladder tissues.
A. MiR-493 expression levels in SV-HUC-1 and bladder cancer cell lines were detected by real-time PCR and normalized to RNU48. Data are presented as mean value ± SD for three independent experiments and compared to the level of miR-493 in SV-HUC-1 cells normalized as 1. B. miR-493 expression level in tissue microarray using in situ hybridization. Staining intensity of in situ hybridization was divided into 0, no staining; 1, positive staining; 2, strong positive staining. C. The expression levels of miR-493 in primary normal bladder and transitional cell carcinoma tissues were examined by real-time PCR. Data are presented as mean values and compared with the level in SV-HUC-1 cells normalized as 1. Bars show mean values.
Localization of miR-493 expression and decreased miRNA-493 expression in bladder cancer tissues
To investigate miR-493 localization in human bladder tissues, we performed in situ hybridization in bladder tissue array (normal and cancer tissues). As shown in Figure 2B, miR-493 expression was significantly lower in bladder cancer tissues than in normal bladder tissues (P = 0.0416). Expression of miR-493 was observed in connective tissues, endothelial cells of blood vessels, and lamina muscularis mucosa in normal bladder tissues (data not shown). In urothelial cell carcinoma, miR-493 expression was lower than in layers of normal urothelium (Supplementary Figure 1). We then compared miRNA-493 expression levels in bladder cancer tissues and normal bladder tissues by real-time PCR to validate the ISH results. Similar to the ISH results, the expression of miR-493 was significantly lower in bladder cancer tissues than in normal bladder tissues. The expression of miR-493 in the normal bladder cell line, SV-HUC-1, was used as reference (expression = 1). Consistent with the ISH results, the miR-493 expression was significantly lower in bladder cancer tissues (P = 0.0080) than in normal tissues (Figure 2C). Regarding miR-107, miR-141 and miR-1290, we did not find any difference of expression between normal bladder tissues and bladder cancer tissues (Supplementary Figure 2).
Evaluation of the functional effects of miR-493 on T24 and J82 cells
As the expression of miR-493 was significantly lower in bladder cancer cell lines and bladder cancer tissues compared to normal bladder cells and tissues, we next focused on the functional effects of miR-493 on bladder cancer cells. MiR-493 and miR control were transiently transfected into T24 and J82 cells. The expression level of miR-493 was significantly increased at 48 hr after transfection (Figure 3A). Cell viability was decreased to 70–80 % in miR-493 transfected cells compared to controls at 4 days after tranfection (P = 0.0286, Figure 3B). Cell motility was also significantly decreased in miR-493 transfected T24 and J82 cells (P = 0.0068 and P < 0.0001, respectively, Figure 3C). Cell migration was also significantly decreased to about 30% in miR-493 transfected T24 and J82 cells (P = 0.0007 and P < 0.0001, respectively, Figure 3D). Cell cycle analysis based on flowcytometry was performed with miR-493 transfected T24 cells. Significantly increased G1 cell cycle arrest in the miR-493 transfectants was observed compared to controls (P = 0.0002, Figure 3E). To see whether control microRNA suppress cancer cells, Mock (without oligo-nucleotide) and Control microRNA were transfected into cancer cells and real-time PCR, Western analysis, migration and wound healing assay were performed. These results did not show significant data (Supplementary Figure 3). To confirm whether miR-493 regulates cell motility, control or miR-493 inhibitors were transfected into SV-HUC-1 cells. MiR-493 inhibitor slightly increased SV-HCU-1 cell motility (Supplementary Figure 4).
Figure 3. Functional effects of miR-493 in transfected T24 and J82 cells.
A. MiR-493 expression levels in bladder cancer cell lines (T24, J82) were detected by using real-time PCR at 48 hr after transfection of miR-493 precursor. Data are presented as mean value ± SD for three independent experiments and compared to the level of miR-493 in SV-HUC-1 cells normalized as 1. B. Cell viability was analyzed by the MTS assay 4 days after transient transfection. C. Wound healing assay with miR-493 transfected cells. At 48 hr after transfection, cells were transferred from 6-well to 24-well plates and further incubated for 24 hr. A wound was formed by scraping and the wound measured after 24 hr (T24 cells) and 72 hr (J82 cells). D. Migration assay with miR-493 transfected cells. At 72 hr after transfection, cells were added into the chamber. Cells were incubated for 4 hr at 37°C in a 5% CO2 tissue culture incubator, no migrated cells were removed from transwell membrane filter inserts using cotton-tipped swab and migrated cells were stained with Hema 3 STAIN SET. Representative photomicrographs are shown at 100x magnification. E. MiR-493 and miR control were transfected into T24 and J82 cells. At 72 hr after transfection, cell cycle assay was carried out by flow cytometry.
