Abstract
The tailed double-stranded DNA bacteriophages, or Caudovirales, constitute ~96% of all the known phages. Although these phages come in a great variety of sizes and morphology, their virions are mainly constructed of similar molecular building blocks via similar assembly pathways. Here we review the structure of tailed double-stranded DNA bacteriophages at a molecular level, emphasizing the structural similarity and common evolutionary origin of proteins that constitute these virions.
Keywords: bacteriophage structure, Caudovirales, tailed double-stranded DNA bacteriophages, virus structure, virus capsids, phage tails, phage proteins
Introduction
Bacteriophages are probably the most abundant entities in the biosphere. The total number of phage particles is estimated to be in the order of 1031, 10 times larger than the estimated number of bacterial cells on Earth.1,2 Among ~5500 phages examined by electron microscopy (EM), ~96% have tails,3 a special organelle designed for the host recognition, cell wall penetration, and genome ejection into the host. The tailed phages constitute the order Caudovirales, which is divided into 3 families: Myoviridae, Siphoviridae, and Podoviridae, based on the tail morphology. Myoviridae phages (e.g., T4, φ92, φKZ) are characterized by long straight contractile tails, Siphoviridae phages (e.g., λ, HK97, SPP1, p2, TP901-1) possess long flexible non-contractile tails, and Podoviridae phages (e.g., φ29, T7, P22) have short, stubby, non-contractile tails (Fig. 1). Of the tailed phages analyzed with electron microscopy, 61% belong to the Siphoviridae family, 25% to the Myoviridae family, and 14% to the Podoviridae family.3,7
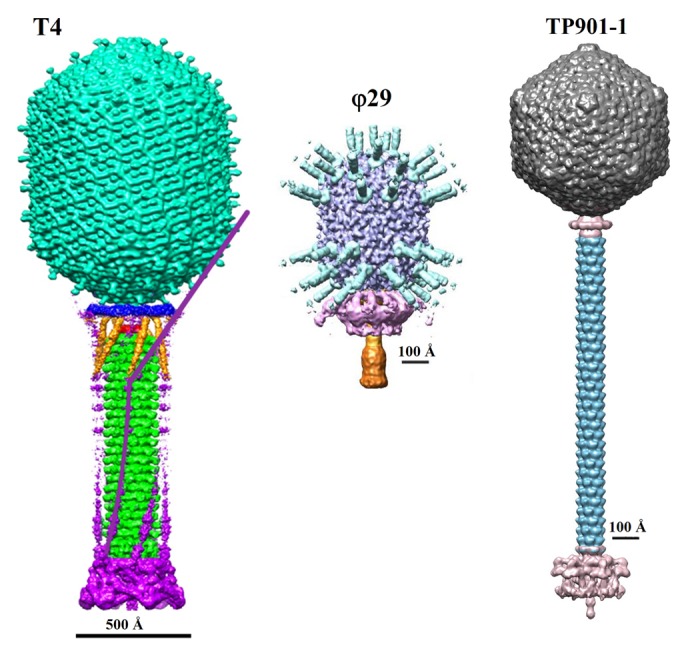
Figure 1. Structure of the Myoviridae phage T4 (left), Podoviridae phage φ29 (middle), and Siphiviridae phage TP901-1 (right). The left panel was reproduced from reference 4, the middle panel from reference 5, and the right panel from reference 6.
During the past decade or so tremendous progress has been made in obtaining 3-dimensional (3D) structures of the phage virions and their protein components. Based on the similar folds of the structural proteins, most likely all Caudovirales had a common evolutionary origin, despite large differences in virion sizes and morphology. Their virions are mainly built from similar molecular building blocks, via similar assembly pathways. Here we describe the structure of Caudovirales phages at a molecular level. We emphasize the structural similarity and evolutionary relationships between proteins which form the phage virions and participate in the virus assembly.
Structure of the Phage Head
Capsids of the tailed phages come in a large variety of sizes ranging in diameter from about 400 to 1700 Å.3 Most of Caudovirales (75%) examined in the electron microscope3 have icosahedral (isometric) capsids, while about 15% of Caudovirales (e.g., φ29 and T4) have prolate heads, which are icosahedrons elongated along the 5-fold axis coincident with the axis of the phage tail. The capsid contains linear double-stranded (ds) DNA chromosome which is packaged to a high density of ~500 g/l and exerts internal pressure of tens of atmospheres on the capsid walls.8-10 The special pentameric vertex of the phage capsids, to which the tail is attached, is occupied by a dodecameric portal protein, also called a connector, which forms a channel for genome packaging during the virion assembly and exit during the infection process.
Capsid assembly
Caudovirales assemble their capsids with the help of internal scaffolding proteins (Fig. 2).11-13 The head assembly typically starts at the portal vertex by copolymerization of the scaffolding proteins and the major capsid protein leading to formation of the capsid precursors called prohead (or procapsid). The prohead consists of the portal protein, the internal scaffolding core, and the outer major capsid protein shell surrounding the core. In comparison to the mature capsid, the prohead has a smaller size, a more rounded shape, and a thicker capsid protein shell.14-17 Most phages encode a separate scaffolding protein which is the main part of the internal core. The scaffolding protein ensures the correct geometry of the capsid. In its absence the capsid protein usually assembles into aberrant structures. Some phages, like HK97 and T5, have N-terminal major capsid protein regions (called Δ domains), which act as scaffolding proteins.14,18,19 The scaffolding proteins show great variability in their sizes and amino-acid sequences. For example, the scaffolding proteins of the bacteriophages P22 and φ29 have 100 and 303 residues, respectively and have no sequence similarity. However, the φ29 scaffolding protein20 and the C-terminal part of the P22 scaffolding protein16,21 contain a similar helix-turn-helix motif, which, in the P22 procapsid, interacts with the major capsid protein shell and the portal. Apart from the scaffolding protein the internal procapsid core may contain several extra proteins. For example, the core of the bacteriophage T4 prohead contains, in addition to ~580 copies of the scaffolding protein, 7 extra proteins with copy numbers ranging from 40 to 370.13,22
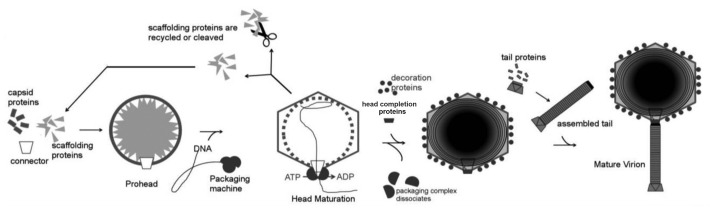
Figure 2. Schematic representation of the bacteriophage assembly. Reproduced from reference 11.
Procapsids of many phages (like T4, HK97, φKZ, and λ) contain head maturation proteases (serine proteases in case of T413,22 and φKZ23) which are activated after prohead assembly and degrade the internal core partially or completely. The digestion products exit the prohead, freeing the space for the genomic DNA. The genome is packaged into the procapsid through the portal vertex by a DNA translocation motor, driven by ATP hydrolysis.24-27 In some phages, like P22 and φ29, the scaffolding protein is not degraded, but removed from the capsid intact during the DNA packaging. The scaffolding protein is probably pushed out by the entering DNA, and exits the capsid via openings in the prohead shell.16 As DNA is being packaged the prohead undergoes large structural rearrangement to become the mature capsid. The capsid maturation involves reorientation and partial refolding of the major protein subunits,14,15 resulting in significant increase (by ~50%) of the capsid volume. In phage λ the expansion occurs when ~30% of the genome has been packaged.28 In phage T4 the expansion can be decoupled from DNA packaging.29 Capsids of many phages attach stabilization and/or decoration proteins to their outer surface during the final stage of maturation (see below).
Once the genome has been packaged, the DNA translocation motor is dislodged from the portal vertex. The capsid assembly is then finalized by attachment of the head completion proteins, which seal the portal gate30 and provide a hub for the tail assembly in podophages or a platform for binding of the preassembled tail in sipho- and myophages.
Genome packaging
Genomes of Caudovirales range in size from about 15 to 500 kilo base pairs.2,31,32 DNA of some phages, contains modified nucleotides, which helps to protect the genomes against nuclease degradation. For instance, chromosomes of T-even phages contain glucosylated hydroxymethylcytosine residues.33
DNA of most Caudovirales, as well as herpesviruses, is synthesized as long head-to-tail multimers, or concatemers, of genome units.24,34 The phage chromosomes are cut from the concatemers by the DNA packaging machine, called the terminase complex because it creates the termini of the virion DNA.24,25,27 The packaging machine of most phages contains 2 proteins termed the large and the small terminases. Subunits of the large terminase, usually contain 2 domains: an ATPase domain and a nuclease domain. The large terminase is responsible for the DNA cleavage and translocation into the procapsid, while the small terminase is involved in the packaging initiation and stimulates the ATPase activity of the large terminase.
The first genome packaging cycle is typically started by the small terminase which specifically recognizes phage DNA and recruits the large terminase to make the initial cut in the DNA concatemer. After the cut, the large and small terminase associated with one end of the cleaved DNA concatemer bind to the portal vertex of a prohead. The large terminase then starts the DNA translocation fueled by ATP hydrolysis. Phage terminases are among the strongest and fastest known molecular machines.35 The T4 packaging motor has an average translocation speed of ~700 bp/sec, and can achieve rates as high as 2000 bp/sec.35,36 The motor can exert forces larger than 60 pN to overcome the pressure generated inside the capsid by the packaged DNA.9,35 At the end of the packaging cycle the large terminase makes another cut to separate the packed phage chromosome from the rest of the DNA concatemer. The motor/DNA complex then dislodges from the filled head and binds to another DNA-free procapsid to start the next packaging cycle.
In λ-like and T7-like phages the packaging starts and stops at specific sites, called cos or pac.34 In λ-like phages the large terminase generates 12-base cohesive ends of the phage chromosomes. The T7 chromosomes have blunt ends and are flanked by direct terminal repeats (terminal redundancies), generated in concert with DNA packaging.
Many phages, like P22, SPP1, use the head-full mechanism for packaging termination.34 The first packaging cycle starts at a specific pac site and continues until the capsid is full. These phages encapsidate more than one complete genome length (typically 102–110%), and, therefore, the second DNA cut occurs at a non-specific site (beyond the pac sequence). Consequently, the following DNA packaging cycles start and end at different sites, resulting in encapsidation of different circularly-permuted chromosomes with terminal redundancy. Phage T4, which also uses the head-full packaging mechanism, has no strict pac sequence. The first cut of it’s DNA concatemer can be made at different points.37,38
Bacteriophage φ29 synthetize monomeric genomes which have covalently linked gp3 proteins attached to the 5′-ends of both DNA strands.39 φ29 does not have a small terminase, and its large terminase does not have nuclease activity.40 Another unusual feature of φ29 is the presence of a viral-encoded structural RNA (called p-RNA) which resides between the portal protein and the terminase during genome packaging.41
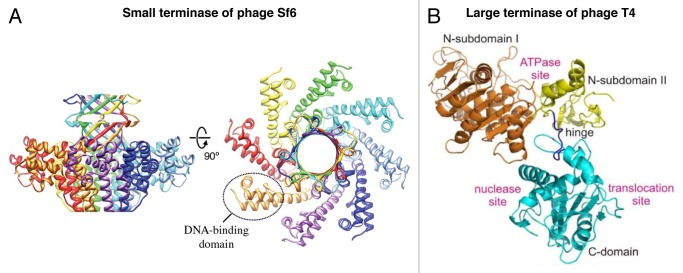
Crystal structures of the small terminases from podophages Sf6 (Fig. 3A)42,44 and P22,45 SPP1-like siphophage SF6,46 and a T4-like myophage 44RR47 have been determined. Comparison of these structures showed that the small terminases have similar organization. Their subunits assemble into vase-shaped oligomers with 8- to 12-fold rotation symmetry possessing a central pore. The small terminases consist of an N-terminal domain, located on the periphery of the vase, the central oligomerization domain, and the C-terminal domain which form the top of the vase and is responsible for interactions with the large terminase. Studies of the SF6, T4 and Sf6 small terminases44,46,47 showed that the N-terminal periphery region is responsible for binding to DNA. In the proposed small terminase-DNA binding models, DNA wraps around the side of the small terminase oligomer. An alternative way for DNA binding to the small terminases would be for DNA to go through the central pore. However it is unlikely because the central pores of the SF6 and Sf6 terminases are too narrow to accommodate the double-stranded DNA helix. Moreover, the small terminases of different phages have different oligomeric state suggesting that the number of subunits and consequently the diameter of the central pore is not critical for the small terminase function.46,47
Figure 3. Structures of the phage terminases. (A) Ribbon diagram of the small terminase of phage Sf6. Different chains of the small terminase octamer are shown in a different colors. (B) Ribbon diagram of the large terminase protein, gp17, of phage T4. The N-terminal ATPase domain of T4_gp17 is subdivided into 2 subdomains marked N-subdomain I and II. N-subdomain I is in gold, N-subdomain II is in yellow, and the C-terminal nuclease domain is in cyan. (A) was reproduced from reference 42, (B) from reference 43.
