Abstract
Background
The clinical significance of anti-T. cruzi low-level reactive samples is incompletely understood. PCR-positive rates and antibody levels among seropositive blood donors in three countries are described.
Methods
Follow-up whole blood and plasma samples were collected from T. cruzi-seropositive donors from 2008-2010 in the US (n=195) and Honduras (n=58). Also 143 samples from Brazil in 1996-2002, originally positive by three serological assays, were available and paired with contemporary follow-up samples from these donors. All samples were retested with the FDA-approved Ortho ELISA. PCR assays were performed on coded sample panels by two laboratories (BSRI and ARC) that amplified kinetoplast minicircle DNA sequences of T. cruzi.
Results
PCR testing at BSRI yielded slightly higher overall sensitivity and specificity (33% and 98%) compared with the ARC lab (28% and 94%). Among seropositive donors, PCR-positive rates varied by country (p<0.0001) for the BSRI laboratory: Brazil (57%), Honduras (32%) and the US (14%). ELISA signal/cutoff (S/CO) ratios were significantly higher for PCR-positive compared to PCR-negative donors (p<0.05 for all comparisons). Additionally, PCR-negative Brazilian donors exhibited greater frequencies of antibody decline over time versus PCR-positive donors (p=0.003).
Conclusion
For all three countries, persistent DNA positivity correlated with higher ELISA S/CO values, suggesting that high-level seroreactivity reflects chronic parasitemia. The higher rate of PCR positivity for Brazilian donors was likely attributable to required reactivity on three assays available a decade ago. Significant S/CO declines in 10% of the PCR-negative Brazilian donors may indicate seroreversion following parasite clearance in the absence of treatment.
Keywords: T cruzi, Chagas disease, PCR, antibodies
Introduction
Diagnosis of T. cruzi infection and Chagas disease is based on serological techniques because parasitological techniques including hemoculture and xenodiagnosis, while extremely specific, are less sensitive and more challenging to perform1. Blood donor screening serologic assays have evolved from indirect hemagglutination (IHA) and indirect immunofluorescence assays (IFA) to enzyme-linked immunosorbent assays (ELISA) and chemiluminescent immunoassays, and confirmatory assays from radioimmunoprecipitation assays (RIPA) to recombinant antigen strip immunoassays2-6.
Currently available T. cruzi antibody screening and diagnostic assays have significantly improved sensitivity and specificity2,3,6. However, one challenge is the detection of low-level seropositive samples that are inconsistently detected by different screening tests 7. The clinical significance of these results for donor health and transfusion-transmission risk is incompletely understood. One possible explanation is that low-level confirmed seroreactive samples represent cases in which the donor has cleared parasitemia in the absence of treatment with consequent decreasing antibodies values over time. If this hypothesis is correct, an association between donor antibody levels and DNA detection, would be observed.
Few studies have evaluated the detection of T. cruzi DNA relative to antibody levels, in part because PCR assays for T. cruzi have been challenging to optimize and standardize due to the low levels of parasitemia and consequently circulating DNA in chronically infected, asymptomatic subjects8. Also, assessment of anti-T. cruzi levels were historically based on IFA or IHA titration analyses9 that are less sensitive than currently available ELISAs.
In this study we compare the results obtained by two laboratories using different PCR protocols on coded sets of samples collected from seropositive blood donors from Brazil, Honduras and the US, as well as blinded seronegative control specimens. All samples were tested by a contemporary ELISA; the ELISA antibody levels as assessed by their signal/cutoff ratios (S/CO) were compared to the PCR results. In addition, plasma aliquots from the index donations from the seropositive Brazilian donors collected approximately 10 years earlier permitted a comparison of current PCR results to the evolution of antibody reactivity over time.
Methods
T. cruzi analytical sensitivity panel
T. cruzi parasites were obtained as epimastigotes grown in LIT medium from stocks provided by the laboratory of Parasitology of the Institute of Tropical Medicine of the University of Sao Paulo. The parasite concentration was determined by direct counting in a hemocytometer chamber. Parasites were spiked into T. cruzi antibody-negative blood to achieve a concentration of 512 parasites/20mL, followed by 2-fold serial dilutions into 20mL volumes of whole blood to yield estimated concentrations of 8, 4, 2, 1 and 0.5 parasites/20mL. Spiked and control unspiked blood samples were mixed with an equal volume of 6M guanidine HCl-0.2M EDTA solution. The samples were immersed in boiling water for 15 min, aliquoted and frozen at −20C. Five replicate 1mL aliquots of each dilution of spiked blood and of the unspiked diluent were coded into a blinded analytical sensitivity panel that was sent to the two PCR laboratories: Blood Systems Research Institute (BSRI) and the American Red Cross Holland Laboratory (ARC).
Clinical samples
Brazil
The REDS-II Chagas Cohort study recruited 499 T. cruzi seropositive blood donors (cases) and 488 seronegative blood donors (controls) who had donated blood in 1996-2002 in Sao Paulo and Montes Claros, Brazil. Frozen plasma from the index donation plasma components, as well as whole blood and plasma samples collected at the time of Chagas Cohort enrollment visits in 2010-2011, were available for 143 of the enrolled seropositive donors from Sao Paulo and were included in this study. In addition, for this study, samples from 45 of the ELISA non-reactive control donors were included. Donors were interviewed and were only included if they did not report previous treatment with benznidazole.