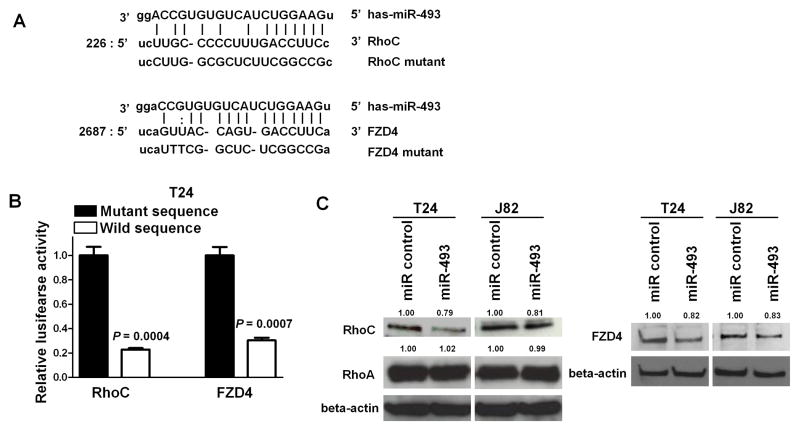
Identification of miR-493 target genes and its effect on their 3′-UTR-Luciferase assays and protein expression
Based on the observation that miR-493 affects cell motility and migration, we searched for target genes of miR-493 related to motility and migration using a target scan algorithm (microRNA.org http://www.microrna.org/microrna/home.do) and identified RhoC and FZD4 as target oncogenes. As shown in Figure 4A, FZD4 mRNA has one potential complimentary miR-493 binding site within its 3′ UTR. RhoC mRNA also has one potential complimentary miR-493 binding site within its 3′UTR. We performed RhoC and FZD4 3′UTR luciferase assays and found that the relative luciferase activities with these sites were significantly decreased in miR-493 transfected T24 cells (P = 0.0004 and P =0.0007, respectively, Figure 4B). With mutated plasmids, there was no significant difference in luciferase activity between controls and mutated miR-493 transfectants. To examine the inhibitory effect of miR-493 on protein expression levels, we did Western analysis 72 hr after miR-493 transfection into T24 and J82 cells. We observed that the protein levels of RhoC and FZD4 in miR-493 transfected T24 and J82 cells were significantly decreased compared to control cells, however miR-493 did not repress protein levels of RhoA (Figure 4C). These results suggest that FZD4 and RhoC are target oncogenes of miR-493.
Figure 4. miR-493 targets RhoC and FZD4 genes.
A. RhoC and FZD4 3′UTR sequence, complementary miR-493 binding sequences, and 3′UTR lusiferase assay. MiR-493 binding site in the RhoC/FZD4 3′UTR predicted by microRNA.org. Upper is miR-493 sequence. Middle is target wild type sequence. Lower is target mutant sequence. B. MiR-493 and 3′UTR vectors with wild type or mutant sequence were co-transfected into T24 cells. Cell lysates were measured for relative luciferase activities at 48 hours after transfection. Levels of luciferase activity were compared to those of cells transfected with 3′UTR vector with mutant type sequence. C. RhoC and FZD4 protein levels in T24 and J82 cells transfected with miR-493 or miR control. At 72 hr after transfcetion, total protein was extracted and analyzed by Western blots. β-Actin was used as a loading control.