Structures of the large terminases of phages T4 (Fig. 3B) and Sf6 have been determined using X-ray crystallography.48,49 In addition crystal structures the large terminase nuclease domains of the phages SPP1, P22, and a T4-like phage RB69 are available.48,50,51 The N-terminal ATPase domains of large terminases have a classical oligonucleotide-binding fold,52 whereas their C-terminal nuclease domains have a ribonuclease H-like fold.53
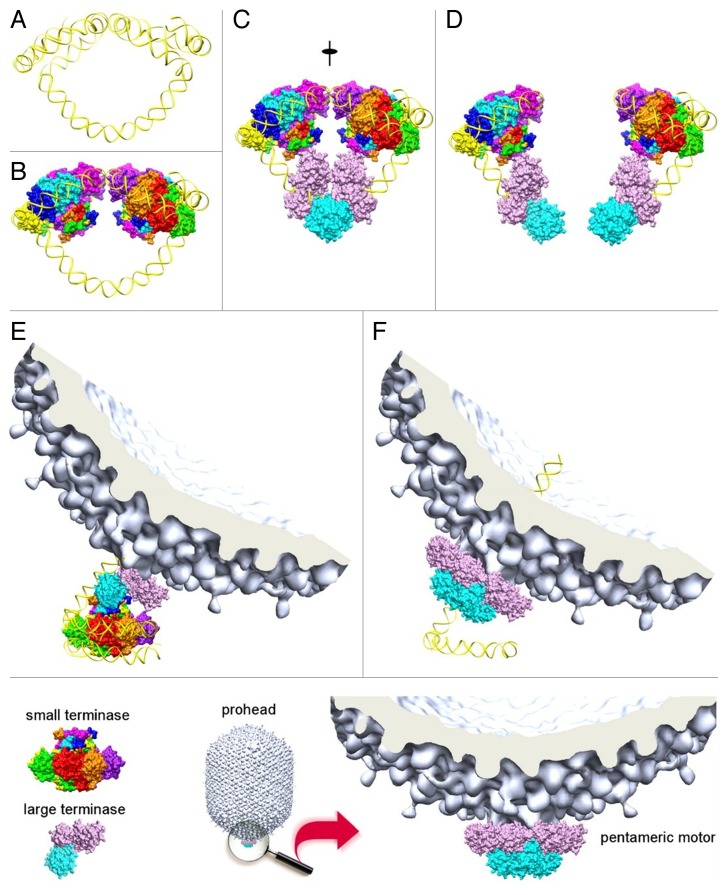
Based on the studies of the T4 large (gp17, 70 kDa) and small (gp16, 18 kDa) terminases, and the cryo-EM reconstruction of the T4 capsid complexed with the large terminase, a model for the packaging initiation and DNA translocation was proposed (Figs. 4 and 5).48 In this model, 2 vase-shaped oligomers of the small terminase and 2 molecules of the large terminase form a packaging initiation complex with the phage DNA concatemer. In the initiation complex, the terminase molecules have a 2-fold-symmetric arrangement that corresponds to the 2-fold symmetry of the DNA molecule at the anticipated cleavage site. The large terminase molecules make cuts on each strand of DNA generating free ends. Then 1 large terminase molecule and 1 small terminase oligomer, associated with 1 end of DNA, bind to a procapsid portal vertex. This process recruits 4 other large terminase molecules to form a pentameric DNA translocation motor. The large terminase molecules attach to the portal vertex by their N-terminal domain in a 5-fold-symmetric fashion, suggesting that they interact not only with the 12-fold symmetric portal protein but also with the 5 major capsid proteins near the portal vertex.
Figure 4. A model for packaging initiation by the small terminase (reproduced from ref. 47). Framed area: The small terminase subunits are represented in rainbow colors. The N-terminal ATPase domain of the large terminase is colored purple and the C-terminal nuclease domain cyan. (A) Packaging initiation sites on the phage genomic DNA. (B) Two small terminase oligomers bound to the initiation sites. (C) Two large terminase molecules are recruited and cleave the DNA backbones associated with the same base pair. The nuclease domain of the large terminase on the left-hand side is obscured by the 2-fold related nuclease domain of the large terminase on the right-hand side. (D) Two free ends are generated on the DNA, and further digestion is prevented by the small terminase. (E) The initiation complex binds to an empty procapsid. (F) Four more large terminase molecules are recruited to assemble the pentameric motor and rapid DNA translocation causes the dissociation of the small terminase.
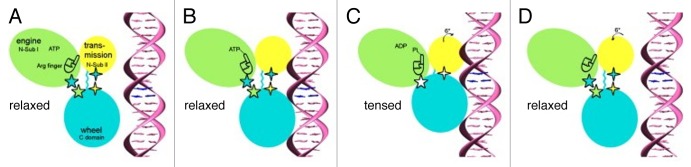
Figure 5. DNA Packaging by the large T4 terminase (reproduced from ref. 48). The figure shows the sequence of events that occur in a single T4 gp17 molecule during packaging. The gp17 N-terminal subdomain I, subdomain II, and C-terminal domain are represented as green, yellow, and cyan ovals, respectively. The 5-pointed stars show the charge interactions between the N-terminal subdomain I and the C-terminal domain. The 4-pointed stars show the charge interaction between the N-terminal subdomain II and the C-terminal domain. The flexible linker between N- and C-terminal domains is represented by a wiggly cyan line. (A) The gp17 C-terminal domain is ready to bind DNA. (B) The C-terminal domain, when bound to the DNA, brings the DNA closer to the N-terminal domain of the same subunit. Conformational change in the N-terminal domain causes Arg162 to be placed into the ATPase active center in preparation for hydrolysis. (C) Hydrolysis of ATP has rotated the N-terminal subdomain II by about 6°, thereby aligning the charge pairs resulting in an electrostatic attraction that moves the C-terminal domain and the DNA 6.8 Å (equivalent to the distance between 2 base pairs) closer to the N-terminal domain and into the capsid. (D) ADP and Pi are released and the C-terminal domain returns to its original position. DNA is released and is aligned to bind the C-terminal domain of the neighboring gp17 subunit.
The proposed DNA packaging model48 assumes that during the translocation process 5 gp17 molecules sequentially bind the DNA and move it into the capsid in 2 base pair steps. Comparison of the gp17 crystal structure with the cryo-EM reconstruction of the prohead-gp17 complex suggested that the gp17 molecules can adopt 2 conformational states: tensed and relaxed (Fig. 5). In the tensed state the ATPase and nuclease domains of gp17 are in close contact, whereas in the relaxed state they are separated by ~7 Å. When ATP binds to one of the gp17 molecules in the relaxed state, its affinity to DNA is increased. Then this gp17 molecule binds to DNA via its C-terminal nuclease domain. ATP hydrolysis switches the conformation of the gp17 molecule from the relaxed to the tensed state. As a result the C-terminal domain moves toward the N-terminal domain and the capsid by ~7 Å, causing the translocation of 2 base pairs of DNA. As the structure of B-form DNA contains 10 base pairs per turn, the translocation of 2 base pairs by 1 gp17 molecules brings the DNA into register with the adjacent gp17 molecule related by 5-fold symmetry. The neighboring gp17 molecule binds to DNA by its C-terminal domain to start the next translocation step. After release of the ATP hydrolysis products, the gp17 molecules return to their relaxed state.
Although the ATPase and nuclease domains of T4 and Sf6 terminases have similar folds, their relative orientation to each other in the crystal structures is different.48,49 Based on the crystal structure of the Sf6 terminase, Zhao et al.49 suggested an alternative mechanism for conversion of the ATP hydrolysis reaction into the physical motion of DNA. Single particle studies of the phage φ29 packaging motor suggest that DNA is translocated in 10 base pair bursts of consisting of 4 2.5 base pair steps.54 Various DNA packaging mechanisms have been proposed to account for non-integer translocation steps for the φ29 motor.40,54,55
Structure of the portal protein
The portal protein forms a conduit through which the DNA enters the capsid, during the virus formation, and exits, during the infection process. The portal protein is an attachment site for the DNA-packaging motor, and later for the head completion proteins.
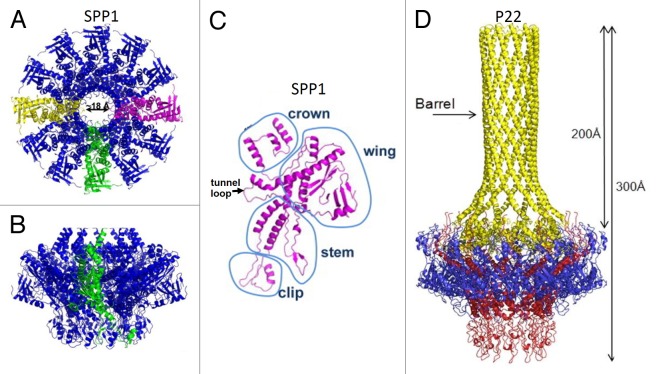
The portals of Caudovirales phages and herpesviruses are turbine-shaped molecules (Fig. 6) containing a central channel for DNA passage (for a recent reviews see refs. 30, 40, 56, and 58). When in the head the portal proteins are always dodecamers. However recombinantly produced portals assemble into oligomers of 11 to 14 subunits.59,60 The portal proteins show large variability in size and amino-acid sequences. For example, the portal subunits of podophages φ29 and P22 have molecular weights of 36 and 83 kDa, respectively. The structures of the portal proteins from phages φ29,57 SPP1,59 and P2261 have been determined using X-ray crystallography. Additionally the crystal structure of a putative portal protein encoded in the Corynebacterium diphtheriae genome has been determined (PDB ID: 3KDR). Despite low sequence homology, all these proteins have similar folds, suggesting that they have evolved from a common ancestor. The surface of the central channel of the portals is mainly negatively charged allowing smooth DNA passage. The monomer of the portal protein can be divided into the clip, stem, wing, and crown regions59 (Fig. 6C). The clip region protrudes to the capsid exterior and is involved in interactions with the large terminase and later with the head closure proteins. The clip is connected to the wing via the stem containing 2 long α helices. The wing and crown regions are located in the capsid interior and contact the packaged DNA in the mature capsids. The sizes of these regions vary significantly among the portals. The C-terminal part of the phage P22 portal forms a ~200 Å-long coil-coiled tube resembling a rifle barrel57 (Fig. 6D), which protrudes almost to the center of the phage capsid.
Figure 6. Structures of the portal proteins of phages SPP1 and P22. (A and B) display the top and side views of the SPP1 portal protein dodecamer, respectively. (C) Structure of the SPP1 portal protein monomer. (D) Side view of the P22 portal protein dodecamer. (A–C) were reproduced from reference 56, (D) from reference 57.
Due to the symmetry mismatch between the 12-fold symmetric portal and the 5-fold vertex it was proposed that the portal would rotate easily with respect to the capsid. In the early models of genome packaging, the DNA translocation was assumed to be coupled with the portal rotation.59,61 However later single-molecule fluorescence spectroscopy of the φ29 portal62 and studies of the T4 connector non-covalently cross-linked to the capsid shell63 showed that the portal does not rotate during the DNA translocation.
The portal appears to play a more significant role than that of a passive conduit. Point mutations in the SPP1 portal protein can affect the initiation of DNA packaging, ATP hydrolysis by the large terminase, and DNA translocation, suggesting that there is cross-talk between the portal and the terminase subunits during packaging.56,59,64,65 Covalent cross-linking of the stem helices, belonging to adjacent subunits of the SPP1 portal, abolishes packaging, suggesting that relative motion of subunits is important for the portal function.66 The SPP1 portal contains long tunnel loops which point into the channel and probably interact with DNA during translocation. Initially it was proposed that these loops grip tightly the DNA and drive it into the capsid.59 However the analogous loops in the P22 portal are too short to grip the DNA.57 Moreover, deletion of the tunnel loops in the φ29 portal did not disrupt the packaging, but rather affected the ability of the phage to retain the encapsidated DNA.67 The potential role of the tunnel loops in retaining the packaged DNA was also observed in the biochemical/bioinformatics study of the phage T4 portal.68
Once the phage chromosome has been encapsidated, a signal is sent to the motor causing the termination of packaging and dissociation of the motor from the portal vertex. In phages that use the head-full packaging mechanism (e.g., P22 and SPP1), the tightly packed DNA exerts a force onto the portal protein inducing conformational changes which are then transmitted to the motor.69,70
After the motor is jettisoned and before the closure of the vertex by the head completion proteins, the genome is retained in the capsid by the portal protein, which probably acts as a valve preventing leakage of DNA.59,67 In the φ29 and SPP1 portals the tunnel loops might be involved in the DNA retention. In the P22 portal the barrel is probably responsible for both the head-full signaling and the retention of DNA. Indeed, comparison of the cryo-EM reconstruction of the P22 capsid69 with the crystal structure of the portal57 showed that the packaged DNA induced significant conformational changes in the portal resulting in compression of the barrel tube. The cryo-EM reconstruction also showed the terminus of the genomic DNA entrapped inside the compressed barrel tube. The P22 portal barrel probably has extra functions in the packaging and ejection processes. Olia et al.57 suggested that during packaging, the barrel undulates in a circular motion while spooling the DNA outwards, onto the capsid shell, in a way reminiscent of a water sprinkler. This allows the capsid to be filled with ordered, circumcentric layers of DNA. Another proposed function of the barrel is the stabilization of the linear momentum of the genome as it exits the capsid during infection.57
Organization of the major capsid protein shell
In the mature Caudovirales capsids, subunits of the major capsid protein form pentameric and hexameric capsomers which are arranged on the capsid surface in accordance with the quasi-equivalence principle, proposed by Caspar and Klug.71 However immature phage capsids show significant departure from the quasi-equivalence and have skewed capsid protein hexamers which are 2-fold rather than 6-fold symmetric.14-17 Distance between the centers of adjacent capsomers in the mature Caudovirales heads is about 140 Å. The arrangement of the major capsid protein subunits in the isometric phage heads is determined by the triangulation number T, which is equal to the number of major capsid protein subunits in the icosahedral asymmetric unit. As the icosahedral symmetry has 60 asymmetric units, the total number of major capsid protein subunits in the phage head equals (60T – 5), where the second term accounts for the absence of the major capsid protein molecules in the portal vertex.