Index donation samples were originally identified as T. cruzi antibody reactive by three donor screening tests used at Fundação Pro-Sangue in 1996-2002: ELISA (Embrabio, Sao Paulo, SP), IFA (BioLab Merieux, Jacarepagua, Rio de Janeiro) and IHA (BioLab Merieux, Jacarepagua, Rio de Janeiro). At the time of cohort follow-up in 2010-2012, 10mL of blood was collected in EDTA-anticoagulated tubes for preparation of plasma aliquots. In addition, a 20mL EDTA-containing whole blood sample was collected for PCR that was immediately mixed with an equal volume of a solution of 6M guanidine HCl-0.2M EDTA. The guanidine-EDTA blood mixture was then maintained at room temperature until boiled for 15 min and divided into aliquots. Aliquots were frozen at −20°C until shipped to the US REDS-II central laboaratory (BSRI) on dry ice, followed by storage at −70°C.
US
The ARC began universal screening of all donations on January 27, 2007 using the FDA licensed Ortho® Trypanosoma cruzi ELISA Test System (Raritan, NJ), an ELISA for the qualitative detection of antibodies to T. cruzi. Samples with an ELISA S/CO of 1.00 or greater were repeat tested in duplicate and considered repeatedly reactive (RR) if one or both of the retests were reactive. All RR donation samples were further tested using a laboratory-developed RIPA10, available through Quest Diagnostics (Chantilly, VA). Reactivity corresponding to surface glycoproteins at 72 and 90 kDa was considered a positive result confirming ELISA antibody reactivity.
Donors testing RR on the Ortho ELISA and confirmed by RIPA were invited to participate in a subsequent study that included the collection of additional samples for follow-up testing including PCR. At follow-up, consenting donors were asked to provide two 5mL whole blood samples collected in EDTA for PCR testing. EDTA-containing whole blood was then added to 10mL of 6M guanidine HCl-0.2M EDTA solution, vortexed and boiled for 10 min before storage at 4°C. Stored material was used for subsequent parasite DNA extraction. Remaining lysates were stored at −70°C. No donor reported having received treatment for T. cruzi infection.
Honduras
The Honduras Red Cross Blood Program recruited 71 T. cruzi seropositive blood donors who had donated blood between January 2007 and October 2009 in the cities of Tegucigalpa and San Pedro Sula, Honduras. Donors were invited to participate when they were informed of their positive reactivity on their blood donation. All donors gave their authorization in writing for the collection of the samples. No donor reported having received treatment for T. cruzi infection.
All donation samples included in this study were screened T. cruzi-antibody reactive by the Weiner lab Chagastest Recombinant version 2 (Rosario, Argentina). , These samples were further tested by the Ortho ELISA at Creative Testing Solutions (Tempe, AZ). Only Ortho ELISA RR samples were considered T. cruzi-antibody confirmed positive. At the time of cohort accrual in 2008-2009, 10mL of blood was collected for preparation of serum aliquots. In addition, 20mL of an EDTA-containing whole blood sample was collected for PCR; such samples were frozen immediately at −30C until shipped to BSRI on dry ice followed by storage at −70C. EDTA-containing whole blood samples were subsequently thawed, mixed with an equal volume of 6M guanidine HCl-0.2M EDTA solution, vortexed, boiled for 10 min, aliquoted (1mL) and stored at −70°C.
Human Subjects Protections
Brazil
The Brazil samples were collected as part of the NHLBI funded REDS-II Chagas Cohort Study. Approvals were obtained from Institutional Review Boards (IRBs)/Ethics Committees at, Hospital das Clinicas, CONEP, University of California at San Francisco, and Westat. This study also was reviewed and received a clinical exemption from the Office of Management and Budget.
US
Donor screening and follow-up procedures, and recruitment materials including informed consent documents were reviewed and approved by the ARC IRB prior to the initiation of the study and were reviewed annually.
Honduras
The routine donor history form used by the Honduras Red Cross Blood Program includes a separate written authorization to conduct additional testing on donor samples for T. cruzi as well as other infectious diseases. IRB approval for this specific study was obtained from the IRB Committee at the Medical School from Universidad Nacional Autónoma de Honduras.
Specificity panel
Both labs tested the specificities of their PCR assays using a blinded panel of whole blood lysates derived from ARC donors including 100 negative control samples (T. cruzi ELISA-nonreactive and RIPA negative) and two positive control samples (T. cruzi seroreactive/RIPA-positive) from the ARC.
Sample processing and PCR testing
BSRI
HemoBind™ Sample Extraction
All blood lysate samples were pulse spun at high speed to bring all the solution to the bottom of the tubes. HemoBind™ buffer 0.5mL was added to the mixture, along with 1uL each of the four capture probes (see below). The mixture was vortexed and incubated at 60°C for 30 min and then spun at the highest speed in a microcentrifuge for one min to bring down condensation and sediments11. The supernatant was transferred to a clean mirocentrifuge tube and heated at 100°C for 5 min. The preparation was microcentrifuged at high speed for one minute and the clean supernatant was transferred to the ten-tube unit cassettes (Gen-Probe, San Diego, CA) containing 3.6uL of magnetic beads (Sera-Mag Magnetic Oligo dT Microparticles [Seradyn Inc., Indianapolis, IN]). After generating a clean sample, the Target Capture protocol for plasma was followed (add ref to GP TC on eSAS).