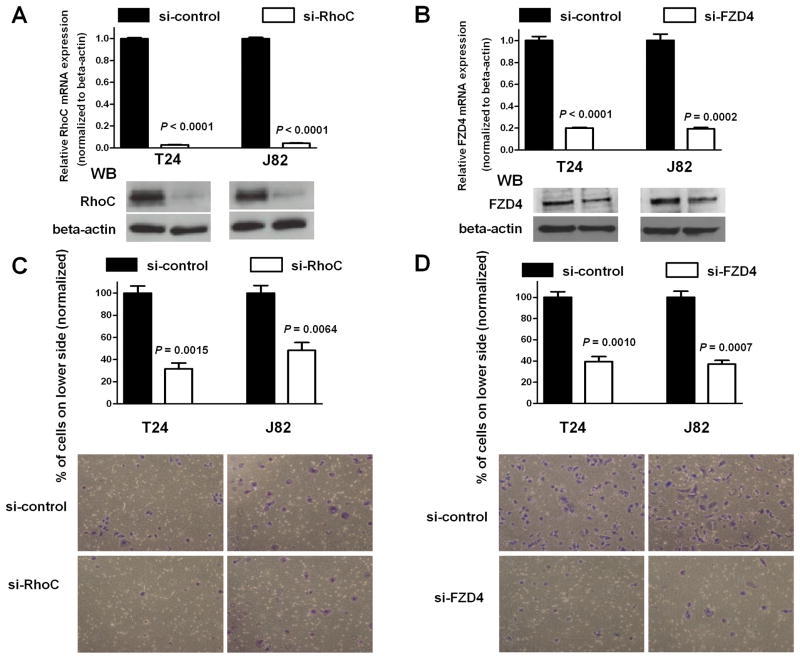
Effects of RhoC or FZD4 siRNA knockdown on bladder cancer cell motility
To analyze whether RhoC affects cell migration of T24 and J82 cells, RhoC siRNA (si-RhoC), FZD4 siRNA (si-FZD4) or control siRNA (si-control) was transfected into T24 and J82 cells. Expression levels of RhoC and FZD4 mRNAs were analyzed using real-time PCR at 48 hr after transfection. Both were significantly decreased to < 10 to 20% in si-RhoC or si-FZD4 transfected T24 and J82 cells (P < 0.0001, respectively, Figure 5A and P < 0.0001 and P = 0.0002, respectively, Figure 5B). Knockdown of RhoC and FZD4 protein expression was also confirmed by Western analysis (Figure 5A and 5B). To determine whether RhoC and FZD4 knockdown affects cell migration, migration assay was done out using T24 and J82 cells. Cell migration was significantly decreased to 35 to 50% in si-RhoC and si-FZD4 transfectants compared with control cells (P = 0.0015 and P = 0.0064, respectively, Figure 5C and P = 0.0010 and P = 0.0007, respectively, Figure 5D). To examine potential off target effects of siRNA, other RhoC and FZD4 siRNAs from a different source were also used (Supplementary Figure 5). As shown in supplementary Figure 4, the results were consistent with Figure 5. These data suggest that the siRNAs used in this study did not induced the off target effects in the siRNA experiments.
Figure 5. Functional effects of RhoC and FZD4 siRNA knockdown on T24 and J82 cells.
A. mRNA and protein expression level in si-control and si-RhoC transfected bladder cancer cells (T24, J82). WB stands for Western blot analysis. B mRNA and protein expression level in si-control and si-FZD4 transfected bladder cancer cells (T24, J82). At 48 hr after transfection, mRNA and protein were analyzed using real-time PCR and Western analysis. RhoC and FZD4 expression were normalized to β-actin.
C and D. Migration assay of si-RhoC and si-FZD4 transfectants. si-RhoC/si-FZD4 or si-control was transfected into T24 and J82 cells. At 72 hr after transfection, cells were added into the chamber. Cells were incubated for 4 hr at 37°C in a 5% CO2 tissue culture incubator, no migrated cells were removed from transwell membrane filter inserts using cotton-tipped swab and migrated cells were stained with Hema 3 STAIN SET. Representative photomicrographs are shown at 100x magnification.