The prolate phage capsids are composed of 2 icosahedral caps and an elongated midsection. The symmetry of the major capsid protein shell in a prolate head is described by 2 triangulation numbers: T for icosahedral caps, and Q for the capsid midsection (for details see refs. 72 and 73). The total number of the major capsid molecules per a prolate head is defined by (30 (T + Q) – 5). These formulae assume that the major capsid protein is present in all pentameric vertices, except the portal vertex; however some phages, like T4, may encode special vertex proteins.74,75
Consistent with the wide range of genome and capsid sizes, a large variety of triangulation numbers have been found in Caudovirales phages studied by electron microscopy. For example, the prolate head of the small podophage φ29 is characterized by the triangulation numbers T = 3 and Q = 5,76,77 whereas the icosahedral capsid of the giant myophage G has T = 52 symmetry (James Conway and Roger Hendrix, unpublished data). However, capsids of many well-studied phages, like HK97,78 P22,16 SPP1,79 λ,80 and T7,81 have T = 7 laevo symmetry.
The major capsid proteins of Caudovirales have a similar fold, despite of large divergence in their sequences. The fold was first described in the bacteriophage Hong Kong 97 (HK97), for which the structure of the capsid was determined using X-ray crystallography (Fig. 7).78 Later, the HK97-fold was found all Caudovirales, with known 3D structures (for examples see refs. 16, 75, 77, 80, 81, and 83–85), as well as in herpesviruses,86 and bacterial enzyme-packaging compartments, called encapsulins.87
Figure 7. Structure of the bacteriophage HK97 major capsid protein shell. (A–C) correspond to the mature head. (D–F) correspond to the prohead. (A and D) display structures of the capsid protein subunits. (B and E) show entire heads and top views of capsid protein hexamers and pentamers. (C) and (F) show side views of capsid protein hexamers. (A) was reproduced from reference 82, (B–F) from ref. 15.
The HK97 major capsid protein, gene product (gp)5, is a 385-residue molecule, which is proteolytically processed during capsid maturation.14 The 103-residue N-terminal region of gp5, acting as a scaffold for capsid assembly, is subsequently cleaved in the maturation process by a head protease, resulting in a 282-residue gp5*, which is present in the mature capsid.
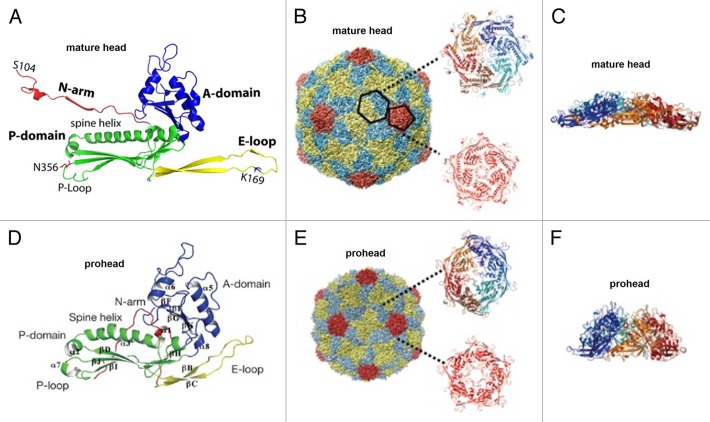
The gp5* protein, is an L-shaped molecule containing 2 distinct domains (Fig. 7A).78,88 In the mature capsid, having T = 7 laevo symmetry, the protein forms pentameric and hexameric capsomers with the axial (A) domains located near the capsomer’s axis, and the peripheral (P) domains located on the capsomer’s periphery. The A domain contains a 4-stranded β sheet and 2 helices; and the P domain comprises a 43 Å long “spine” helix packed against a 3-stranded antiparallel β sheet. In addition to the core formed by domains A and P, the gp5* molecule contains a 50 Å-long extended (E) loop, and a 60 Å-long N-terminal arm.
The inter-subunit interactions in the mature HK97 capsid are extremely extensive.78,88 Each gp5* molecules is in contact with 9 other molecules. The N-arm and E-loop of gp5* molecules show remarkable acrobatic quaternary associations spanning across neighboring subunits within the same capsomer and interacting with subunits in adjacent capsomers. The HK97 capsid is further stabilized by formation of covalent bonds between gp5* subunits belonging to different capsomers. The side chain of Lys169, located in the E loop of 1 gp5* subunit, forms the isopeptide bond with the side chain of Asn356 located in the P domain of another gp5* molecule. Thus each gp5* molecule in the capsid is covalently bound to 2 other molecules belonging to different capsomers, resulting in 420 isopeptide bonds per capsid. The gp5* cross-linking results in the formation of covalently-linked protein circles, which, in turn, are organized topologically into a chainmail-like structure. Such cross-linking is specific for phage HK97 and is not found in other well-studied phages.
Mature Caudovirales capsids have rather thin protein shells because their major capsid protein subunits are oriented roughly tangentially to the capsid surface. For example, the thickness of the HK97 gp5* shell is only 18 Å.78 However the prohead shells are thicker than the shell of the mature virus due to roughly radial orientation of the major capsid protein subunits. Interactions between the major capsid protein subunits are much weaker in the prohead compared with the mature capsid. Studies of the capsid assembly intermediates of the HK9714,15 (Fig. 7) and P2216 phages showed that capsid proteins in the prohead and mature head have significant differences in their tertiary structure. In the prohead, the capsid proteins are in a strained higher energy state, characterized by the bent spine helix and the twisted P-domain β sheet (Fig. 7D).15,16
The mature capsids protein shells are remarkably stable and can sustain high pressure (~60 atm, in case of φ299) created inside the head by the encapsulated DNA. Although the core of the major capsid protein and its organization into the capsid shell is similar in all Caudovirales, many phages have additional domains compared with the HK97 core. These domains lying on the capsid surface appear in cryo-EM reconstructions as additional protrusions (or bumps) compared with the rather smooth HK97 capsid surface. The extra domains are often involved in inter-subunit interactions reinforcing the capsid.
For example, the capsid protein of phage P22 has a ~150-residue ED (extra density) domain which is inserted into domain A of the HK97-like core.16,89 In the capsid this domain is involved in both inter-capsomer and intra-capsomer interactions. The ED domain is important for the fold of the capsid protein monomer, and also plays an important role during the capsid maturation.89 The domain possesses an extended D-loop stabilizing the immature and mature capsids through interactions with a neighboring capsomer.16
Bacteriophage φ29 capsid protein has an additional C-terminal domain with an immunoglobulin (BIG2-like) fold.77 This extra domain stabilizes the capsid by bridging neighboring capsomers and also provides the attachment sites for the head fibers.77,90
Phage T4 major capsid protein gp23*, and its special pentameric vertex protein, gp24*, have a globular (~60-residue) domain inserted through the E-loop. This domain probably interacts with adjacent molecules, located within the same capsomers, thus stabilizing the capsomer structure.75,91
The capsid protein of bacteriophage T7 is made in 2 forms: 10A and 10B.92 The latter form, has 53 additional residues at the C-terminus, and is produced via a translational frameshift. About one tenth of the subunits in the heads are of the longer protein. The frameshift does not provide additional stability to the T7 capsid; however it is conserved in the related phage T3.92
Another strategy for capsid reinforcement is the use of special “cement” proteins. An example of such protein is gpD of phage λ.80,93 GpD (109 residues) is a monomer in solution, but forms trimers when bound to the capsid surface. The gpD trimers sit on quasi-3-fold axes reinforcing inter-capsomer interactions. The positions of the gpD trimers correspond to the positions of the covalent cross-links in HK97. Phage λ capsid lacking gpD cannot sustain the pressure of the full-length genome, but can pack 82% of the genomic DNA.94
Bacteriophage ε15 has a capsid surface protein gp10 (111 residues) which staples adjacent capsomers.95 Gp10 molecules located near local 2-fold axes form tight dimers, further reinforcing the capsid. Gp10 has the jellyroll-like fold, which is very common for capsids of icosahedral viruses, but was not previously found in Caudovirales capsids.
Phage T4 capsid contains the small outer capsid protein, Soc, which binds at the interface between hexameric capsomers and clamps 2 capsomers together (Fig. 8).96 Further reinforcement of the T4 capsid is provided by trimeric interaction of Soc molecules at the quasi-3-fold axes passing between hexameric capsomers. The Soc protein is dispensable in normal conditions; however it helps to stabilize the capsids against extremes of pH and temperature. Both T4_Soc and λ_gpD attach to the capsid surface at the final stage of their maturation. All 3 cement proteins mentioned above reinforce inter-capsomer interactions. However they have different folds and use different sites for capsid attachment.
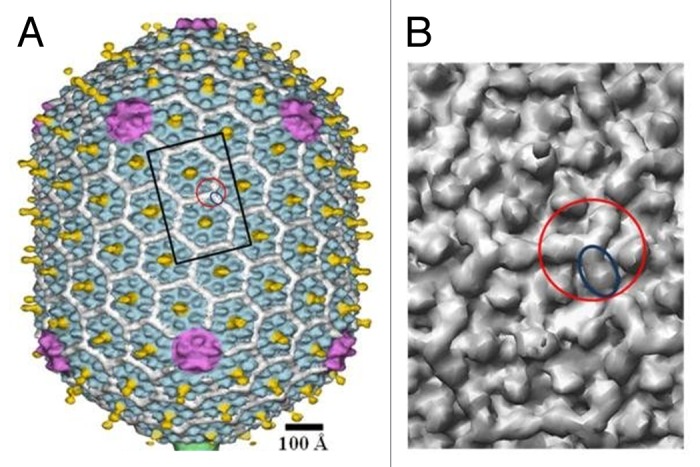
Figure 8. Structure of the bacteriophage T4 head (T = 13 / Q = 20 symmetry). (A) Shaded surface representation of the cryo-EM reconstruction74 viewed perpendicular to the 5-fold axis. The major capsid protein, gp23, is shown in blue, the special vertex protein, gp24, is in magenta, Soc is in white, Hoc is in yellow, and the portal vertex is in green. (B) Close up view of 2 gp23 capsomers corresponding to the rectangle outlined in black in (A). One Soc monomer is outlined by a blue ellipse; one Soc trimer is outlined by a red circle. The figure was modified from reference 96.
Some bacteriophages contain capsid decorative proteins which have marginal effect on the capsid stability and protrude far from the capsid surface. The example of such protein is the highly immunogenic outer capsid protein (Hoc) of T4-like phages (Fig. 8). T4 Hoc monomers (404 residues) attach to the capsid at the center of each hexameric capsomer with a total of 155 molecules per capsid.74 Each Hoc molecule contains 4 domains, 3 of which have immunoglobulin-like folds.97-99 Immunoglobulin domains are present on the surfaces of ~25% of Caudovirales100,101 indicating that these are beneficial for the phage life cycle. The presence of a large number of immunoglobulin domains probably makes the virion surface “sticky” and help the phage to bind reversibly to different surfaces. For example, the phage can attach to bacteria (without infecting it) and use it as a vehicle to travel to different locations. Immunoglobulin domains may also help the virus to bind to surfaces of eukaryotic organisms and therefore be advantageous for phages propagating on symbiotic bacteria.102 It was recently demonstrated103 that Hoc helps the T4 virions to adhere to metazoan mucosal surfaces via interactions with mucin glycoproteins. As mucus layers provide favorable habitats for bacteria, such attachment likely benefits the phage via more frequent interactions with its potential hosts.103 Furthermore, Barr et al.103 stated that the increased concentration of lytic phages on mucosal surfaces provides a metazoan immune defense affected by phage lysis of incoming bacterial pathogens.
Other examples of lengthy protruding capsid proteins are gp8.5 of phage φ29 (280 residues), and gp12 of phage SPP1 (64 residues). Unlike Hoc, these proteins are trimeric. SPP1_gp12 binds at the centers of capsomers,79 while φ29_gp8.5 binds at quasi-3-fold positions.77,90
Internal capsid proteins
Most Caudovirales phages contain internal proteins inside their capsid. The proteins can be dispersed within the capsid interior or form ordered structures which occupy a substantial fraction of the capsid volume.104 Some of these proteins (“pilot proteins”) are injected into the host cell during infection prior or along with the phage DNA.