T. cruzi-specific DNA target-capture and amplification
Capture of T. cruzi nucleic acids was achieved by specific binding to 20mer oligonucleotide, including a combination of at least four capture primers, which then bound to magnetic beads. The capture is done by hybridization of T. cruzi minicircle DNA to specific capture probes that contain poly A tails which bind to magnetic beads with covalently bound poly T (Sera-Mag Magnetic Oligo dT Microparticles [Seradyn]). Once hybridized, beads were captured by magnets in an enhanced semi-automated system (eSAS; Novartis, Emeryville CA) and washed twice. The following sequences were used as capture probes12: (CaptureTc_121) AAAAAAAAAAAAAAAAAAAAAAAAAAAATAATGTACGGGKGAGATGCATGA; (CaptureTc_122). GGTTCGATTGGGGTTGGTGTAATATAAAAAAAAAAAAAAAAAAAAAAAAAA; (CaptureTc-S35)AAAAAAAAAAAAAAAAAAAAAAAAAATAATGTACGGGKGAGATGCATG; (CaptureTc-S36)GGGTTCGATTGGGGTTGGTGTAAAAAAAAAAAAAAAAAAAAAAAAA.
The captured minicircle DNA targets were eluted from the magnetic beads with 100uL of Solution A (0.1M KCl, 0.01M Tris Base,0.0025M MgCl26H2O (pH 8.3)) and B (0.01M Tris (pH8.3), 2.5 mM MgCl26H2O, 1% Tween-20, 1% NP40) , by heating to 100°C for 5 minutes. Twenty-five uL of DNA was added to 50uL of BSRI PCR mix and amplified by real-time PCR using the following primers: (Tc-S36) GGGTTCGATTGGGGTTGGTGT13; (Tc_S35/A) TAATGTACGGGKGAGATGCATGA (shortened from original Tc_121 sequence)12. PCR amplification was performed on the Applied Biosystems’ 7500 for 45 cycles at the following settings: 10 min at 95°C, followed by 45 cycles of 30 sec at 95°C, 30 sec at 64°C, 45 se at 72°C. A dissociation step (melting curve analysis) was performed after completion of thermal cycling. Products dissociating with one or two peaks within the range of 80-82°C were considered positive if the cycle threshold (Ct) was less than 40 cycles. A total of 1mL of guanidium-EDTA lysed whole blood was processed in four 0.25 mL replicates as above and amplified. Interpretation of results was as follows: 1) positive if at least two of the four replicates crossed the Ct at less than 40 cycles and the amplicons were specific products based on dissociation analysis; 2) negative if none or one of the four replicates yielded specific product.
ARC
A total of 200uL of each sample was extracted three times, using the Qiagen QIAamp DNA Blood Mini kit and eluted in 100uL of DNA-free water. A total of 5uL of the extracted DNA was amplified using Applied Biosystems SYBR reaction and the primers S35 (AAA TAA TGT ACG GGK GAG ATG CAT GA) and S36 (GGG TTC GAT TGG GGT TGG TGT) that target the T. cruzi minicircle DNA. Triplicate PCR reactions for each extraction (total of 9 reactions) were carried out in an Applied Biosystem 7500 at a 63°C annealing temperature for 40 cycles with a dissociation cycle profile (Tm) of 60°-95°C. A PCR reaction was considered positive if the Tm was between 73.5°-76°C. A sample was considered positive if at least three of the nine replicates were positive, and if at least two were derived from a different extraction. Samples were considered negative if at least eight of the replicates were negative. All the other possible results were considered inconclusive, and these samples were further tested using a Taq-Man Real-Time PCR assay.
The Taq-Man Real-Time PCR assay amplifies a satellite parasite DNA sequence, as previous described by Prion et al.14. PCR was carried out using the Applied Biosystems’ Universal Taq with primers Cruzi1 (AST CGG CtG ATC GTT TTC GA) and Cruzi2 (AAT TCC TCC AAG CAG CGG ATA) and probe Cruzi3 (6FAM CAC ACA CtG GAC ACC AAMGBNFQ). Triplicates were performed using 5ul of the extracted DNA. A sample was considered positive if at least one of the triplicates yielded an amplicon signal below 40 cycles.
Serological testing
All samples from the US and Honduras were tested by the Ortho ELISA as previously described. For the Brazilian samples, the index plasma unit samples and follow-up plasma samples were tested in triplicate in the same run to minimize S/CO run-to-run variability.
Data analysis
All data analyses used SAS (SAS 9.2, SAS Institute Inc., Cary, NC). Analysis of EIA levels in relation to PCR positivity was restricted to samples that were Ortho ELISA RR at follow-up; BSRI PCR results were used for this analysis. Chi-square tests were used to compare the PCR positivity rates by country (for Table 1), by high or low level seroreactivity (ELISA S/CO ratios equal to or greater than 4.0 vs. less than 4.0), and by level of antibody decline or increase from the time of index to enrollment sample collection. Wilcoxon two-sample tests were used to compare the ELISA S/CO values between PCR positive and PCR negative samples within the specific country (for Figure 2),
Table 1.
Rates of PCR positivity among seropositive (cases) and seronegative (control) blood donors, sorted by country and laboratory performing the PCR assays.
| Cases * | BSRI PCR+ (%) |
Total Tested at BSRI |
ARC PCR+ (%) |
Total Tested at ARC |
|---|---|---|---|---|
| Brazil | 81 (57) | 143 | 46 (32) | 143 |
| Honduras | 9 (32) | 28 | 7 (25) | 28 |
| US | 24 (14) | 172 | 39 (24) | 163 |
| Total | 114 (33) | 343 | 92 (28) | 334 *** |
| Controls ** | ||||
| Brazil | 0 | 45 | 0 | 45 |
| Honduras | 0 | 30 | 5 (17) | 30 |
| US | 2 (9) | 23 | 4 (17) | 23 |
| Total | 2 (2) | 98 | 9 (9) | 98 |
Brazilian index samples were reactive by IHA, IFA and ELISA. US samples were reactive by the Ortho ELISA and confirmed by RIPA. Honduran samples screened with the Weiner ELISAs and confirmed by the Ortho ELISA.