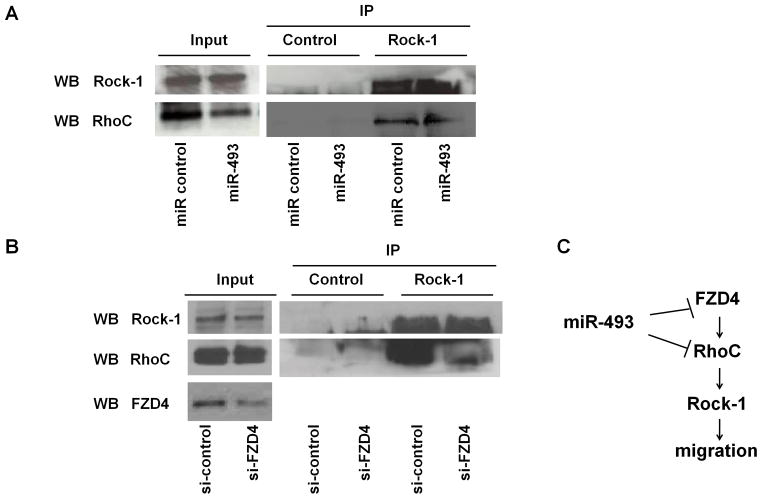
Effect of miR-493 on the binding of RhoC toRock1 in bladder cancer cells
To analyze whether over-expression of miR-493 affects the binding of RhoC to Rock-1, immunoprecipitation was carried out at 72 hr after transfection in T24 cells. We found that the binding of RhoC to Rock-1 in miR-493 transfected T24 cells was decreased compared to miR control transfectans (Figure 6A). We also analyzed the binding of RhoC to Rock-1 after FZD4 siRNA trasfection. At 72 hr after transfection, the binding was decreased in siFZD4 transfected cells (Figure 6B). The schematic representation of the possible role of miR-493 in the FZD4/RhoC signaling pathway is shown in Figure 6C.
Figure 6. Functional effects of Rock-1 and RhoC after miR-493 and FZD4 knockdown.
A. Binding of RhoC to Rock-1 after miR-493 trasfection. At 72 hr after transfection, cell lysate was immunoprecipitated by anti-Rock-1 antibody and immunoblotted with anti-RhoC antibody. IP stands for immunoprecipitation. WB stands for Western blot analysis. B. Binding of RhoC to Rock-1 after FZD4 siRNA transfection. At 72 hr after transfection, cell lysate was immunoprecipitated by anti-Rock-1 antibody and immunoblotted by anti-RhoC antibody. C. A possible role of miR-493 in the FZD4/RhoC signaling pathway.
Discussion
There have been several miRNA studies related to clinical bladder cancer and most have involved microarray screening (11–15). We used a miR microarray service to compare miRNA expression levels in a normal uroepithelium cell line (SV-HUC-1) and bladder cancer cell lines (T24, J82 and TCCSUP). Based on the miRNA microarray data, we initially chose 10 microRNAs whose expression was significantly higher (5 microRNAs) or lower (5 microRNAs) compared to the normal bladder cell line. Among 10 microRNAs, the expression of miR-493 and miR-141 was lower in bladder cancer cell lines compared to the normal cell line which is consistent with previous microRNA microarray results however there are no reports regarding the functional role of miR-493 in bladder cancer.
Thus we focused on miR-493 as a potentially new candidate tumor suppressor miRNA in bladder cancer. Additionally we confirmed the miR-493 expression level in clinical bladder cancer tissues using ISH and real-time PCR. The expression of miR-493 was significantly lower in bladder cancer tissues compared to normal bladder tissues. Based on these results, we hypothesized that miR-493 may play an important role as a tumor suppressor in bladder cancer. To test this hypothesis, we performed functional analyses (MTS, migration, wound healing and cell cycle) to look at miR-493 function using miR-493 transfected cells. As expected, miR-493 over-expression inhibited cell proliferation in bladder cancer cells (T24 and J82). Cell migration and motility were also dramatically decreased after miR-493 transfection. These results suggest that miR-493 may function as a tumor suppressor and play an important role in inhibition of cell growth and motility of bladder cancer cells.
As a next step, we also used microRNA org. to identify target oncogenes of miR-493, and identified potential candidate target oncogenes related to invasion and migration. TargetScan also showed RhoC and FZD4 as miR-493 target genes (Supplmentary Figure 6). We performed 3′UTR lucifease assay, and observed that luciferase activity was decreased after co-transfection of miR-493 and a 3′UTR vector containing the RhoC/FZD4 miR-493 target sequence. RhoC and FZD4 protein expression were also significantly down-regulated in miR-493 transfected T24 and J82 cells indicating that RhoC and FZD4 are direct targets of miR-493.