For example, phage T4 capsid contains ~40 copies of the gpalt protein (75 kDa), which are probably located near or on the portal protein.13,104 Gpalt is an ADP-ribosyltransferase which modifies the host’s RNA polymerase resulting in preferential recognition of phage early promoters.105 T4 also contains the gp2 protein, which is likely associated with the ends of the phage DNA, protecting them from exonucleotic degradation.106 In addition, T4 capsid contains more than 1000 molecules of 3 small internal proteins (8–20 kDa), which are dispersed within the DNA.13,104 One of these proteins (IPI) helps to protect the phage genome from degradation by the host’s endonucleases.107,108
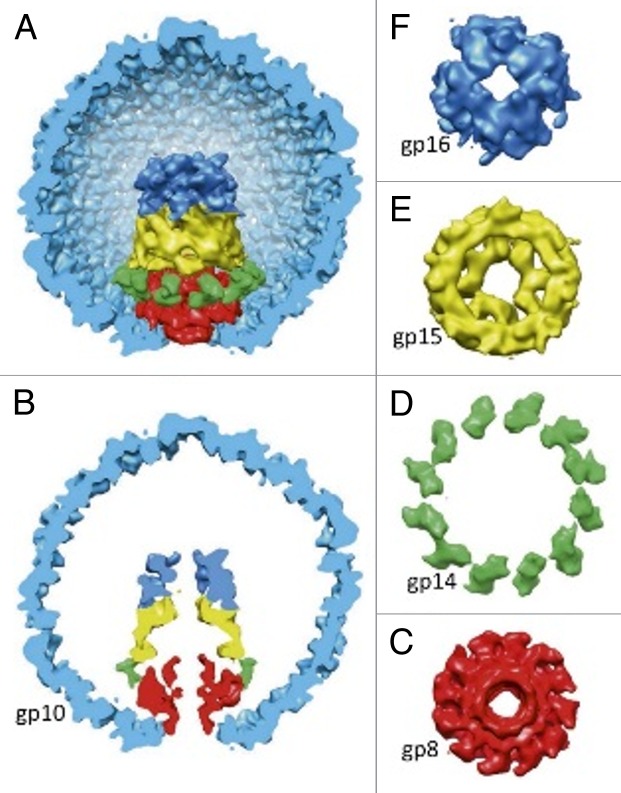
Capsids of T7-like phages contain internal cores assembled on the portal protein.81 The cores have a central channel for DNA passage. In T7, the core is composed of 12 copies of gp14, 8 copies of gp15, and 4 copies of gp16 (Fig. 9). During the infection process these proteins exit the capsid and form the extension of the phage tail that spans the host’s membranes and the periplasmic space.109 Inside the capsid, the core proteins are arranged in 3 layers with 12-, 8-, and 4-fold symmetry, respectively. In different phage particles these layers have different combinations of relative orientations, resulting in structural polymorphism of T7 virions.81 Therefore the classical cryo-EM asymmetric reconstruction, which assumes that all particles have identical structures, cannot resolve structural details of the core. However, the structure of the core was determined using the focused asymmetric reconstruction approach (Fig. 9).81 The axis of the core was found to be slightly tilted relatively to the 5-fold axis of the capsid. Based on this observation, it was suggested81 that during DNA packaging the core precesses around the capsid axis to assists DNA spooling.
Figure 9. Structure of the bacteriophage T7 internal capsid core. The portal protein, gp8, is in red. The core proteins gp14, gp15, and gp16 are in green, yellow, and blue, respectively. (A and B) display side views of the core. (C and D) show top views of the core layers. (F) displays the top view of the portal protein. The figure was reproduced from reference 81.
The podophage N4 capsid contains ~4 copies of viral RNA polymerase, gp50 (380 kDa). The gp50 protein is injected into the host during infection. The cryo-EM studies of N4 virion showed that gp50 is localized above the portal protein and forms the base of the internal core.110
The capsid of giant bacteriophage φKZ contains a large internal protein body which is ~1050 Å long and ~240 Å wide.111 The axis of the body is tilted by ~22° relative to the phage axis. The 2 ends of the internal body are anchored on opposing hexameric capsomers on either side of the capsid.111 The inner body contains 100–200 copies of each of 5 major proteins, as well as a number of minor proteins.104 The axis of the DNA spool inside the φKZ capsid appears to coincide with the inner body axis,111,112 suggesting that the body plays a role in the DNA organization during packaging.
Structure of the packaged DNA
Phage chromosomes are packed to near crystalline density of ~500 g/l. Asymmetric cryo-EM reconstructions of phage capsids are not able to visualize the structure of whole chromosomes, suggesting that the path of DNA molecule inside the capsid varies among individual phage particles. Full phage capsids, show local hexagonal packing of DNA helices with an inter-helix distance of 25–28 Å, depending on phage species.113 Several models have been proposed for phage DNA organization.104 For the phages containing an internal protein core, like T7,81 K1E,84 P-SPP7,114 the coaxial inverse spool model115 is the most widely accepted (Fig. 10A and B). In this model the part of DNA which enters the capsid first is located near the capsid protein shell while the last portion of DNA is in the central region of the capsid. The axis of the DNA spool is aligned with the axis of the capsid core. The asymmetric reconstruction of the podophage C1, which does not have an internal capsid core, is also consistent with the spool model.85 A spool model was also proposed for phage T4, although the axis of the spool was perpendicular to the axis of the tail.117
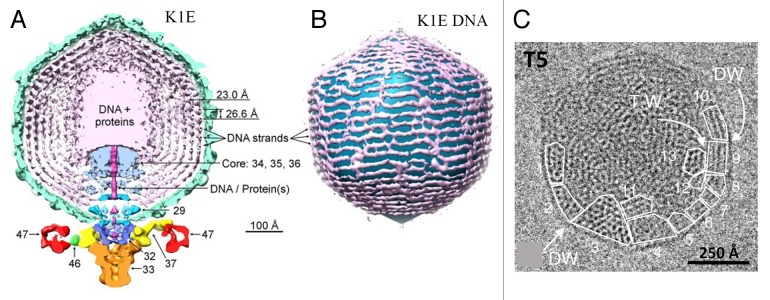
Figure 10. Structure of the packaged DNA within phage capsids. (A and B) Organization of DNA in phage K1E, whose capsid contains an internal protein core. (A) displays a 75 Å thick slice containing the central section of the K1E cryo-EM map. Several DNA strands in the outermost layers are marked with black dots to emphasize their hexagonal packing. (B) shows the outermost layer of DNA, most proximal to the inner capsid wall. The observed DNA pattern is consistent with the coaxial spool model. (C) Organization of DNA in the T5 capsid which does not have the internal core. A cryo-EM image is shown of a T5 capsid. The hexagonal DNA monodomains are underlined, and positions of dislocation walls (DW) and twist walls (TW) are shown. The scale bar represents 250 Å. (A and B) were reproduced from reference 84, (C) from reference 116.
The spool model, however, does not always apply to the capsids without internal cores.113 Cryo-EM analysis of T5 capsids showed that in the full heads the DNA is organized into a set of hexagonal domains separated by defect walls (twist, bent, and dislocation walls) to form a 3D lattice (Fig. 10C).113,116 In domains, located near the capsid protein shell, the directions of the DNA helices are parallel to the capsid walls. The twist walls, separating the domains, are generally parallel to the capsid walls, whereas the translocation walls are radial to the capsid surface. Analysis of T5 phages which ejected different parts of their genome showed that DNA fills the capsid uniformly irrespective of the length of the genome still present in the capsid. During ejection the structure of the DNA becomes liquid crystalline and then isotropic.113,116
The head completion proteins
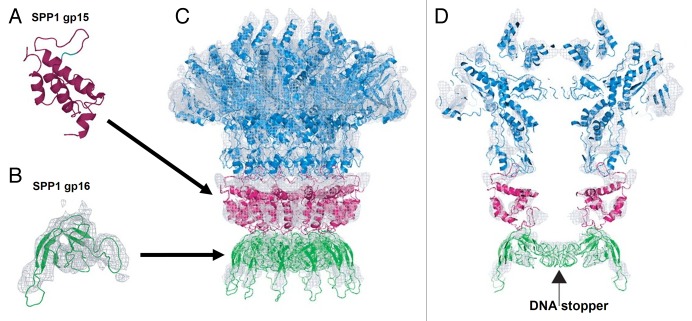
The Sipho- and Myoviridae phages typically contain 2 head completion proteins each of which assembles into a ring below the portal (Fig. 11).30 In the mature virion the 2 rings are sandwiched between the portal protein and the tail. Mutant capsids lacking the head completion proteins cannot bind tails and lose their DNA. In the Siphoviridae phages SPP1 and HK97, the ring closest to the portal is formed by dodecamers of gp15 (102 residues)118 and gp6 (108 residues),119 respectively. The SPP1_gp15 (Fig. 11A) and HK97_gp6 proteins have a similar fold characterized by a 3-helix bundle stabilized by a small hydrophobic core. A similar fold was found in the podophage P22 gp4 protein which also forms dodecameric rings below the portal,57 and in a putative neck protein YqbG, of a Myoviridae prophage found in the B. subtilis genome. Hence the helix-bundle fold seems to be very common for the phage neck proteins located immediately below the portal. However the bacteriophage λ protein gpW (68 residues), which had been predicted to bind to the portal, has a different fold.120
Figure 11. Structure of the bacteriophage SPP1 tail completion proteins. (A and B) display ribbon diagrams of SPP1_gp15 and SPP1_gp16 proteins, respectively. (C and D) show the portal protein (blue) and 2 rings formed by the tail completion proteins below the portal (magenta and green). Reproduced from reference 118.
The family of proteins which form the second ring below the portal, that binds to the phage tail, is represented by gp16 of SPP1 (Fig. 11B, 109 residues),118 gpFII of phage λ (117 residues),121 and XkdH of the Myoviridae prophage PBSX, found in the B. subtilis genome. These proteins have a conserved core formed by a β-sandwich. Similar organization of the core was found in a very large group of phage tail proteins including the tail-tube and the central baseplate proteins (see below), suggesting their common evolutionary origin.122,123
The head completion proteins are characterized by the presence of large unstructured regions. These proteins are usually monomers in solution and do not form complexes when mixed with other phage proteins. Oligomerization of these proteins in solution may be detrimental for production of infectious virions.124 However, the 2 rings of the head completion proteins assemble rapidly on the portal vertex after termination of DNA packaging, to seal the portal and create the interface for tail attachment.30,118,125 The interaction of proteins with other proteins within the assembling virion results in the folding of unstructured regions. Such conformational switching upon binding to assembly intermediates is a common mechanism for the control of sequential incorporation of proteins into the phage particle.
Lhuillier et al.118 suggested a mechanism for the viral DNA gatekeeping in phage SPP1, in which the premature leakage of DNA is prevented by the stopper protein gp16. The central channel in the gp16 dodecamer is closed by 12 loops coming from different gp16 subunits and forming a parallel inter-subunit β-sheet.
Structure of the Phage Tails
Bacteriophage tails are fascinating molecular machines created to recognize the host cells, penetrate the cell envelope barrier and deliver DNA into the cytoplasm. The Caudovirales tails have very different size and morphology with lengths ranging from ~100 Å, in some podophages to ~8000 Å in some siphoviruses.7,126 The Podoviridae tails usually assemble on the capsid portal vertex whereas the tails of Siphoviridae and Myoviridae phages assemble via an independent pathway and associate with the capsid at the final stage of virion assembly.
Long tails of the Siphoviridae and Myoviridae phages
Siphoviridae and Myoviridae tails consist of the tail tip complex, which is responsible for the host recognition and initiation of the infection process, the tail tube, which makes a conduit for genomic DNA, and the terminator proteins, which terminate the tail assembly and create the binding interface for head attachment (Fig. 12).127,128 The tail of Myoviridae phages also contains a contractile sheath surrounding the tail tube.128,129 The tail tip complex has different size and morphology in different phages. Phages which use protein receptors for cell binding usually have conical tail tips (e.g., SPP1130 and λ), whereas phages, using polysaccharide receptors usually have elaborate baseplates at the distal end of the tail (e.g., lactococcal phages TP901-16,131 and p2132,133). Furthermore phages usually have side tail fibers or spikes, attached to the periphery of the tail tip complex, as well as a central tail spike.
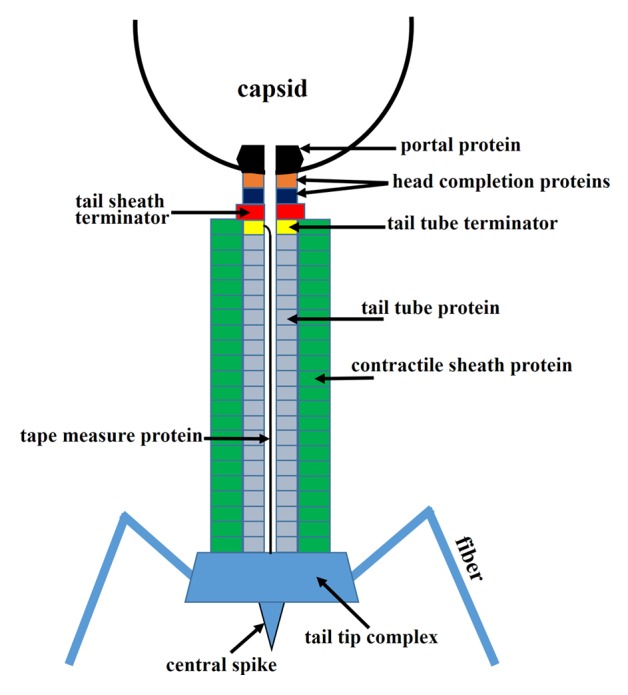
Figure 12. Schematic representation of a Myoviridae phage tail and neck. Siphoviridae phages would be missing the tail sheath (green) and the tail sheath terminator protein (red).