Honduran and US samples were originally reactive but considered negative following confirmatory testing (Ortho ELISA for Honduras, and RIPA for US). The samples from Brazil were negative by the three screening tests (IHA, IFA and ELISA) ten years previously, as well as by the Ortho ELISA performed in the US.
Results were not available for 9 samples.
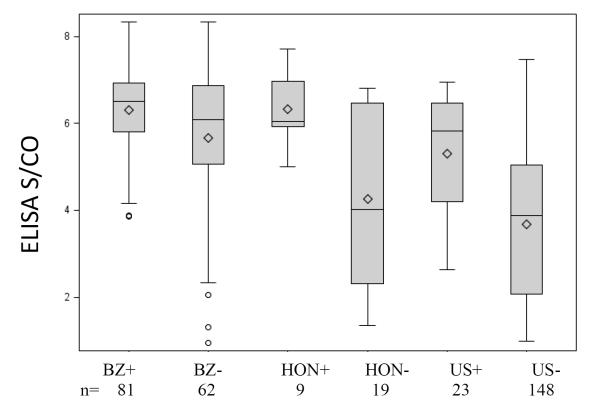
Figure 2. Follow-up sample ELISA S/CO values sorted by country and BSRI PCR results.
Box and whiskers plots were constructed using ELISA S/CO values for seropositive samples that were confirmed positive. Mean ELISA S/CO values were different between PCR-positive and PCR-negative samples within the different countries. Brazil PCR positive mean S/CO was 6.30 (SD = 1.01) which was higher than the Brazil PCR negative mean S/CO ratio of 5.65 (SD = 1.64; p=0.03). Honduras PCR positive mean S/CO was 6.33 (SD = 0.99) which was higher than the Honduras PCR negative mean S/CO ratio of 4.27 (SD = 2.00; p=0.04). US PCR positive mean S/CO was 5.30 (SD = 1.33) which was higher than the US PCR negative mean S/CO ratio of 3.68 (SD=1.75; p<0.0001). BZ+ indicates Brazil samples that were positive via the BSRI assay; BZ-, Brazil samples that were BSRI PCR negative; Hon+, Honduras samples that were BSRI PCR positive; Hon-, Honduras samples that were BSRI PCR negative; US+, US samples that were BSRI PCR positive; US-, US samples that were BSRI PCR negative. Top and bottom edges of the boxes in the plots represent the 25th and 75th percentiles of the distributions; horizontal line drawn within the box is the median; diamond is the mean; vertical lines, or whiskers, extend to the smallest data value still within 1.5 interquartile range (IQR) of the lower quartile and to the greatest data value still within 1.5 IQR of the upper quartile; circles are data values more extreme than the whiskers.
Results
Performance of the PCR assays on the analytical sensitivity and specificity panels
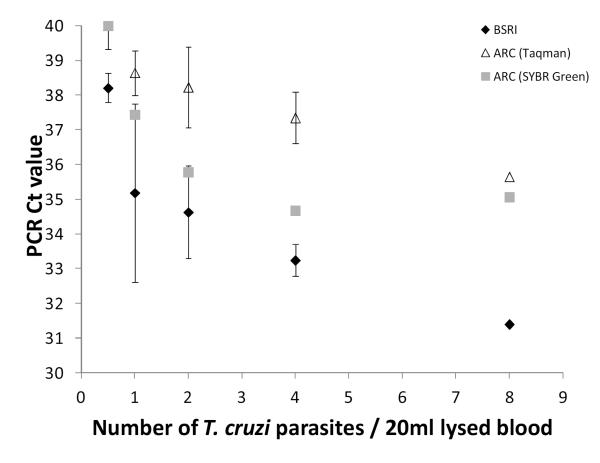
To evaluate the performance of the PCR techniques by the two laboratories, an analytical sensitivity panel was prepared containing five replicates of samples containing between 0.5 and 8 parasites spiked into 20mL of whole blood. Both PCR assays were able to detect one T. cruzi organism/20mL of spiked blood (each T. cruzi epimastigotes contains a single kinetoplast that has ~15,000 DNA minicircles, which were dispersed in the whole blood lysate following lysis, boiling and vortexing). However, while the BSRI PCR assay detected the one copy/20mL spiked samples at 35 cycles, the ARC SYBR Green and Taq-Man techniques detected one parasite copy/20 mL at cycles 37 and 39, respectively (Figure 1). Furthermore, only the BSRI PCR assay was able to detect the terminal dilution containing an estimated 0.5 T. cruzi organisms/20mL of spiked blood (BSRI PCR Ct = 38; ARC SYBR and Taq-Man PCR Cts > 40). All five replicates of unspiked blood in the analytical sensitivity panel tested non-reactive by all assays in both laboratories.
Figure 1. Relative analytical sensitivities of the BSRI and ARC PCR assays.
Five replicates of samples containing between 0.5 and 8 parasites spiked into 20mL of whole blood were analyzed by the two labs. Resulting Ct values were plotted against the number of parasites. Black diamonds represent average Ct values from BSRI assay; white triangles, average Ct values from ARC Taqman assay; gray boxes, average Ct values from ARC SYBR Green assay; bars, standard deviations. Results were negative on four coded replicates of non-spiked blood used as diluent by both the BSRI and ARC PCR assays (not shown).
