Abstract
Background
Apical potassium channels regulate ion transport in airway epithelial cells and influence air surface liquid (ASL) hydration and mucociliary clearance (MCC). We sought to identify whether genetic variation within genes encoding airway potassium channels is associated with chronic rhinosinusitis (CRS).
Methods
Single nucleotide polymorphism (SNP) genotypes for selected potassium channels were derived from data generated on the Illumnia HumanHap550 BeadChip or Illumina Human610-Quad BeadChip for 828 unrelated individuals diagnosed with CRS and 5,083 unrelated healthy controls from the Children's Hospital of Philadelphia (CHOP). Statistical analysis was performed with set-based tests using PLINK, and corrected for multiple testing.
Results
Set-based case control analysis revealed the gene KCNMA1 was associated with CRS in our Caucasian subset of the cohort (598 CRS cases and 3,489 controls; p = 0.022, based on 10,000 permutations). In addition there was borderline evidence that the gene KCNQ5 (p = 0.0704) was associated with the trait in our African American subset of the cohort (230 CRS cases and 1,594 controls). In addition to the top significant SNPs rs2917454 and rs6907229, imputation analysis uncovered additional genetic variants in KCNMA1 and in KCNQ5 that were associated with CRS.
Conclusions
We have implicated two airway epithelial potassium channels as novel susceptibility loci in contributing to the pathogenesis of CRS.
Introduction
Chronic rhinosinusits (CRS) is a spectrum of clinical disease characterized by persistent inflammation of the nasal cavity and paranasal sinuses [1]. Historically, CRS was considered an extension of acute bacterial sinusitis and the role of bacterial infection was considered primary in the development of the disease. Recent research and clinical focus has shifted from a primary infectious etiology (in most cases) to chronic mucosal inflammation as the primary underlying dysfunction in CRS [2]. Despite advances in our understanding of the cellular and molecular changes in the nasal mucosa of patients with CRS, the underlying pathophysiology remains incompletely understood and likely includes many different pathophysiologic mechanisms that result from a complex underlying genetic predisposition that is subsequently influenced by environmental effects.
Multiple clinical observations of the cause of chronic mucosal inflammation drive the rationale for current medical and surgical treatment of the disease and include anatomic osteomeatal complex obstruction, allergy associated inflammation, osteitis, MCC dysfunction, and advanced microbial defense mechanisms such as biofilm formation [3]. Despite our attempts to classify patients based on clinical and radiographic findings, most cases of CRS are currently considered idiopathic and our treatment algorithms are variable in their efficacy for a given patient [4].
Classic studies of heritability for CRS have not been performed, however a genetic basis of disease has been suspected for many reasons [5]. Familial aggregation of disease points to an underlying genetic basis and many reports have shown unusually high prevalence of CRS within families [6]–[8]. Patients with CRS are also more likely to report a positive family history [9]. Observations of the sinonasal manifestations of primary ciliary dyskinesia and cystic fibrosis (CF) raise the possibility that the development of CRS may have a common genetic etiology. Others have shown an increased prevalence of CRS in carriers of the cystic fibrosis transmembrane conductance regulator (CFTR) mutation [10], [11]. Further, there is evidence that the inflammatory profiles in CRS are similar to allergic rhinitis and asthma, two complex diseases with a well-established genetic basis [12].
Cystic fibrosis is the classic example of a monogenic disease producing a CRS phenotype. In the case of CF, the almost universal and often severe sinus disease that develops has been long recognized [13]. In CF patients, changes in the CFTR gene on chromosome 7 result in dysfunctional ion transport of chloride across the apical membrane of exocrine glandular cells, including airway epithelium, and subsequent ASL dehydration develops [13], [14]. This environment is susceptible to chronic bacterial colonization and infection which triggers a host response and leads to a severe phenotype of CRS. The presence of a single gene defect causing ion dysregulation that leads to CRS is a striking observation. We are interested in elucidating whether this observation has implications for the early molecular changes in the non-CF adult patient who develops CRS.