In the Siphoviridae and Myoviridae tails, the tail tip complex assembles first and serves as a platform for polymerization of the tail tube protein subunits. The tail tip complex attaches the tape measure, or ruler, protein, which determines the length of the tail tube.127,134,135 The tape measure protein is extended through the central channel of the tail tube as it is being built. The polymerizing tail tube protein molecules form a stack of hexameric rings on top of the tail tip complex,136 which encircle the tape measure protein. When the end of the growing tube approaches the end of the ruler protein, the polymerization is stopped by attachment of the hexameric ring of the tail tube terminator protein to the top of the tube,136 and, probably, to the N-terminal part of the ruler protein. Myoviridae phages possess a contractile tail sheath which assembles around the tail tube. The polymerization of the tail sheath protein also starts at the tail tip complex (the baseplate) and is propagated to the end of the tube. The sheath subunits arrange into a stack of hexameric rings, rotated relative to each other, thus creating a 6-start helix.129,137 The polymerization of the sheath is terminated by the hexameric sheath terminator protein which binds on top of the tail tube terminator and interacts with the last ring of the sheath (Fig. 12).4 In the Myoviridae phages the sheath terminator protein creates the interface for head attachment, whereas in the Siphoviridae phages, which do not have the sheath, the capsid binding interface is formed by the tube terminator protein.
The tape measure protein
The tape measure protein is extended along the assembled tail tube. The exact stoichiometry of the tape measure protein is unknown, although the copy number was estimated to be 6–7 for phage λ.138 Secondary structure predictions suggest that the protein is mostly α-helical through its entire length. The length of the tail tube in different phage species agrees approximately with the length of the ruler protein if it is modeled as a continuous extended helix. During the infection process a signal should be transmitted from the tail tip complex to the neck proteins to open the portal vertex for DNA exit. As the tape measure protein spans the entire tail tube, it is likely to be involved in the signal transmission. Alternatively the signal may be transmitted through the tail tube protein.130 The tape measure protein should exit the tail tube during infection before the DNA is ejected. In Siphoviridae phages the tape measure protein may be involved in the creation of a channel for DNA translocation through the cell wall.127
The tail tube protein
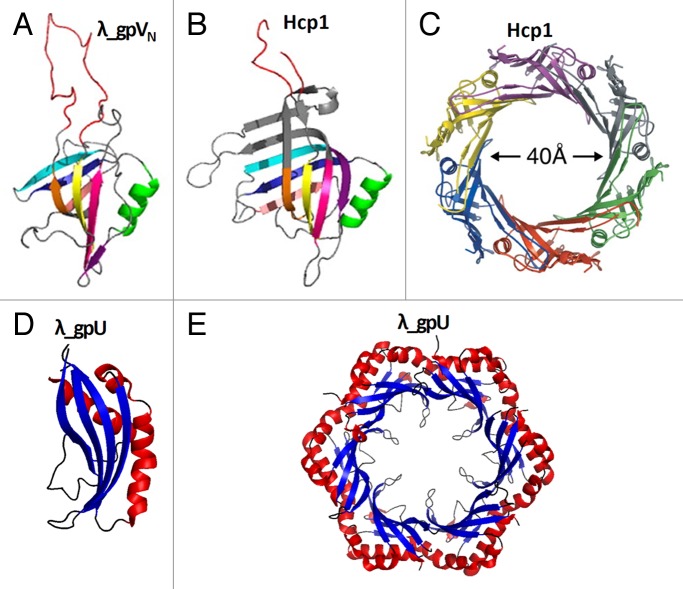
The tail tube proteins usually do not polymerize until they encounter the tail tip complex. In solution these proteins typically contain many flexible loops, impeding the production of well-diffracting crystals. The structures of the 2 domains of the tail tube protein, gpV, of phage λ were determined using nuclear magnetic resonance (NMR) spectroscopy.139,140 The N-terminal domain of gpV (gpVN, residues 1–160) is necessary and sufficient to make the λ tail tube (Fig. 13A),139 whereas the C-terminal immunoglobulin-like domain (residues 161–246) plays an accessory role, probably helping the virus to attach reversibly to different surfaces. The N-terminal domain is composed of 2 antiparallel sheets, which form a twisted β-sandwich, flanked by an α-helix. This structure is similar to the tail tube protein, XkdM, from the PBSX prophage of B. subtilis, which encodes Myoviridae particles, indicating the common evolutionary origin of the Siphoviridae and Myoviridae tail tubes. The gpV N-terminal domain is also similar to the tube proteins, Hcps, from the bacterial secretion system VI, which is structurally related to phage tails (Fig. 13B and C).139,142 The Hcp proteins form hexameric rings of ~90 Å external diameter and ~40 Å height, with the inner pore having a ~40 Å diameter. These dimensions are close to those of the tail tube protein rings in phages, allowing modeling of homologous phage tail tube structures.139
Figure 13. Tail tube and tail tube terminator proteins. (A) Structure of the N-terminal domain of the bacteriophage λ tail tube protein, gpV. This domain is necessary and sufficient for tail tube formation. (B) Structure of the tube protein Hcp1 from the bacterial secretion system VI. (C) Hexameric ring of Hcp1. (D) Structure of the tail tube terminator protein gpU of phage λ. (E) A hexameric ring of gpU. (A and B) were reproduced from reference 139. (C) was reproduced from reference 141.
The tail tube protein has structural similarity to a large family of Siphoviridae and Myoviridae proteins, including the tail terminator proteins, the head completion proteins forming the head-tail binding interface, and the proteins located in the center of the phage baseplate (see below, and refs. 122, 123, and 142). All these proteins have probably evolved from a common ancestor. In phage genomes the genes, encoding these proteins, are usually located close to each other suggesting that they appeared by gene duplication, followed by mutation and selection.
Unlike most other phages, T5 has a 3-fold- rather than 6-fold-symmetric tail tube.143 Its tail tube protein is about twice as big as other tail tube proteins. Therefore it is possible that the T5 tail tube protein has 2 domains with similar folds, and that 3 copies of the protein form pseudohexameric rings in the tail tube.127,143 Such pseudo-hexameric rings are formed by the baseplate hub proteins (see below).
The tail terminator proteins
The tail terminator proteins are critical components for the phage assembly. Tails lacking the terminator proteins cannot attach to the heads. In the absence of the tail terminator proteins the tail tube (and the contractile sheath in Myoviridae phages) can polymerize (though at a slower rate) beyond the length determined by the tape measure protein resulting in aberrant tails of different sizes. The structure of tail tube terminator protein, gpU (131 residues), of phage λ has been determined using NMR and X-ray crystallography (Fig. 13D and E).136,144 In one of the crystal forms gpU forms hexameric rings, as is presumably in the phage particle (Fig. 13E). The fold of gpU is defined by an antiparallel, 4-stranded β-sheet that is decorated on one side by 3 α-helices.144 Although the overall fold of gpU is somewhat different from that of the tail tube protein, gpV, a similar portion of the tertiary structure was detected in both proteins, suggesting their common evolutionary origin.122 Myophage T4 has 2 terminator proteins: gp3, which (like gpU of phage λ) stops the polymerization of the tail tube, and gp15, which terminates the polymerization of the contractile sheath.145 In the T4 virion, gp3 (175 residues) and gp15 (272 residues) make hexameric rings, with the gp3 ring located on top of the tail tube and the gp15 ring docked on top of gp3. Although the gp15 protein is twice as large as gpU of phage λ, the structure of the central part of the gp15 hexamer is similar to that of gpU.4 In comparison with gpU, the gp15 hexamer has additional structural elements located in its periphery, designed to interact with the contractile sheath.4
The contractile sheath structure
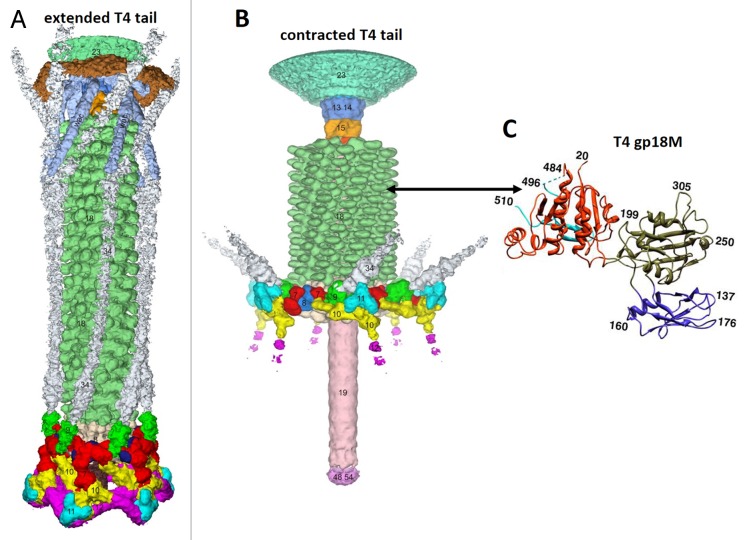
The bacteriophage T4 tail is the most extensively studied contractile system.128,129,137,146,147 The structure of the T4 tail before and after contraction has been determined using cryo-EM (Fig. 14A and B).137,147 The T4 tail sheath is composed of 138 subunits of the gp18 protein (659 residues), organized into a 6-start right-handed helix. In the extended tail, the sheath is ~240 Å wide and ~925 Å long, with the pitch and twist of the helix being 40.6 Å and 17.2°, respectively. Because the tail tube density is smooth in the T4 cryo-EM reconstructions, it is not possible to conclude if it has the same helical symmetry as the sheath. The extended sheath represents a metastable high-energy structure akin to an extended spring. Before and after the contraction, the sheath is attached to the baseplate at one end and to the neck at the other end. As the sheath contracts the tail tube (with the cell-puncturing tip) is pushed through the center of the baseplate and penetrates into the cell wall. The sheath contraction causes dramatic rearrangement of the gp18 subunits. Each subunit moves by ~50 Å away from the tail axis and rotates by ~45°.128,129,146 The sheath contraction drastically changes the environment of gp18 subunits within the sheath, resulting in stronger inter-subunit interactions. The contracted sheath has a length of 420 Å, and a width of 330 Å. It is also a 6-start helix with the pitch and twist of 16.4 Å and 32.9°, respectively.
Figure 14. Structure of the bacteriophage T4 tail in the extended (A) and contracted (B) conformations. Constituent proteins are shown in different colors. The contractile tail sheath is shown in green. (C) Structure of the tail sheath protein mutant, gp18M, containing 3 out of 4 domains of gp18. Domain I is in blue, domain II is in olive green, domain III is in orange red. Residues 454–470 and the last 27 C-terminal residues of gp18M are shown in cyan. (A) was reproduced from reference 137, (B) from reference 147, and (C) from reference 146.
The T4 tail sheath subunit, gp18, consists of 4 domains. The structure of the gp18 mutant containing domains I–III has been determined using X-ray crystallography (Fig. 14C).146 The domain organization of gp18 can be described as a series of insertion domains, similar to a set of Russian Dolls. The domain I in gp18 is an insertion of domain II, which is itself an insertion of domain III, which is an insertion of domain IV. Such domain structure suggests that domain IV is the most ancient part of the protein and that other insertion domains were added later during evolution. Domain IV which is mostly composed of the C-terminal part of the protein interacts with the tail tube in the extended tail. Domains II and III and IV are involved in the interaction with other gp18 subunits within the extended and contracted sheath, while domain I is located at the periphery of the sheath and is exposed into the solvent. Fitting of the gp18 structure into the cryo-EM reconstructions of the extended and contracted tails showed that gp18 subunits move as rigid bodies during the tail contraction without refolding or significant change in the relative orientations of the domains.
Apart from T4 gp18, structures of 2 putative tail sheath proteins, DSY3957 and LIN1278, encoded by prophages have been determined by a structural genomics consortium.148 Those structures contain the C-terminal domain IV, absent in the gp18 crystal structure. In addition, a partial structure of the tail sheath protein from the giant bacteriophage φKZ has been determined.148 Despite the low sequence similarity, these proteins have similar structures, indicating that the sheath proteins of tailed phages have evolved from a common ancestor. In the DSY3957 crystal structure the N-terminal arm (residues 15–25) of the protein is a part of a β-sheet in the C-terminal domain of a symmetry related molecule. Based on this observation, Leiman and Shneider128 proposed that in the phage sheaths such chain swapping cross-links neighboring subunits stacked on top of each other, thus helping to maintain the sheath integrity.
The phage tail-tip complexes
Bacteriophage baseplates vary in size and complexity. The most extensively studied Myoviridae baseplate is that of phage T4 (Fig. 15A), which is composed of ~140 polypeptide chains of at least 16 different proteins (for a recent review see ref. 129). The T4 baseplate attaches 6 long tail fibers and 6 short tail fibers to its periphery. The long tail fibers, with a length of ~1450 Å, bind reversibly to the E.coli lipopolysaccharide (LPS) and/or OmpC molecules, and serve for primary host recognition.151 Upon binding of the long tail fibers a signal is send to the baseplate causing the 6 short tail fibers to extend and bind irreversibly to the LPS. On attachment, the baseplate conformation switches from the dome-shaped to the star-shaped which, in turn, triggers the contraction of the tail sheath. The structures of T4 baseplate in the dome-shaped and star-shaped conformations have been determined using cryo-EM (Fig. 14A and B; Fig. 15A),137,147 and structures of 9 baseplate proteins have been determined using X-ray crystallography.129 Comparison of the X-ray structures showed that some proteins, located in the T4 baseplate periphery (gp10, gp11, and gp12) evolved from a common primordial fold via gene duplication,152 suggesting an evolutionary mechanism for developing complex baseplates.