To estimate specificity, both laboratories tested a blinded analytical specificity panel comprised of blood lysates derived from 100 T. cruzi seronegative donor samples and two seropositive donor samples. ARC reported 97 out of 100 seronegative samples as PCR negative (97% specificity). BSRI reported 97 out of 99 seronegative samples (1 sample did not yield interpretable results) as PCR negative (98% specificity).
PCR results among seronegative and seropositive blood donors in Brazil, Honduras, and US
Aliquots of 20mL blood lysates from blood donors from Brazil, Honduras and the US were subjected to PCR testing at BSRI and ARC. A total of 431 samples were tested by PCR in both labs (1 Brazil and 11 US samples did not have valid PCR results from both labs). Concordant PCR positive and negative results were obtained by ARC and BSRI in 69 (16%) and 186 (66%) samples, respectively. There were 44 (10%) samples that tested BSRI positive/ARC negative and 33 (7%) samples that tested ARC positive/BSRI negative.
Among the 343 seropositive donors, PCR testing at BSRI yielded 33% positivity and ARC yielded 28% positivity (Table 1). Ninety-eight samples were included in the donor panels that were derived from donors who were negative by screening or confirmatory serological assays; these are referred to as “controls” in Table 1. Nine (9%) of these controls were reported as positive by the ARC and two (2%) positive by BSRI, with no overlap in these false-positive cases. When combined with the results from the blinded specificity panel, ARC PCR testing yielded an overall specificity of 94% (186/198) and BSRI PCR testing yielded an overall specificity of 98% (193/197).
The rate of PCR positivity for seropositive donors as assessed by BSRI’s PCR varied by country (p<0.0001). PCR positivity for seropositive Brazilian donors (57%) was higher than the PCR positivity rate for samples from seropositive Honduran donors (32%; p= 0.02). PCR positivity for seropositive Honduran donors (32%) was higher than the PCR positivity rate for samples from seropositive US donors (14%; p= 0.02). PCR positivity for seropositive Brazilian donors (57%) was also higher than the PCR positivity rate for samples from seropositive US donors (14%; p<0.0001). The rates were statistically indistinguishable by country for ARC (p=0.15); 32% for Brazilian, 25%‘ for Honduran, and 24% for US seropositive donor samples.
Figure 2 presents the Ortho ELISA antibody reactivity levels (S/CO) for PCR-positive and negative samples, based on results of the BSRI PCR assay, by country; similar results were observed when the ARC PCR assay was used (data not shown). The overall mean S/CO values of the Brazilian donor samples was higher than for the Honduran and US samples, probably due to the selection criteria for enrollment that required reactivity on three Brazilian screening and confirmatory assays available a decade ago. Nevertheless, for all three countries, mean S/CO values were consistently higher among PCR-positive (6.30 for Brazil, 6.33 for Honduras, and 5.30 for US) as compared to PCR-negative samples (5.65 for Brazil, 4.27 for Honduras, and 3.68 for US) (p=0.03, p=0.04, and p<0.0001 respectively). Overall, there were 101 confirmed seropositive samples with PCR results that had S/CO values below 4.0 and 242 with S/CO values equal to or greater than 4.0. The PCR positivity rate was significantly lower in the low-level seroreactive group (5/101; 5 %) compared to the high level seroreactive group (109/242; 45%) (p<0.0001).
Change in antibody levels over time among PCR-positive and PCR-negative seropositive donors
The change over time in S/CO values for the 143 Brazilian donors for which index donation and 10-year follow-up samples were available for serological testing were analyzed (Table 2). The BSRI PCR results were used for these analyses since the sensitivity and specificity of the BSRI PCR assay were higher than the ARC PCR assays. Of note, the index blood donation plasma samples had two factors that could influence antibody reactivity: a) the plasma was obtained from a whole blood bag that was diluted by approximately 15% (~400 mL of collected blood with 65mL anticoagulant), and b) the plasma unit bags and resulting aliquots had been stored at −20C for over 10 years. Consequently, for 77% of the cases that were PCR positive and 62% of the cases that were PCR negative, S/CO values were higher with the follow-up sample (undiluted EDTA plasma maintained at −80C) compared to the index donation plasma bag sample. However, significant (>1 S/CO unit) antibody declines could still be detected in six (10%) of the 62 PCR-negative cases, and one of those samples became negative (S/CO = 0.90) by the Ortho ELISA assay, whereas none of the PCR positive cases has this level of decline of seroreactivity (Table 2). The PCR-negative group had a greater level of antibody decline as compared to the PCR-positive group (6/62 versus 0/81, respectively; p=0.003).
Table 2.
ELISA S/CO change between the index donation and follow-up samples from Brazilian seropositive donors according to BSRI PCR results.
| Level of Change in Antibody Reactivity (Index donation S/CO – Follow-up S/CO) |
PCR Positive (N=81) Number (%) |
PCR Negative (N=62) Number (%) |
|---|---|---|
| Increase greater than 1 | 16 (20%) | 4 (6%) |
| Increase less than 1, but greater than no change | 46 (57%) | 35 (56%) |
| No change or decrease less than 1 | 19 (24%) | 17 (27%) |
| Decrease greater than or equal to 1 | 0 | 6 (10%) |
Discussion
The widely accepted concept for the natural history of Chagas disease is that after acute infection all individuals remain infected for life1,15,16. Although widely cited and the basis for recommending treatment of all seropositive subjects17,18, to our knowledge this concept has never been formally proven. Seroreversion in the absence of treatment has been previously described as a rare event19-21 and also documented among untreated controls in benznidazole clinical trials 22,23, but the generally accepted explanation for these “rare cases” of possible seroreversion has been lack of reproducibility of the original serological assays.
