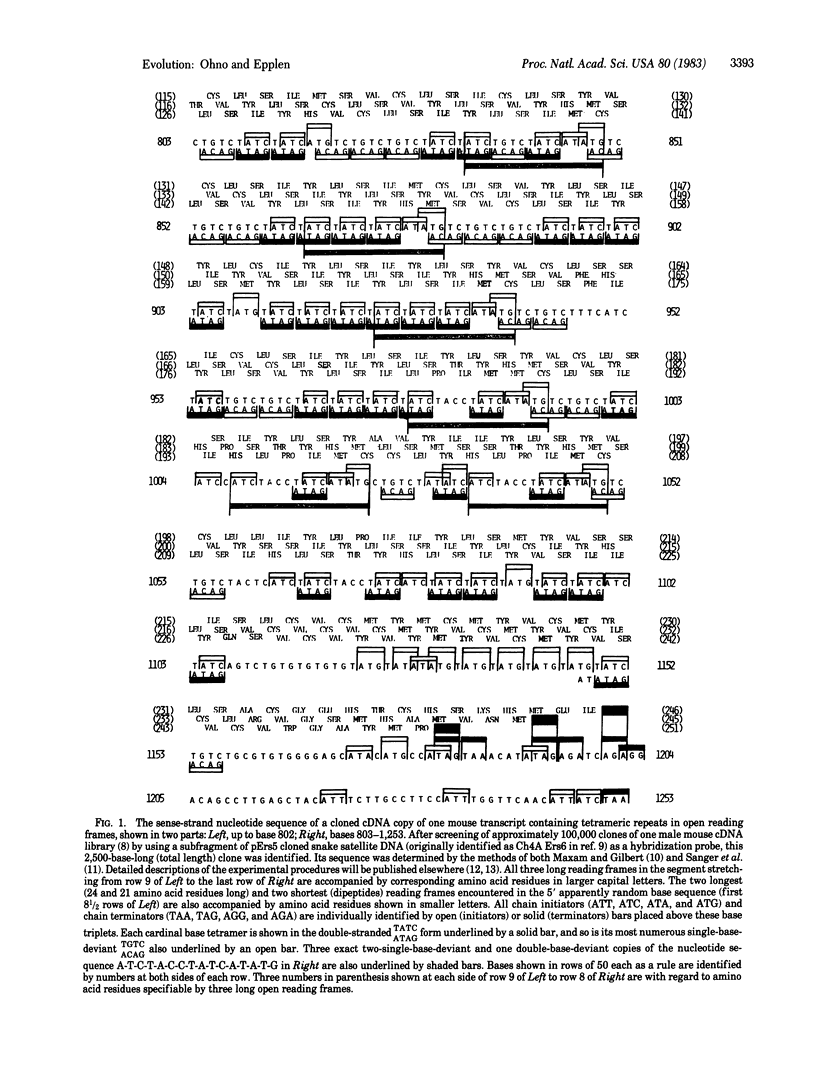
Abstract
Even if the prebiotic self-replication of nucleic acids and the subsequent emergence of primitive, enzyme-independent tRNAs are accepted as plausible, the origin of life by spontaneous generation still appears improbable. This is because the just-emerged primitive translational machinery had to cope with base sequences that were not preselected for their coding potentials. Particularly if the primitive mitochondria-like code with four chain-terminating base triplets preceded the universal code, the translation of long, randomly generated, base sequences at this critical stage would have merely resulted in the production of short oligopeptides instead of long polypeptide chains. We present the base sequence of a mouse transcript containing tetranucleotide repeats conserved during evolution. Even if translated in accordance with the primitive mitochondria-like code, this transcript in its three reading frames can yield 245-, 246-, and 251-residue-long tetrapeptidic periodical polypeptides that are already acquiring longer periodicities. We contend that the first set of base sequences translated at the beginning of life were such oligonucleotide repeats. By quickly acquiring longer periodicities, their products must have soon gained characteristic secondary structures--alpha-helical or beta-sheet or both.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bridson P. K., Orgel L. E. Catalysis of accurate poly(C)-directed synthesis of 3'-5'-linked oligoguanylates by Zn2+. J Mol Biol. 1980 Dec 25;144(4):567–577. doi: 10.1016/0022-2836(80)90337-x. [DOI] [PubMed] [Google Scholar]
- Crick F. H. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Epplen J. T., McCarrey J. R., Sutou S., Ohno S. Base sequence of a cloned snake W-chromosome DNA fragment and identification of a male-specific putative mRNA in the mouse. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3798–3802. doi: 10.1073/pnas.79.12.3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T. H. Mitochondrial codes and evolution. Nature. 1983 Jan 6;301(5895):19–20. doi: 10.1038/301019a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ohno S., Matsunaga T., Epplen J. T., Itakura K., Wallace R. B. Identification of the 45-base-long primordial building block of the entire class I major histocompatibility complex antigen gene. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6342–6346. doi: 10.1073/pnas.79.20.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Matsunaga T., Wallace R. B. Identification of the 48-base-long primordial building block sequence of mouse immunoglobulin variable region genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1999–2002. doi: 10.1073/pnas.79.6.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L. E. Evolution of the genetic apparatus. J Mol Biol. 1968 Dec;38(3):381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- Reyes A. A., Johnson M. J., Schöld M., Ito H., Ike Y., Morin C., Itakura K., Wallace R. B. Identification of an H-2Kb-related molecule by molecular cloning. Immunogenetics. 1981;14(5):383–392. doi: 10.1007/BF00373318. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Singh L., Jones K. W. Sex reversal in the mouse (Mus musculus) is caused by a recurrent nonreciprocal crossover involving the x and an aberrant y chromosome. Cell. 1982 Feb;28(2):205–216. doi: 10.1016/0092-8674(82)90338-5. [DOI] [PubMed] [Google Scholar]
- Singh L., Purdom I. F., Jones K. W. Conserved sex-chromosome-associated nucleotide sequences in eukaryotes. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):805–814. doi: 10.1101/sqb.1981.045.01.099. [DOI] [PubMed] [Google Scholar]
- Sümegi J., Wieslander L., Daneholt B. A hierarchic arrangement of the repetitive sequences in the Balbiani ring 2 gene of Chironomus tentans. Cell. 1982 Sep;30(2):579–587. doi: 10.1016/0092-8674(82)90254-9. [DOI] [PubMed] [Google Scholar]
- Yazaki A., Ohno S. Recurrence of 49-base decamers, nonomers, and octamers within mouse C mu gene of Ig heavy chain and its primordial building block. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2337–2340. doi: 10.1073/pnas.80.8.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]