It has been reported that RhoC protein is over-expressed in bladder tumor tissues compared to bladder nontumor tissues and higher RhoC protein expression in tumors was associated with clinico-pathological factors (grade and pT), with poorer disease-free survival and overall survival than low expression (17). Although there have been no reports about RhoC function in bladder cancer, in hepatocellular carcinoma cells RhoC is essential for invasion and migration but not proliferation and apoptosis (18). This is consistent with our finding that cell migration in T24 and J82 RhoC knockdown was significantly decreased compared to control siRNA. However cell growth in T24 and J82 RhoC knockdowns was not significantly different compared to control siRNA (data not shown). This cell growth inhibition result is consistent with RhoC function in hepatocellular carcinoma cells (18). It has been reported that RhoC is essential for invasion in some cancers (19–24) and RhoC expression is significantly correlated with poor prognosis (25–27). FZD4 knockdown decreased the binding of RhoC to Rock-1. We have also previously reported that miR-584 targets Rock-1 (16). As high Rock-1 expression in bladder tumors was associated with poor disease-free survival and overall survival than low expression, functional RhoC and Rock-1 in bladder cancer may be important to malignancy progression.
It has also been reported that miR-138 directly targets RhoC and Rock-II in tongue squamous cell carcinoma and inhibits cell migration and invasion (28). Our microarray data shows that miR-138 expression in TCCUP cells is significantly lower than SV-HUC-1, but not in T24 and J82 cells (Table S1).
Our study showed that miR-493 inhibited FZD4 and RhoC and this pathway is involved in Wnt-PCP pathway. Wnt signaling pathways include: [1] Wnt-beta-catenin, [2] Wnt-PCP [3] Wnt-Ca2+ and these signaling pathways have been reported to be associated with cancer progression and poor prognosis (29). In bladder cancer, Ras oncogene activation plays a very important role in bladder cancer progression (7). Interestingly Ahmad et al have recently reported that Ras pathway activation cooperates with Wnt-beta catenin signaling to drive urothelial cell carcinoma (30). There are few reports regarding other Wnt signaling pathways (Wnt-PCP or Wnt-Ca2+) in bladder cancer. In conclusion, our study suggests that miR-493 may be a new tumor suppressive microRNA inhibiting cell invasion and migration by blocking FZD4 and RhoC in bladder cancer, implicating the Wnt-PCP pathway in bladder carcinogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript.
This study was supported by Grants R01CA130860, R01CA138642, R01CA111470, T32DK007790 from the NIH, VA Research Enhancement Award Program (REAP), and Merit Review grants.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Pollard C, Smith SC, Theodorescu D. Molecular genesis of non-muscle-invasive urothelial carcinoma (NMIUC) Expert Rev Mol Med. 2010;12:e10. doi: 10.1017/S1462399410001407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proctor I, Stoeber K, Williams GH. Biomarkers in bladder cancer. Histopathology. 2010;57:1–13. doi: 10.1111/j.1365-2559.2010.03592.x. [DOI] [PubMed] [Google Scholar]
- 4.Pasin E, Josephson DY, Mitra AP, Cote RJ, Stein JP. Superficial bladder cancer: an update on etiology, molecular development, classification, and natural history. Rev Urol. 2008;10:31–43. [PMC free article] [PubMed] [Google Scholar]
- 5.Stenzl A, Cowan NC, De Santis M, Kuczyk MA, Merseburger AS, Ribal MJ, et al. Treatment of Muscle-invasive and Metastatic Bladder Cancer: Update of the EAU Guidelines. Eur Urol. 2011;59:1009–1018. doi: 10.1016/j.eururo.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Liebert M, Seigne J. Characteristics of invasive bladder cancers: histological and molecular markers. Semin Urol Oncol. 1996;14:62–72. [PubMed] [Google Scholar]
- 7.Wallerand H, Reiter RR, Ravaud A. Molecular targeting in the treatment of either advanced or metastatic bladder cancer or both according to the signalling pathways. Curr Opin Urol. 2008;18:524–532. doi: 10.1097/MOU.0b013e3283097889. [DOI] [PubMed] [Google Scholar]