Blood donor screening is a setting in which the concept that individuals remain infected for life carries important consequences. With contemporary donor screening assays, which are significantly more sensitive than older tests, approximately 30% of all individuals with confirmed positive results have low antibody titers which may not be detected by different screening tests3,24 . For those who believe that Chagas infection is lifelong, the development of more sensitive antibody screening assays has been considered a major advance for donor screening, clinical diagnostics and Chagas disease research16,18. Another direct consequence of this increase in sensitivity of screening tests is that parallel or serial application of screening tests for Chagas diagnosis, as WHO recommends, requires use of comparably sensitive assays. Furthermore, based on inconsistent detection of borderline reactive samples the FDA required that manufacturers of recently licensed T. cruzi donor screening assays in the US decrease the cut-off threshold values, resulting in detection of even lower level seroreactivity.
Early studies using PCR techniques to detect T. cruzi parasitemia were very optimistic, indicating a sensitivity of 100%12. Subsequent studies, however, were unable to reproduce these results, with rates of PCR positivity among seropositive individuals (detected by relatively insensitive serological assays) ranging from 20% to 70%8. These discrepancies may be due to differences in populations studied (country, diagnostic vs donor screening settings, acute or chronic disease status, age or time since last exposure, etc.); parasite strains present in the samples/population; or the PCR techniques employed (parasite genomic or kinetoplast DNA sequences, primers, volume of blood collected and processed, extraction procedures, and procedures taken to avoid amplicon contamination). Since the studies were generally focused on one population of seropositive subjects, no firm conclusions could be reached regarding the rate of DNA positivity as an indicator of parasitemia or association with antibody reactivity.
The lack of reliable assays to detect parasite or parasite DNA in peripheral blood has been a major obstacle to understanding the clinical significance of weakly seroreactive samples. Our study addressed this problem by first comparing the performance of two PCR assays, initially using a panel of samples spiked with known numbers of T. cruzi, followed by a large set of clinical specimens collected in three different countries. In the initial validation process, both techniques seemed to perform well. One parasite copy per 20mL blood was the lowest concentration that the ARC procedure was able to detect; the detection level of one parasite copy per 20mL of processed blood was achieved at Ct values of 37 to 39, near the cutoff value of the assay (Ct=40). In contrast, the BSRI assay detected samples spiked with 0.5 parasites per 20mL at a Ct value of 38 and detected one parasite per 20mL at 35 Cts (5 Cts below the assay Ct cutoff threshold of 40), providing excellent discrimination between target DNA signal and background. However, the clinical sensitivities between the two PCR assays were similar at 33% for BSRI and 28% for the ARC (although some of the discordant positive samples detected by the ARC lab could have been false positives given the 94% specificity of that test). Increased analytic sensitivity by the BSRI assay may be due to the five-fold greater volume of extracted DNA that was amplified per sample in the BSRI assay compared to the ARC assays (25uL vs 5uL of DNA, respectively). This higher input was possible through the use of the Hemobind extraction method developed for processing lysed whole blood samples and the magnetic bead target-capture method that has been optimized by Gen-Probe for routine NAT screening assays (i.e., HIV, HBV, HCV and WNV). The extraction and target capture approach that concentrates the parasite target DNA and also decreases human background DNA in the purified nucleic acid sample subjected to PCR may also be responsible for the improved specificity of the BSRI PCR assay.
We showed that higher antibody levels tended to be associated with higher rates of PCR reactivity, independent of the country in which the seropositive donors were identified. The overall higher rate of PCR positivity for the Brazilian donor samples was likely attributable to higher level of seroreactivity in these donor samples, which was due to the selection criteria that required reactivity on three Brazilian screening and confirmatory antibody assays available a decade ago. Using contemporary ELISAs, these Brazilian samples had much higher S/CO values compared to seropositive donor samples from the US or Honduras.
Of note, three of the six cases demonstrating an S/CO decline had index sample S/CO values below 4.0, which was much more commonly seen in the US and Honduran samples than in the pre-selected Brazilian cases. Low S/CO values were associated with a much lower rate of PCR positivity than observed in donors with S/CO values above 4.0 (5% vs 45%, respectively). In contrast, for the majority of PCR-negative Brazilian donor samples, seroreactivity was relatively high at baseline and stable or increased over time, suggesting active infection that PCR assays were not able to detect.
We hypothesize that follow-up of a larger group of donors with low-level seroreactivity, such as seen in 1/3 of US and Honduran as well as Brazilian donors screened with contemporary ELISAs22, would document moderate rates of seroreversion. We are planning follow-up studies of US and Brazilian donors with such low-level reactivity to confirm this hypothesis, and thus support the premise that such donors have resolved infection as demonstrated by persistent absence of parasite DNA combined with declining antibody reactivity reflecting lack of ongoing antigenic stimulation. This would result in revision of the concept for the natural history of Chagas disease to perhaps reflect that observed for malaria, where in the absence of repeat exposures, particularly early in life, infections usually resolve spontaneously or following treatment and patients serorevert over time, returning to normal baseline levels25,26.
Demonstrating resolution of infection in the absence of treatment has many implications including the need to develop confirmatory assays/algorithms that can differentiate active from non-active infection based on a combination of antibody and PCR results. This would allow improved donor counseling and reentry of donors who serorevert and test PCR negative. Lastly, the findings of this study suggest that recommendation of treatment with current drugs that have serious side effects and are only moderately efficacious may not be appropriate for individuals with the potential for resolved infections (PCR-negative and low-level seroreactivity)...