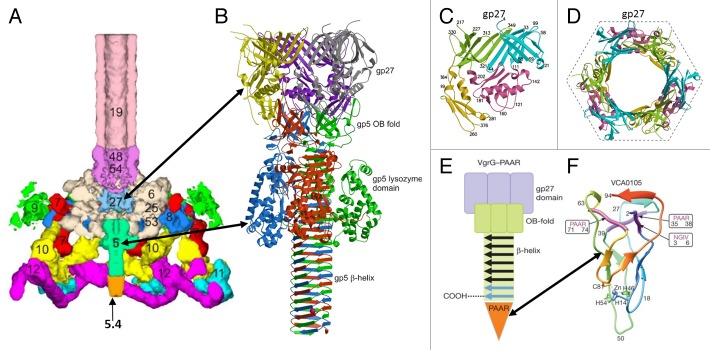
Figure 15. Structure of the bacteriophage T4 baseplate and cell-puncturing device. (A) Cross-section view of the T4 baseplate. Constituent proteins are shown in different colors and are identified with their corresponding gene names. (B) Structure of the gp5-gp27 trimer is shown as a ribbon diagram in which each chain is shown in a different color. (C) Domains of gp27. The 2 homologous domains are colored in light green and cyan. These domains are similar to the tail-tube proteins. (D) The pseudohexameric ring formed by the tail-tube like domains of the gp27 trimer. (E) Schematic representation of a spike from the bacterial secretion system VI. The spike consists of the VgrG and PAAR-repeat proteins. (F) Structure of the PAAR-repeat protein. (A) was reproduced from reference 149. (B–D) were reproduced from reference 129. (E and F) were reproduced from reference 150.
Most of the T4 baseplate structure, obeys the 6-fold symmetry. However, the central hub of the baseplate is formed by the trimeric gp27 protein. The gp27 molecule (391 residues) consists of 4 domains, 2 of which have a fold resembling that of the tail-tube proteins (Fig. 15B–D). In the baseplate these domains form a pseudo-hexameric ring, analogous to the tail-tube protein hexamers. The gp27 homologs, forming pseudo-hexameric rings, were also found in non-contractile Siphoviridae tails (see below and ref. 132), suggesting that such a baseplate hub structure is common in phages. The gp27 protein binds the central tail spike made by a trimer of gp5 protein.153 T4 gp5 (575 residues) has 3 domains (Fig. 15B). The N-terminal domain has an oligosaccharide/oligonucleotide-binding (OB) fold, the central domain has a lysozyme-like fold, and the C-terminal domain forms an intertwined β-helix. The tip of the gp5 β-helix is capped by a monomeric protein, recently identified as gp5.4 (97 residues), which sharpens the central spike.150 During the infection process the (gp5)3-gp5.4 spike punctures the cell membrane and the lysozyme domain of gp5 digests the peptidoglycan in the E. coli periplasm.
Similar organization of the central tail spike was observed in the bacterial secretion system VI.142,150 This system is responsible for translocation of toxic effector molecules, allowing predatory bacteria to kill prokaryotic and eukaryotic prey cells. The secretion system VI VgrG proteins correspond to both gp27 and gp5 of T4, although without the lysozyme domain (Fig. 15E). The tip of the VgrG β-helix is capped by a spike sharpening protein, characterized by PAAR sequence repeats, and containing a zinc atom. In the secretion system VI, the VgrG and/or PAAR-repeat proteins can be fused with effector domains which have cytotoxic effects (Fig. 15E and F).150
The central cell-puncturing spikes of phages P2 and φ92154 have a structure similar to that of gp5 of T4, without the lysozyme domain, although there are differences in how the 3 monomer chains intertwine to form the β-helix. The P2 and φ92 phages do not encode a separate monomeric protein which binds to the very tip of the spike. Instead their gp5 homologs have a C-terminal tip-sharpening extension characterized by the presence of an iron ion.154
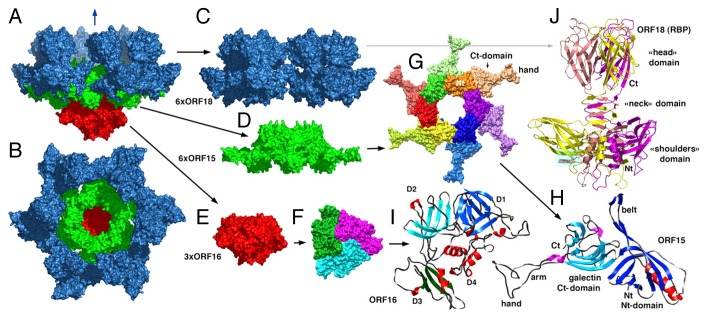
The best studied Siphoviridae baseplates are those of phages p2 and TP901-1 infecting Gram-positive Lactococcus lactis.131,132 Both these phages use carbohydrate molecules as receptors for attachment to the host. The p2 baseplate (Fig. 16) is composed of 3 different proteins: Dit (ORF15, 298 residues), Tal (ORF16, 398 residues), and the receptor binding protein (ORF18, 264 residues).132 The Dit protein forms hexameric rings below the tail tube. The central part of the Dit hexamer, made by the N-terminal domains (residues 1–132) of 6 monomers, is very similar to the tail tube protein hexamers (Fig. 16G). The C-terminal domains of 6 Dit molecules, attach trimers of the receptor binding protein, resulting in 18 copies of the receptor binding subunits per phage.
Figure 16. Crystal structure of the phage p2 baseplate and its components. (A) View of the baseplate surface; ORF15 (Dit) is in green, ORF16 (Tal) is in red, and ORF18 (receptor binding protein) is in blue. The blue arrow indicates the position of the quasi 6-fold axis and points toward the rest of the phage tail and the capsid. (B) The baseplate has been rotated by 90° around the horizontal axis. The central channel formed by ORF15 hexamer is closed by the ORF16 trimeric dome. (C) The arrangement of ORF18 as 6 trimers. (D) Hexameric ORF15 (Dit). (E) Trimeric ORF16 (Tal). (F) View of ORF16 trimer rotated 180° relative to baseplate (B) with a different color for each subunit. (G) ORF15 hexamer is viewed in the same orientation as in (B). Each subunit, as well as the N- and C-terminus domains, have a different color. The central channel is ∼40 Å wide. (H) Ribbon view of the ORF15 (Dit) subunit. (I) Ribbon view of ORF16 (Tal) subunit. The 4 domains have been identified by D 1 to 4 and different colors of the β-strands. These domains correspond to those identified in gp27 from phage T4. (J) Ribbon view of the receptor-binding protein ORF18 trimer, with a different color for each chain. Reprinted from reference 132.
The trimeric Tal protein (ORF16) attaches to the bottom of the Dit ring. The fold of Tal is similar to that of T4 gp27 (Fig. 16E, F, and I). Like gp27, the Tal monomer is composed of 4 domains, 2 of which have the tail-tube-like fold. Contrary to T4, the p2 baseplate does not have any central tail spike attached to the Tal protein.
The p2 baseplate has 2 conformational states: the “rest” state and the “active” state. The switching from the rest to the active state is triggered by Ca2+ ions, which are mandatory for p2 infection. In the rest conformation the host-recognition domains of the receptor binding proteins point toward the phage head. In the presence of Ca2+ the receptor-binding protein trimers rotate by ~200°, and acquire the “down” orientation, presenting their binding sites to the host. The baseplate activation also results in significant conformational changes in the Tal trimer, which opens up to allow the DNA passage.132
The baseplate of the phage TP901-1 contains a hexameric ring of Dit which attaches 6 trimers of the baseplate upper protein (BppU) to its periphery.131 Each BppU monomer, in turn, attaches a trimer of the receptor binding protein, resulting in 54 copies of the receptor binding subunits per phage. Thus BppU serves as an adaptor between Dit and the receptor-binding protein, allowing TP901-1 to expose 3 times more receptor-binding molecules on its baseplate compared with phage p2. Contrary to p2, the TP901-1 baseplate does not require activation by Ca2+ ions. It permanently stays in the “infection-ready” conformation with the receptor binding proteins suitably oriented for attachment to the host surface.131 The Dit ring attaches the trimeric Tal protein. The TP901-1 Tal, containing 918 residues, is much bigger, compared with that of phage p2. The N-terminal part of the TP901-1 Tal is similar to that of p2 and to the gp27 protein of phage T4, while its C-terminal part forms the central tail spike and contains a predicted peptidoglycan-degrading domain.
Proteins analogous to Dit were also found in the Gram-positive infecting phage SPP1155 and in the Gram-negative infecting phage T5,143,156 which do not possess baseplates, but have tapered tail tip complexes. Phage T5 encodes a separate protein analogous to p2_Tal and T4_gp27,143,156 which binds to Dit and attaches a long central tail spike, whereas phage SPP1 has a Tal/gp27-like region at the N-terminus of its central fiber protein.157
The central part of the tail tip complex has a common architecture in the long- tailed phages. A hexameric Dit-like protein, attached to the tail tube, serves as a binding site for proteins located at the periphery of the baseplate/tail-tip complex. A Tal/gp27-like trimer attaches below the Dit hexamer. Two domains of Tal/gp27-like protein form a pseudo-hexameric ring, which acts as an adaptor between the 6-fold-siymmetric Dit and the 3-fold-symmetric central tail spike.
The accumulated structural data demonstrate that the tail tube-like fold is a crucial building block of Siphoviridae and Myoviridae virions found in several proteins with different functions. The stack of the tail tube-like rings starts with the Tal/gp27-like protein, continues with the Dit-like protein, then with the tail tube, the tail terminator protein(s), and finishes with the ring of the head completion protein, on top of the tail. All these proteins probably have a common evolutionary origin.122,123
Structure of the peripheral part of the baseplate/tail-tip complexes vary in size and complexity, depending on specificity of phage-host interactions. However the receptor binding- proteins of phages, have common features. They are all trimeric and bind the phage through their N-terminal part. The chains of subunits in the trimer are usually intertwined. The phage receptor binding proteins typically have a high β-structure content, and contain β-helical regions.
Organization of the short Podoviridae tails
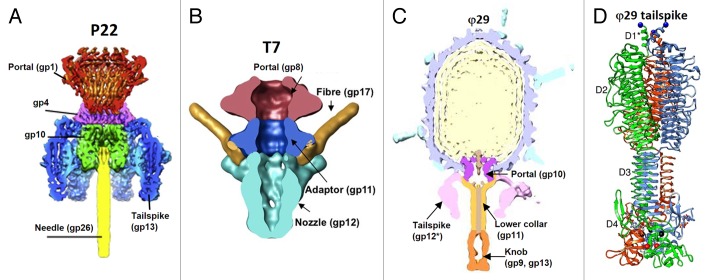
Three-dimensional structures of several Podoviridae phages (φ29,158,159 C1,85 P22,16,160 T7,66,81 ε15,83,95,161 N4,110 K1E,84 K1-5,84 P-SSP7114) were determined using cryo-EM. Despite variation in size and shape, the Podoviridae tails have a common structural organization. They contain a central tubular structure to which 6 or 12 appendages or fibers are attached (Fig. 17). The tubular part is typically composed of 2 different proteins. One of these proteins, which is bound to the portal vertex, and forms the upper part of the tube is usually a dodecamer. The example of such protein, for which the X-ray structure is available, is gp4 of phage P22 (166 residues).57 The core of the protein is formed by a 4-helix bundle and, as mentioned above, is similar to the head completion proteins of the long-tailed phages SPP1 (Fig. 11A), and HK97.118,119 The dodecamers, located below the portal vary significantly in size and shape. The P22 gp4 and the corresponding protein, gp11 from phage T7 (196-residues) form ring-like structures below the portal (Fig. 17A and B), whereas the analogous protein, gp11 of phage φ29 is bigger (293 residues) and makes a funnel-like structure, termed the lower collar (Fig. 17C). However, the similar position of these proteins in the virions, the dodecameric state, and high predicted α-helical content suggests that the T7_gp11162 and φ29_gp11 proteins may be structurally related to P22_gp4 and, consequently, to the head completion proteins of the long-tailed phages.
Figure 17. Tails of Podoviridae phages. (A) Cross-section view of the bacteriophage P22 tail-portal complex. The portal protein (gp1) is in red, the gp4 dodecamer is in magenta, the gp10 hexamer is in green, the tail spikes (trimers of gp13) are in blue, and the cell-puncturing needle (trimer of gp26) is in yellow. The barrel of the portal protein is not shown. Reproduced from reference 160. (B) Cross-section view of the bacteriophage T7 tail-portal complex. The portal, gp8, is in dark pink, the dodecamer of gp11 is in blue, the hexamer of gp12 is in cyan, parts of the fibers (gp17) are in gold. Reproduced from reference 162. (C) Central section of the bacteriophage φ29 cryo-EM reconstruction. The portal protein, gp10, is in magenta, the lower collar, gp11, is in yellow, the knob (gp9, gp13) is in orange. Reproduced from reference 5. (D) Ribbon diagram of the phage φ29 tail spike, gp12. The 3 polypeptide chains of the trimer are shown in blue, green, and orange, respectively. The structure contains domain 4 which is removed by an autocatalytic mechanism and is not present in the virion. Reproduced from reference 163.
The lower part of the tail tube is usually formed by a hexameric protein. The example of such a protein is gp12 of phage C1, whose structure has been determined by X-ray crystallography.85 The C1_gp12 (573 residues) protein is composed of 4 domains, with the N-terminal domain, binding to the dodecameric upper-tube protein. The N-terminal domain C1_gp12 (120 residues) has a fold similar to the tail-tube proteins of the Siphoviridae and Myoviridae phages showing evolutionary ties between the short and long phage tails.85
Podophages may contain different proteins bound at the end of their tail tubes. For example, phage φ29 has 2 copies of the peptidoglycan-degrading protein, gp13 (365 residues) at the very tip of its tail.5 Phage P22 contains a 240-Å-long cell-puncturing needle, attached to its tail tube (Fig. 17A).164 The needle made by the trimeric protein gp26 (233 residues) is mainly a coiled-coil structure. A part of the needle is inserted into the tube and serves as a plug to prevent a premature DNA leakage.