- 8.Netto GJ, Epstein JI. Theranostic and prognostic biomarkers: genomic applications in urological malignancies. Pathology. 2010;42:384–394. doi: 10.3109/00313021003779145. [DOI] [PubMed] [Google Scholar]
- 9.Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38:257–68. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737–47. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 11.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 13.Baffa R, Fassan M, Volinia S, O’Hara B, Liu CG, Palazzo JP, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 14.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–60. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 15.Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z, et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer. 2010 Jun 14; doi: 10.1002/ijc.25509. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Ueno K, Hirata H, Shahryari V, Chen Y, Zaman MS, Singh K, et al. Tumour suppressor microRNA-584 directly targets oncogene Rock-1 and decreases invasion ability in human clear cell renal cell carcinoma. Br J Cancer. 2011;104:308–15. doi: 10.1038/sj.bjc.6606028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamai T, Tsujii T, Arai K, Takagi K, Asami H, Ito Y, et al. Significant association of Rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin Cancer Res. 2003;9:2632–41. [PubMed] [Google Scholar]
- 18.Wang W, Wu F, Fang F, Tao Y, Yang L. Inhibition of invasion and metastasis of hepatocellular carcinoma cells via targeting RhoC in vitro and in vivo. Clin Cancer Res. 2008;14:6804–12. doi: 10.1158/1078-0432.CCR-07-4820. [DOI] [PubMed] [Google Scholar]
- 19.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 20.Hakem A, Sanchez-Sweatman O, You-Ten A, Duncan G, Wakeham A, Khokha R, et al. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–9. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikoma T, Takahashi T, Nagano S, Li YM, Ohno Y, Ando K, et al. A definitive role of RhoC in metastasis of orthotopic lung cancer in mice. Clin Cancer Res. 2004;10:1192–200. doi: 10.1158/1078-0432.ccr-03-0275. [DOI] [PubMed] [Google Scholar]
- 22.Faried A, Faried LS, Kimura H, Nakajima M, Sohda M, Miyazaki T, et al. RhoA and RhoC proteins promote both cell proliferation and cell invasion of human oesophageal squamous cell carcinoma cell lines in vitro and in vivo. Eur J Cancer. 2006;42:1455–65. doi: 10.1016/j.ejca.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Iiizumi M, Bandyopadhyay S, Pai SK, Watabe M, Hirota S, Hosobe S, et al. RhoC promotes metastasis via activation of the Pyk2 pathway in prostate cancer. Cancer Res. 2008;68:7613–20. doi: 10.1158/0008-5472.CAN-07-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam M, Lin G, Brenner JC, Pan Q, Merajver SD, Hou Y, et al. RhoC expression and head and neck cancer metastasis. Mol Cancer Res. 2009;7:1771–80. doi: 10.1158/1541-7786.MCR-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, Yang LY, Huang GW, Lu WQ, Yang ZL, Yang JQ, et al. Genomic analysis reveals RhoC as a potential marker in hepatocellular carcinoma with poor prognosis. Br J Cancer. 2004;90:2349–55. doi: 10.1038/sj.bjc.6601749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horiuchi A, Imai T, Wang C, Ohira S, Feng Y, Nikaido T, et al. Up-regulation of small GTPases, RhoA and RhoC, is associated with tumor progression in ovarian carcinoma. Lab Invest. 2003;83:861–70. doi: 10.1097/01.lab.0000073128.16098.31. [DOI] [PubMed] [Google Scholar]
- 27.Suwa H, Ohshio G, Imamura T, Watanabe G, Arii S, Imamura M, et al. Overexpression of the rhoC gene correlates with progression of ductal adenocarcinoma of the pancreas. Br J Cancer. 1998;77:147–52. doi: 10.1038/bjc.1998.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang L, Liu X, Kolokythas A, Yu J, Wang A, Heidbreder CE, et al. Downregulation of the Rho GTPase signaling pathway is involved in the microRNA-138-mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer. 2010;127:505–12. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad I, Patel R, Liu Y, Singh LB, Taketo MM, Wu XR, et al. Ras mutation cooperates with β-catenin activation to drive bladder tumourigenesis. Cell Death Dis. 2011;2:e124. doi: 10.1038/cddis.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.