ACKNOWLEDGMENTS
The Retrovirus Epidemiology Donor Study-II (REDS-II), International Component (Brazil) is the responsibility of the following persons:
Blood Centers:
Fundação Pró-Sangue/Hemocentro São Paulo (São Paulo): Ester C. Sabino, Cesar de Almeida-Neto, Alfredo Mendrone-Jr, Ligia Capuani, Nanci A Salles
Hemominas (Belo Horizonte, Minas Gerais): Anna Bárbara de Freitas Carneiro-Proietti, Fernando Augusto Proietti, Claudia Di Lorenzo Oliveira, Carolina Miranda
Fundação Hemope (Recife, Pernambuco): Divaldo de Almeida Sampaio, Silvana Ayres Carneiro Leão, Maria Inês Lopes
DataWarehouse:
University of São Paulo (São Paulo): João Eduardo Ferreira,Márcio Oikawa, Pedro Losco Takecian
US Investigators:
Blood Systems Research Institute and University of California at San Francisco: M.P. Busch, E.L. Murphy, B. Custer, T. Gonçalez
Coordinating Center: Westat, Inc.: J. Schulman, M. King, K. Kavounis
National Heart, Lung, and Blood Institute, NIH:
S.A. Glynn.
Footnotes
The authors certify that they do not have a conflict of interest or financial involvement related to this manuscript.
References
- 1.Rassi A, Jr., Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375:1388–402. doi: 10.1016/S0140-6736(10)60061-X. [DOI] [PubMed] [Google Scholar]
- 2.Chang CD, Cheng KY, Jiang LX, Salbilla VA, Haller AS, Yem AW, Bryant JD, Kirchhoff LV, Leiby DA, Schochetman G, Shah DO. Evaluation of a prototype Trypanosoma cruzi antibody assay with recombinant antigens on a fully automated chemiluminescence analyzer for blood donor screening. Transfusion. 2006;46:1737–44. doi: 10.1111/j.1537-2995.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- 3.Otani MM, Vinelli E, Kirchhoff LV, del Pozo A, Sands A, Vercauteren G, Sabino EC. WHO comparative evaluation of serologic assays for Chagas disease. Transfusion. 2009;49:1076–82. doi: 10.1111/j.1537-2995.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 4.Saez-Alquezar A, Sabino EC, Salles N, Chamone DF, Hulstaert F, Pottel H, Stoops E, Zrein M. Serological confirmation of Chagas’ disease by a recombinant and peptide antigen line immunoassay: INNO-LIA chagas. J Clin Microbiol. 2000;38:851–4. doi: 10.1128/jcm.38.2.851-854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah DO, Chang CD, Cheng KY, Salbilla VA, Adya N, Marchlewicz BA, Kirchhoff LV. Comparison of the analytic sensitivities of a recombinant immunoblot assay and the radioimmune precipitation assay for the detection of antibodies to Trypanosoma cruzi in patients with Chagas disease. Diagn Microbiol Infect Dis. 2010;67:402–5. doi: 10.1016/j.diagmicrobio.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Tobler LH, Contestable P, Pitina L, Groth H, Shaffer S, Blackburn GR, Warren H, Lee SR, Busch MP. Evaluation of a new enzyme-linked immunosorbent assay for detection of Chagas antibody in US blood donors. Transfusion. 2007;47:90–6. doi: 10.1111/j.1537-2995.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 7.Custer B, Agapova M, Bruhn R, Cusick R, Kamel H, Tomasulo P, Biswas H, Tobler L, Lee TH, Caglioti S, Busch M. Epidemiologic and laboratory findings from 3 years of testing United States blood donors for Trypanosoma cruzi. Transfusion. 2012 doi: 10.1111/j.1537-2995.2012.03569.x. [DOI] [PubMed] [Google Scholar]
- 8.Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, Mejia Jaramillo AM, Cura C, Auter F, Veron V, Qvarnstrom Y, Deborggraeve S, Hijar G, Zulantay I, Lucero RH, Velazquez E, Tellez T, Sanchez Leon Z, Galvao L, Nolder D, Monje Rumi M, Levi JE, Ramirez JD, Zorrilla P, Flores M, Jercic MI, Crisante G, Anez N, De Castro AM, Gonzalez CI, Acosta Viana K, Yachelini P, Torrico F, Robello C, Diosque P, Triana Chavez O, Aznar C, Russomando G, Buscher P, Assal A, Guhl F, Sosa Estani S, DaSilva A, Britto C, Luquetti A, Ladzins J. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis. 2011;5:e931. doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leiby DA, Rentas FJ, Nelson KE, Stambolis VA, Ness PM, Parnis C, McAllister HA, Jr., Yawn DH, Stumpf RJ, Kirchhoff LV. Evidence of Trypanosoma cruzi infection (Chagas’ disease) among patients undergoing cardiac surgery. Circulation. 2000;102:2978–82. doi: 10.1161/01.cir.102.24.2978. [DOI] [PubMed] [Google Scholar]
- 10.Kirchhoff LV, Gam AA, Gusmao RA, Goldsmith RS, Rezende JM, Rassi A. Increased specificity of serodiagnosis of Chagas’ disease by detection of antibody to the 72- and 90-kilodalton glycoproteins of Trypanosoma cruzi. J Infect Dis. 1987;155:561–4. doi: 10.1093/infdis/155.3.561. [DOI] [PubMed] [Google Scholar]