Recent work has been performed examining the role of various potassium channels and their relationship to ion transport in bronchial epithelial cells. Potassium channels are known to be functional at both the basolateral and apical membrane of airway epithelium [15]. The collective function of these channels is to mediate potassium currents of diverse electrophysiological and regulatory properties. The role of basolateral potassium channels to regulate membrane potential and maintain an electrochemical driving force for transepithelial ion and liquid transport is well established [16]. Understanding the role of apical potassium channels and exploring the possibility of utilizing them as a therapeutic target for diseases characterized by abnormal ion transport is an emerging field [17]. An exciting clinical example of the potential of ion transport modulation is the recent success of the CFTR potentiating agent, ivacaftor, in the treatment of CF. This investigational, orally administered drug augments CFTR function and has been shown to produce sustained improvement in multiple clinical outcome measures in patients with CF while possessing an acceptable safety profile [18]–[20].
Three main classes of apical epithelial potassium channels have been identified in respiratory epithelium. These include large conductance Ca2+-activated and voltage-dependent 6-TMD (KCa and Kv), 2-pore 4-TMD (K2P), and inward rectified 2-TMD potassium channels (Kir) [21]. Evidence suggests that all three classes may have the ability to modulate ion transport at the epithelial border, and subsequently impact air surface hydration, and therefore MCC. Large conductance, Ca2+-activated, and voltage-dependent BK channels have specifically been shown to contribute to an apical loop current favoring apical chloride efflux and maintenance of ASL hydration [22]. Members of the Kir family have been shown to impact trans-epithelial transport of sodium and chloride ions in addition to modulating amiloride-sensitive sodium channel (ENaC) and CFTR expression [23]. Other studies have shown that K2P channels are expressed and functionally active at the apical membrane of the airway epithelium [17].
The aim of this study is to identify whether genetic variation within genes encoding known airway potassium channel genes is associated with chronic sinusitis in a pediatric population.
Methods
Study subjects
In our study, 828 unrelated individuals diagnosed with CRS and 5,083 unrelated controls without CRS were included. Following genotyping, principal component analysis was conducted using EIGENSTRAT [24], in which 230 CRS cases and 1,594 controls were identified as African Americans; 598 CRS cases and 3,489 controls were identified as Caucasians. Further analyses were performed in these two ethnicity groups separately.
Patients included in this study had a history of CRS diagnosed by a Pediatric Otolaryngologist at CHOP using previously published criteria [25] that included history and physical exam findings consistent with CRS. Common symptoms of CRS included rhinorrhea, nasal congestion, facial pain, and post nasal drip. The duration of symptoms was greater than 12 weeks in all patients. Anterior rhinoscopy or flexible nasal endoscopy was performed in all patients. While not required for the diagnosis of CRS in children, CT imaging of the sinuses was used as an ancillary test in 46.2% of patients to confirm the diagnosis of CRS. All patients underwent allergy and immunological testing when clinically indicated as well as exclusion of CF when appropriate. Further phenotype refinement was performed to exclude CF patients as described below.
All recruited individuals filled in detailed questionnaires of medical history, medication and demographic data. Data were stored in a fully-integrated electronic medical record of the study subjects. Parental DNA was available for approximately 20% of the study participants. The institutional review board (IRB) of the Children's Hospital of Philadelphia specifically approved this study. Adults (>18 years) included in this study provided informed written consent using consent guidelines/procedures approved by the IRB of the Children's Hospital of Philadelphia. Written consent for children/minors included in the study was obtained from appropriate legal guardians with strict adherence to consent guidelines/procedures approved by the IRB of the Children's Hospital of Philadelphia.
SNP genotyping
DNA extraction from whole blood and SNP genotyping was performed at the Center for Applied Genomics, CHOP. CRS cases and control samples were genotyped on Illumnia HumanHap550 BeadChip or Illumina Human610-Quad BeadChip, which share 535,752 common SNP markers. Because we are interested in airway potassium channel genes, we focused on the surprisingly large collection of potassium channel genes that have been identified in the literature [21]. Since important transcriptional or translational regulators could reside upstream or downstream of a gene, we extracted genotyping information on SNPs which are located within 20 kb upstream and downstream of the tested potassium channel genes.