Trimeric tail fibers or tail spikes attach to the side of the tail tube of Podoviridae phages. For example, phage T7 has 6 tail fibers,109,162,165 which interact with the 6-fold-symmetric and 12-fold symmetric tube proteins (Fig. 17B), whereas phage φ29 has 12 spindle-shaped tail spikes attached via the 12-fold-symmetric tube protein (Fig. 17C and D). Usually phage infection is initiated by reversible binding of the tail spikes/fibers to specific host molecules. Many phages use long extended glycolpolymers for initial reversible binding. Tail spikes of such phages typically have enzymatic activity allowing digestion of the glycolpolymers, which helps the phage to move closer to the cell membrane. Subsequently the phage binds irreversibly to the cell surface with its tail tube properly positioned for infection. This irreversible binding triggers conformational changes in the virion, eventually resulting in the tail extension and formation of a channel between the capsid and the host cytoplasm through which the viral genome is injected.109
Structures of the tail spike from Podophages P22,166,167 φ29,163 HK620,168 K1F,169,170 have been determined using X-ray crystallography. They are all trimers with the N-terminal part responsible for attachment to the virion. The structures of the spikes are dominated by the β-helical fold. The phage φ29 tail spike protein, gp12, is synthetized as a 854-residue molecule (Fig. 17D). The 163-residue C-terminal domain of φ29_gp12 acts as a chaperon for trimer assembly, and is subsequently removed by an auto-catalytic mechanism.163 The cleaved protein consists of 3 domains with the N-terminal domain attached to the virion, the second domain responsible for reversible binding to and degradation of the cell wall teichoic acids, and the third domain responsible for the irreversible attachment to the cell.163 Binding of the third domains of the tail spikes to cell receptors probably fixes the phage particle on the cell wall, so that the tail can degrade the peptidoglycan layer with the help of the gp13 enzyme, located at the tail tube tip. The tail tip then reaches the cell membrane, and the phage forms a conduit for genome injection.
Conclusion
Due to progress in cryo-electron microscopy and high-throughput X-ray crystallography, numerous structures of phage virions and their protein components have been determined. These structures show that tailed double-stranded DNA phages have a common evolutionary origin. Their capsids, built of similar HK97-like capsid protein folds, contain similar dodecameric portals. Caudovirales use similar molecular motors to package their genomic DNA. A common fold was also recognized in the phage proteins forming rings below the portal vertex. In Siphoviridae and Myoviridae phages the tail tube proteins, the proteins forming the central parts of the tail tip complexes or baseplates, the tail terminator proteins, and the head completion proteins that make the tail binding interface, have structural similarity and thus have most likely evolved from a common ancestor.
Although a great deal of structural data has been accumulated on Caudovirales phages, dynamic processes of phage absorption to the host and infection are still far from being well understood. Future studies are likely to provide more structural data on phage-receptor interactions, transmission of the infection-initiation signals from the tail tips along the tails, and formation of conduits for the DNA injection into host cells. Hence structural studies of phages is likely to remain a rewarding research topic.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge support provided by the National Institutes of Health grant AI081726 and the National Science Foundation grant MCB-1014547 to M.G.R.
Citation: Fokine A, Rossmann MG. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 2014; 4:e28281; 10.4161/bact.28281
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/28281
References
- 1.Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–11. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–43. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 4.Fokine A, Zhang Z, Kanamaru S, Bowman VD, Aksyuk AA, Arisaka F, Rao VB, Rossmann MG. The molecular architecture of the bacteriophage T4 neck. J Mol Biol. 2013;425:1731–44. doi: 10.1016/j.jmb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang Y, Morais MC, Cohen DN, Bowman VD, Anderson DL, Rossmann MG. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage phi29 tail. Proc Natl Acad Sci U S A. 2008;105:9552–7. doi: 10.1073/pnas.0803787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bebeacua C, Lai L, Vegge CS, Brøndsted L, van Heel M, Veesler D, Cambillau C. Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1. J Virol. 2013;87:1061–8. doi: 10.1128/JVI.02836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ackermann HW. Bacteriophage observations and evolution. Res Microbiol. 2003;154:245–51. doi: 10.1016/S0923-2508(03)00067-6. [DOI] [PubMed] [Google Scholar]
- 8.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc Natl Acad Sci U S A. 2003;100:9292–5. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 10.Evilevitch A, Gober JW, Phillips M, Knobler CM, Gelbart WM. Measurements of DNA lengths remaining in a viral capsid after osmotically suppressed partial ejection. Biophys J. 2005;88:751–6. doi: 10.1529/biophysj.104.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aksyuk AA, Rossmann MG. Bacteriophage assembly. Viruses. 2011;3:172–203. doi: 10.3390/v3030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prevelige PE, Fane BA. Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Adv Exp Med Biol. 2012;726:325–50. doi: 10.1007/978-1-4614-0980-9_14. [DOI] [PubMed] [Google Scholar]
- 13.Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–70. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrix RW, Johnson JE. Bacteriophage HK97 capsid assembly and maturation. Adv Exp Med Biol. 2012;726:351–63. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- 15.Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–50. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011;108:1355–60. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preux O, Durand D, Huet A, Conway JF, Bertin A, Boulogne C, Drouin-Wahbi J, Trévarin D, Pérez J, Vachette P, et al. A two-state cooperative expansion converts the procapsid shell of bacteriophage T5 into a highly stable capsid isomorphous to the final virion head. J Mol Biol. 2013;425:1999–2014. doi: 10.1016/j.jmb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995;247:636–47. doi: 10.1016/S0022-2836(05)80144-5. [DOI] [PubMed] [Google Scholar]
- 19.Huet A, Conway JF, Letellier L, Boulanger P. In vitro assembly of the T=13 procapsid of bacteriophage T5 with its scaffolding domain. J Virol. 2010;84:9350–8. doi: 10.1128/JVI.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–6. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PE, Jr., Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J Mol Biol. 2000;297:1195–202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- 22.Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam JD, ed. Molecular Biology of Bacteriophage T4. Washington, D.C.: American Society for Microbiology, 1994:218-58 [Google Scholar]
- 23.Thomas JA, Black LW. Mutational analysis of the Pseudomonas aeruginosa myovirus KZ morphogenetic protease gp175. J Virol. 2013;87:8713–25. doi: 10.1128/JVI.01008-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun S, Rao VB, Rossmann MG. Genome packaging in viruses. Curr Opin Struct Biol. 2010;20:114–20. doi: 10.1016/j.sbi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feiss M, Rao VB. The bacteriophage DNA packaging machine. Adv Exp Med Biol. 2012;726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- 26.Speir JA, Johnson JE. Nucleic acid packaging in viruses. Curr Opin Struct Biol. 2012;22:65–71. doi: 10.1016/j.sbi.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–81. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 28.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–22. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao VB, Black LW. DNA packaging of bacteriophage T4 proheads in vitro. Evidence that prohead expansion is not coupled to DNA packaging. J Mol Biol. 1985;185:565–78. doi: 10.1016/0022-2836(85)90072-5. [DOI] [PubMed] [Google Scholar]
- 30.Tavares P, Zinn-Justin S, Orlova EV. Genome gating in tailed bacteriophage capsids. Adv Exp Med Biol. 2012;726:585–600. doi: 10.1007/978-1-4614-0980-9_25. [DOI] [PubMed] [Google Scholar]
- 31.Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–8. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Hendrix RW. Jumbo bacteriophages. Curr Top Microbiol Immunol. 2009;328:229–40. doi: 10.1007/978-3-540-68618-7_7. [DOI] [PubMed] [Google Scholar]
- 33.Lehman IR, Pratt EA. On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J Biol Chem. 1960;235:3254–9. [PubMed] [Google Scholar]
- 34.Casjens SR, Gilcrease EB. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol. 2009;502:91–111. doi: 10.1007/978-1-60327-565-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith DE. Single-molecule studies of viral DNA packaging. Curr Opin Virol. 2011;1:134–41. doi: 10.1016/j.coviro.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–73. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black LW, Rao VB. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv Virus Res. 2012;82:119–53. doi: 10.1016/B978-0-12-394621-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H, Black LW. DNA requirements in vivo for phage T4 packaging. Virology. 1998;242:118–27. doi: 10.1006/viro.1997.9019. [DOI] [PubMed] [Google Scholar]
- 39.Meijer WJ, Horcajadas JA, Salas M. Phi29 family of phages. Microbiol Mol Biol Rev. 2001;65:261–87. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morais MC. The dsDNA packaging motor in bacteriophage ø29. Adv Exp Med Biol. 2012;726:511–47. doi: 10.1007/978-1-4614-0980-9_23. [DOI] [PubMed] [Google Scholar]
- 41.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–74. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci U S A. 2010;107:1971–6. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao VB, Black LW. Structure and assembly of bacteriophage T4 head. Virol J. 2010;7:356. doi: 10.1186/1743-422X-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao H, Kamau YN, Christensen TE, Tang L. Structural and functional studies of the phage Sf6 terminase small subunit reveal a DNA-spooling device facilitated by structural plasticity. J Mol Biol. 2012;423:413–26. doi: 10.1016/j.jmb.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy A, Bhardwaj A, Datta P, Lander GC, Cingolani G. Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure. 2012;20:1403–13. doi: 10.1016/j.str.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Büttner CR, Chechik M, Ortiz-Lombardía M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, et al. Structural basis for DNA recognition and loading into a viral packaging motor. Proc Natl Acad Sci U S A. 2012;109:811–6. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci U S A. 2012;109:817–22. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–62. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Zhao H, Christensen TE, Kamau YN, Tang L. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc Natl Acad Sci U S A. 2013;110:8075–80. doi: 10.1073/pnas.1301133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, Antson AA. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009;10:592–8. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy A, Cingolani G. Structure of p22 headful packaging nuclease. J Biol Chem. 2012;287:28196–205. doi: 10.1074/jbc.M112.349894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250:194–9. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 53.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–50. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. Mechanochemistry of a viral DNA packaging motor. J Mol Biol. 2010;400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Oliveira L, Tavares P, Alonso JC. Headful DNA packaging: bacteriophage SPP1 as a model system. Virus Res. 2013;173:247–59. doi: 10.1016/j.virusres.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 57.Olia AS, Prevelige PE, Jr., Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuervo A, Carrascosa JL. Viral connectors for DNA encapsulation. Curr Opin Biotechnol. 2012;23:529–36. doi: 10.1016/j.copbio.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 59.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–94. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78:12668–71. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–50. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental test of connector rotation during DNA packaging into bacteriophage phi29 capsids. PLoS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 64.Isidro A, Henriques AO, Tavares P. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology. 2004;322:253–63. doi: 10.1016/j.virol.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 65.Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J Biol Chem. 2006;281:21914–23. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- 66.Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J Biol Chem. 2007;282:18907–13. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- 67.Grimes S, Ma S, Gao J, Atz R, Jardine PJ. Role of φ29 connector channel loops in late-stage DNA packaging. J Mol Biol. 2011;410:50–9. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Padilla-Sanchez V, Gao S, Kim HR, Kihara D, Sun L, Rossmann MG, Rao VB. Structure-function analysis of the DNA translocating portal of the bacteriophage t4 packaging machine. J Mol Biol. 2014;426:1019–38. doi: 10.1016/j.jmb.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang J, Lander GC, Olia AS, Li R, Casjens S, Prevelige P, Jr., Cingolani G, Baker TS, Johnson JE. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–5. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 71.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/SQB.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 72.Moody MF. Geometry of phage head construction. J Mol Biol. 1999;293:401–33. doi: 10.1006/jmbi.1999.3011. [DOI] [PubMed] [Google Scholar]
- 73.Prasad BV, Schmid MF. Principles of virus structural organization. Adv Exp Med Biol. 2012;726:17–47. doi: 10.1007/978-1-4614-0980-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci U S A. 2004;101:6003–8. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163–8. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–7. doi: 10.1016/S0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–59. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 78.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–33. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 79.White HE, Sherman MB, Brasilès S, Jacquet E, Seavers P, Tavares P, Orlova EV. Capsid structure and its stability at the late stages of bacteriophage SPP1 assembly. J Virol. 2012;86:6768–77. doi: 10.1128/JVI.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci U S A. 2013;110:6811–6. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang RK, Khayat R, Lee KK, Gertsman I, Duda RL, Hendrix RW, Johnson JE. The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation. J Mol Biol. 2011;408:541–54. doi: 10.1016/j.jmb.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–4. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 84.Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Mühlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J Mol Biol. 2007;371:836–49. doi: 10.1016/j.jmb.2007.05.083. [DOI] [PubMed] [Google Scholar]