- 11.Lee TH, Kleinman SH, Wen L, Montalvo L, Todd DS, Wright DJ, Tobler LH, Busch MP. Distribution of parvovirus B19 DNA in blood compartments and persistence of virus in blood donors. Transfusion. 2011;51:1896–908. doi: 10.1111/j.1537-2995.2010.03035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avila HA, Pereira JB, Thiemann O, De Paiva E, DeGrave W, Morel CM, Simpson L. Detection of Trypanosoma cruzi in blood specimens of chronic chagasic patients by polymerase chain reaction amplification of kinetoplast minicircle DNA: comparison with serology and xenodiagnosis. J Clin Microbiol. 1993;31:2421–6. doi: 10.1128/jcm.31.9.2421-2426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virreira M, Torrico F, Truyens C, Alonso-Vega C, Solano M, Carlier Y, Svoboda M. Comparison of polymerase chain reaction methods for reliable and easy detection of congenital Trypanosoma cruzi infection. Am J Trop Med Hyg. 2003;68:574–82. doi: 10.4269/ajtmh.2003.68.574. [DOI] [PubMed] [Google Scholar]
- 14.Piron M, Fisa R, Casamitjana N, Lopez-Chejade P, Puig L, Verges M, Gascon J, Gomez i Prat J, Portus M, Sauleda S. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop. 2007;103:195–200. doi: 10.1016/j.actatropica.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Marin-Neto JA, Cunha-Neto E, Maciel BC, Simoes MV. Pathogenesis of chronic Chagas heart disease. Circulation. 2007;115:1109–23. doi: 10.1161/CIRCULATIONAHA.106.624296. [DOI] [PubMed] [Google Scholar]
- 16.Tarleton RL, Reithinger R, Urbina JA, Kitron U, Gurtler RE. The challenges of Chagas Disease-- grim outlook or glimmer of hope. PLoS Med. 2007;4:e332. doi: 10.1371/journal.pmed.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bern C, Montgomery SP, Herwaldt BL, Rassi A, Jr., Marin-Neto JA, Dantas RO, Maguire JH, Acquatella H, Morillo C, Kirchhoff LV, Gilman RH, Reyes PA, Salvatella R, Moore AC. Evaluation and treatment of chagas disease in the United States: a systematic review. JAMA. 2007;298:2171–81. doi: 10.1001/jama.298.18.2171. [DOI] [PubMed] [Google Scholar]
- 18.Lescure FX, Le Loup G, Freilij H, Develoux M, Paris L, Brutus L, Pialoux G. Chagas disease: changes in knowledge and management. Lancet Infect Dis. 2010;10:556–70. doi: 10.1016/S1473-3099(10)70098-0. [DOI] [PubMed] [Google Scholar]
- 19.Dias JC, Dias E, Martins-Filho OA, Vitelli-Avelar D, Correia D, Lages E, Prata A. Further evidence of spontaneous cure in human Chagas disease. Rev Soc Bras Med Trop. 2008;41:505–6. doi: 10.1590/s0037-86822008000500014. [DOI] [PubMed] [Google Scholar]
- 20.Francolino SS, Antunes AF, Talice R, Rosa R, Selanikio J, de Rezende JM, Romanha AJ, Dias JC. New evidence of spontaneous cure in human Chagas’ disease. Rev Soc Bras Med Trop. 2003;36:103–7. doi: 10.1590/s0037-86822003000100014. [DOI] [PubMed] [Google Scholar]
- 21.Zeledon R, Dias JC, Brilla-Salazar A, de Rezende JM, Vargas LG, Urbina A. Does a spontaneous cure for Chagas’ disease exist? Rev Soc Bras Med Trop. 1988;21:15–20. doi: 10.1590/s0037-86821988000100003. [DOI] [PubMed] [Google Scholar]
- 22.de Andrade AL, Zicker F, de Oliveira RM, Almeida Silva S, Luquetti A, Travassos LR, Almeida IC, de Andrade SS, de Andrade JG, Martelli CM. Randomised trial of efficacy of benznidazole in treatment of early Trypanosoma cruzi infection. Lancet. 1996;348:1407–13. doi: 10.1016/s0140-6736(96)04128-1. [DOI] [PubMed] [Google Scholar]
- 23.Viotti R, Vigliano C, Lococo B, Bertocchi G, Petti M, Alvarez MG, Postan M, Armenti A. Long-term cardiac outcomes of treating chronic Chagas disease with benznidazole versus no treatment: a nonrandomized trial. Ann Intern Med. 2006;144:724–34. doi: 10.7326/0003-4819-144-10-200605160-00006. [DOI] [PubMed] [Google Scholar]
- 24.Sabino EC, Salles NA, Sarr M, Barreto AM, Oikawa M, Oliveira CD, Leao SC, Carneiro-Proietti AB, Custer B, Busch MP. Enhanced classification of Chagas serologic results and epidemiologic characteristics of seropositive donors at three large blood centers in Brazil. Transfusion. 2010;50:2628–37. doi: 10.1111/j.1537-2995.2010.02756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss GE, Traore B, Kayentao K, Ongoiba A, Doumbo S, Doumtabe D, Kone Y, Dia S, Guindo A, Traore A, Huang CY, Miura K, Mircetic M, Li S, Baughman A, Narum DL, Miller LH, Doumbo OK, Pierce SK, Crompton PD. The Plasmodium falciparum-specific human memory B cell compartment expands gradually with repeated malaria infections. PLoS Pathog. 2010;6:e1000912. doi: 10.1371/journal.ppat.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soe S, Khin Saw A, Htay A, Nay W, Tin A, Than S, Roussilhon C, Perignon JL, Druilhe P. Premunition against Plasmodium falciparum in a malaria hyperendemic village in Myanmar. Trans R Soc Trop Med Hyg. 2001;95:81–4. doi: 10.1016/s0035-9203(01)90342-6. [DOI] [PubMed] [Google Scholar]