- 85.Aksyuk AA, Bowman VD, Kaufmann B, Fields C, Klose T, Holdaway HA, Fischetti VA, Rossmann MG. Structural investigations of a Podoviridae streptococcus phage C1, implications for the mechanism of viral entry. Proc Natl Acad Sci U S A. 2012;109:14001–6. doi: 10.1073/pnas.1207730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–70. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sutter M, Boehringer D, Gutmann S, Günther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–47. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 88.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol. 2003;334:885–99. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 89.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiang Y, Rossmann MG. Structure of bacteriophage phi29 head fibers has a supercoiled triple repeating helix-turn-helix motif. Proc Natl Acad Sci U S A. 2011;108:4806–10. doi: 10.1073/pnas.1018097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fokine A, Battisti AJ, Kostyuchenko VA, Black LW, Rossmann MG. Cryo-EM structure of a bacteriophage T4 gp24 bypass mutant: the evolution of pentameric vertex proteins in icosahedral viruses. J Struct Biol. 2006;154:255–9. doi: 10.1016/j.jsb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 92.Condron BG, Atkins JF, Gesteland RF. Frameshifting in gene 10 of bacteriophage T7. J Bacteriol. 1991;173:6998–7003. doi: 10.1128/jb.173.21.6998-7003.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Plückthun A, Wlodawer A. Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat Struct Biol. 2000;7:230–7. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 94.Sternberg N, Weisberg R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J Mol Biol. 1977;117:733–59. doi: 10.1016/0022-2836(77)90067-5. [DOI] [PubMed] [Google Scholar]
- 95.Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, et al. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proc Natl Acad Sci U S A. 2013;110:12301–6. doi: 10.1073/pnas.1309947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qin L, Fokine A, O’Donnell E, Rao VB, Rossmann MG. Structure of the small outer capsid protein, Soc: a clamp for stabilizing capsids of T4-like phages. J Mol Biol. 2010;395:728–41. doi: 10.1016/j.jmb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bateman A, Eddy SR, Mesyanzhinov VV. A member of the immunoglobulin superfamily in bacteriophage T4. Virus Genes. 1997;14:163–5. doi: 10.1023/A:1007977503658. [DOI] [PubMed] [Google Scholar]
- 98.Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J Virol. 2011;85:8141–8. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sathaliyawala T, Islam MZ, Li Q, Fokine A, Rossmann MG, Rao VB. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol Microbiol. 2010;77:444–55. doi: 10.1111/j.1365-2958.2010.07219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fraser JS, Maxwell KL, Davidson AR. Immunoglobulin-like domains on bacteriophage: weapons of modest damage? Curr Opin Microbiol. 2007;10:382–7. doi: 10.1016/j.mib.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 101.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol. 2006;359:496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 102.Dabrowska K, Switała-Jeleń K, Opolski A, Górski A. Possible association between phages, Hoc protein, and the immune system. Arch Virol. 2006;151:209–15. doi: 10.1007/s00705-005-0641-7. [DOI] [PubMed] [Google Scholar]
- 103.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Black LW, Thomas JA. Condensed genome structure. Adv Exp Med Biol. 2012;726:469–87. doi: 10.1007/978-1-4614-0980-9_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Depping R, Lohaus C, Meyer HE, Rüger W. The mono-ADP-ribosyltransferases Alt and ModB of bacteriophage T4: target proteins identified. Biochem Biophys Res Commun. 2005;335:1217–23. doi: 10.1016/j.bbrc.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 106.Lipinska B, Rao AS, Bolten BM, Balakrishnan R, Goldberg EB. Cloning and identification of bacteriophage T4 gene 2 product gp2 and action of gp2 on infecting DNA in vivo. J Bacteriol. 1989;171:488–97. doi: 10.1128/jb.171.1.488-497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bair CL, Rifat D, Black LW. Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J Mol Biol. 2007;366:779–89. doi: 10.1016/j.jmb.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rifat D, Wright NT, Varney KM, Weber DJ, Black LW. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J Mol Biol. 2008;375:720–34. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–9. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Choi KH, McPartland J, Kaganman I, Bowman VD, Rothman-Denes LB, Rossmann MG. Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J Mol Biol. 2008;378:726–36. doi: 10.1016/j.jmb.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu W, Thomas JA, Cheng N, Black LW, Steven AC. Bubblegrams reveal the inner body of bacteriophage φKZ. Science. 2012;335:182. doi: 10.1126/science.1214120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fokine A, Battisti AJ, Bowman VD, Efimov AV, Kurochkina LP, Chipman PR, Mesyanzhinov VV, Rossmann MG. Cryo-EM study of the Pseudomonas bacteriophage phiKZ. Structure. 2007;15:1099–104. doi: 10.1016/j.str.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 113.Leforestier A. Polymorphism of DNA conformation inside the bacteriophage capsid. J Biol Phys. 2013;39:201–13. doi: 10.1007/s10867-013-9315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–6. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–80. doi: 10.1016/S0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 116.Leforestier A, Livolant F. The bacteriophage genome undergoes a succession of intracapsid phase transitions upon DNA ejection. J Mol Biol. 2010;396:384–95. doi: 10.1016/j.jmb.2009.11.047. [DOI] [PubMed] [Google Scholar]
- 117.Earnshaw WC, King J, Harrison SC, Eiserling FA. The structural organization of DNA packaged within the heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978;14:559–68. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- 118.Lhuillier S, Gallopin M, Gilquin B, Brasilès S, Lancelot N, Letellier G, Gilles M, Dethan G, Orlova EV, Couprie J, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci U S A. 2009;106:8507–12. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, et al. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J Mol Biol. 2010;395:754–68. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 120.Maxwell KL, Yee AA, Booth V, Arrowsmith CH, Gold M, Davidson AR. The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold. J Mol Biol. 2001;308:9–14. doi: 10.1006/jmbi.2001.4582. [DOI] [PubMed] [Google Scholar]
- 121.Maxwell KL, Yee AA, Arrowsmith CH, Gold M, Davidson AR. The solution structure of the bacteriophage lambda head-tail joining protein, gpFII. J Mol Biol. 2002;318:1395–404. doi: 10.1016/S0022-2836(02)00276-0. [DOI] [PubMed] [Google Scholar]
- 122.Cardarelli L, Pell LG, Neudecker P, Pirani N, Liu A, Baker LA, Rubinstein JL, Maxwell KL, Davidson AR. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc Natl Acad Sci U S A. 2010;107:14384–9. doi: 10.1073/pnas.1005822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev. 2011;75:423–33. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cardarelli L, Maxwell KL, Davidson AR. Assembly mechanism is the key determinant of the dosage sensitivity of a phage structural protein. Proc Natl Acad Sci U S A. 2011;108:10168–73. doi: 10.1073/pnas.1100759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rao VB. A virus DNA gate: zipping and unzipping the packed viral genome. Proc Natl Acad Sci U S A. 2009;106:8403–4. doi: 10.1073/pnas.0903670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ackermann H-W. Classification of Bacteriophages. In: Calendar RL, ed. The Bacteriophages. New York, NY: Oxford University Press, 2006:8-16 [Google Scholar]
- 127.Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:115–42. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- 128.Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 129.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Plisson C, White HE, Auzat I, Zafarani A, São-José C, Lhuillier S, Tavares P, Orlova EV. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J. 2007;26:3720–8. doi: 10.1038/sj.emboj.7601786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Veesler D, Spinelli S, Mahony J, Lichière J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci U S A. 2012;109:8954–8. doi: 10.1073/pnas.1200966109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sciara G, Bebeacua C, Bron P, Tremblay D, Ortiz-Lombardia M, Lichière J, van Heel M, Campanacci V, Moineau S, Cambillau C. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci U S A. 2010;107:6852–7. doi: 10.1073/pnas.1000232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bebeacua C, Tremblay D, Farenc C, Chapot-Chartier MP, Sadovskaya I, van Heel M, Veesler D, Moineau S, Cambillau C. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J Virol. 2013;87:12302–12. doi: 10.1128/JVI.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abuladze NK, Gingery M, Tsai J, Eiserling FA. Tail length determination in bacteriophage T4. Virology. 1994;199:301–10. doi: 10.1006/viro.1994.1128. [DOI] [PubMed] [Google Scholar]
- 135.Katsura I, Hendrix RW. Length determination in bacteriophage lambda tails. Cell. 1984;39:691–8. doi: 10.1016/0092-8674(84)90476-8. [DOI] [PubMed] [Google Scholar]
- 136.Pell LG, Liu A, Edmonds L, Donaldson LW, Howell PL, Davidson AR. The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J Mol Biol. 2009;389:938–51. doi: 10.1016/j.jmb.2009.04.072. [DOI] [PubMed] [Google Scholar]
- 137.Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat Struct Mol Biol. 2005;12:810–3. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- 138.Casjens SR, Hendrix RW. Locations and amounts of major structural proteins in bacteriophage lambda. J Mol Biol. 1974;88:535–45. doi: 10.1016/0022-2836(74)90500-2. [DOI] [PubMed] [Google Scholar]
- 139.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A. 2009;106:4160–5. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pell LG, Gasmi-Seabrook GM, Morais M, Neudecker P, Kanelis V, Bona D, Donaldson LW, Edwards AM, Howell PL, Davidson AR, et al. The solution structure of the C-terminal Ig-like domain of the bacteriophage λ tail tube protein. J Mol Biol. 2010;403:468–79. doi: 10.1016/j.jmb.2010.08.044. [DOI] [PubMed] [Google Scholar]
- 141.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–30. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–9. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zivanovic Y, Confalonieri F, Ponchon L, Lurz R, Chami M, Flayhan A, Renouard M, Huet A, Decottignies P, Davidson AR, et al. Insights into bacteriophage T5 structure from analysis of its morphogenesis genes and protein components. J Virol. 2014;88:1162–74. doi: 10.1128/JVI.02262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Edmonds L, Liu A, Kwan JJ, Avanessy A, Caracoglia M, Yang I, Maxwell KL, Rubenstein J, Davidson AR, Donaldson LW. The NMR structure of the gpU tail-terminator protein from bacteriophage lambda: identification of sites contributing to Mg(II)-mediated oligomerization and biological function. J Mol Biol. 2007;365:175–86. doi: 10.1016/j.jmb.2006.09.068. [DOI] [PubMed] [Google Scholar]
- 145.Zhao L, Kanamaru S, Chaidirek C, Arisaka F. P15 and P3, the tail completion proteins of bacteriophage T4, both form hexameric rings. J Bacteriol. 2003;185:1693–700. doi: 10.1128/JB.185.5.1693-1700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–9. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–29. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 148.Aksyuk AA, Kurochkina LP, Fokine A, Forouhar F, Mesyanzhinov VV, Tong L, Rossmann MG. Structural conservation of the myoviridae phage tail sheath protein fold. Structure. 2011;19:1885–94. doi: 10.1016/j.str.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–93. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 150.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–3. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci U S A. 2010;107:20287–92. doi: 10.1073/pnas.1011218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG. Evolution of bacteriophage tails: Structure of T4 gene product 10. J Mol Biol. 2006;358:912–21. doi: 10.1016/j.jmb.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 153.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–7. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 154.Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG. Phage pierces the host cell membrane with the iron-loaded spike. Structure. 2012;20:326–39. doi: 10.1016/j.str.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 155.Veesler D, Robin G, Lichière J, Auzat I, Tavares P, Bron P, Campanacci V, Cambillau C. Crystal structure of bacteriophage SPP1 distal tail protein (gp19.1): a baseplate hub paradigm in gram-positive infecting phages. J Biol Chem. 2010;285:36666–73. doi: 10.1074/jbc.M110.157529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Flayhan A, Vellieux FM, Lurz R, Maury O, Contreras-Martel C, Girard E, Boulanger P, Breyton C. Crystal structure of pb9, the distal tail protein of bacteriophage T5: a conserved structural motif among all siphophages. J Virol. 2014;88:820–8. doi: 10.1128/JVI.02135-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Goulet A, Lai-Kee-Him J, Veesler D, Auzat I, Robin G, Shepherd DA, Ashcroft AE, Richard E, Lichière J, Tavares P, et al. The opening of the SPP1 bacteriophage tail, a prevalent mechanism in Gram-positive-infecting siphophages. J Biol Chem. 2011;286:25397–405. doi: 10.1074/jbc.M111.243360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tang J, Olson N, Jardine PJ, Grimes S, Anderson DL, Baker TS. DNA poised for release in bacteriophage phi29. Structure. 2008;16:935–43. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J. 2006;25:5229–39. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lander GC, Khayat R, Li R, Prevelige PE, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure. 2009;17:789–99. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–6. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, González-García VA, Garcia-Doval C, Castón JR, Valpuesta JM, van Raaij MJ, Martín-Benito J, et al. Structural characterization of the bacteriophage T7 tail machinery. J Biol Chem. 2013;288:26290–9. doi: 10.1074/jbc.M113.491209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xiang Y, Leiman PG, Li L, Grimes S, Anderson DL, Rossmann MG. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell. 2009;34:375–86. doi: 10.1016/j.molcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope–penetrating needle. Nat Struct Mol Biol. 2007;14:1221–6. doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- 165.Garcia-Doval C, van Raaij MJ. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc Natl Acad Sci U S A. 2012;109:9390–5. doi: 10.1073/pnas.1119719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Steinbacher S, Baxa U, Miller S, Weintraub A, Seckler R, Huber R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc Natl Acad Sci U S A. 1996;93:10584–8. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science. 1994;265:383–6. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- 168.Barbirz S, Müller JJ, Uetrecht C, Clark AJ, Heinemann U, Seckler R. Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol Microbiol. 2008;69:303–16. doi: 10.1111/j.1365-2958.2008.06311.x. [DOI] [PubMed] [Google Scholar]
- 169.Schulz EC, Schwarzer D, Frank M, Stummeyer K, Mühlenhoff M, Dickmanns A, Gerardy-Schahn R, Ficner R. Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF. J Mol Biol. 2010;397:341–51. doi: 10.1016/j.jmb.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 170.Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol. 2005;12:90–6. doi: 10.1038/nsmb874. [DOI] [PubMed] [Google Scholar]