CFTR analysis
It is well known that CFTR has many subtle variants that contribute to pediatric rhinosinusitis. As such, CF is a significant confounding factor that needs to be cautiously identified and removed in a non CF CRS genetics study. We sought to identify all CF patients in our study population and exclude them from subsequent analysis. CF cases within our initial study population were identified by searching the EMR for the ICD code 277. Newborn screening for CF is routine in Pennsylvania and surrounding states. This test is highly sensitive. When combined with the high specificity of sweat chloride testing we are able to identify the overwhelming majority of CF cases in our population. We initially identified an additional 47 CF CRS cases that met study criteria and performed a genome-wide association study (GWAS) that included both CF and non CF CRS populations as described above. Allelic tests implemented in PLINK [26] were used to examine the genotype-phenotype association. In the Caucasian cohort, SNPs located close to or within the CFTR gene were highly significant in this GWAS analysis. However, such association was remarkably ablated when the CF CRS patients were removed (Table S1). This supports that CF is a significant confounding factor that needs to be cautiously identified and removed in a non CF CRS genetics study. The loss of CFTR association with CRS is strong evidence for accurate phenotype identification of the subsequent non CF CRS population that underwent further analysis. Of note, SNPs in the CFTR gene did not show significant association with the CRS phenotype in our African American cohort, with only one SNP with a p-value<0.05 (rs11978434, p-value = 0.0294).
Statistical analysis
We excluded SNP markers which had a call rate <95%, minor allele frequency <0.01 or significant deviation from Hardy-Weinberg equilibrium (P<0.0001). Allelic tests were performed for each SNP using PLINK [26]. Set-based tests were carried out for 44 genes using PLINK. The two most significant independent SNPs (r2 threshold = 0.1) with p-value<0.01 were selected for each set. The mean of the selected SNP statistics was used to represent the statistics of the gene set. Permutations of case/control status were performed 10,000 times to obtain the null distribution of the statistics for each gene set under the hypothesis of no genetic association. Empirical p-values for each gene set were generated by comparing the observed test statistics and the ones based on permutations [26]–[28]. Multiple-testing corrected p-values were generated based on Bonferroni correction for 44 tests. In our imputation analysis via software package IMPUTE2 [29], [30], we used the 1000 Genome Phase I integrated variant set as reference panel (http://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html). For association testing on the imputed variants, we performed missing data likelihood score tests with the SNPTEST v2 package [29]. An estimation of phenotypic variance explained by independent SNPs was performed with GCTA software [31]. We tested whether there is any interaction between independent SNPs in genes KCNMA1 and KCNQ5 via epistasis test implemented in PLINK [26].
Results
There were 230 CRS cases and 1,594 controls in the African American cohort and 598 cases and 3,489 controls in the Caucasian group. Mean ages were similar between groups. Overall, 69% of African American and 60% of Caucasian CRS cases were diagnosed with asthma; 48% of the cases in the African American cohort and 62% of the cases in the Caucasian cohort had atopic syndrome. CRS cases were more likely to have a clinical diagnosis of otitis media. Both CRS groups were also more likely than controls to have undergone placement of tympanostomy tubes or surgical intervention for CRS-either functional endoscopic sinus surgery or adenoidectomy (Table 1). P-values<0.005 in both Caucasian and African American cohorts for the above comparisons between cases and controls.
Table 1. Demographics and clinical features of study groups.
| African American Cohort | Caucasian Cohort | |||
| Cases | Controls | Cases | Controls | |
| Age (years) | 11.7±5.2 | 9.5±5.8 | 12.1±4.7 | 10.8±5.8 |
| Otitis Media (%) | 48.26 | 21.59 | 50.92 | 9.81 |
| Ear Tube (%) | 15.22 | 0.63 | 25.96 | 1.49 |
| Sinus Surgery (%) | 29.57 | 4.39 | 42.21 | 6.0 |
| FESS (%) | 1.30 | 0 | 7.20 | 0.20 |
| Adenoidectomy (%) | 28.70 | 4.39 | 39.70 | 5.82 |
For Age, mean ± standard deviation is shown.
The complete list of analyzed genes encoding airway potassium channels is shown in Table 2. To determine if any genes were associated with the diagnosis of CRS we performed a PLINK set-based analysis as described above. In the Caucasian cohort, the locus yielding the strongest signal was at the KCNMA1 gene, with an empirical p-value = 0.022 after multiple testing correction (Table 3).
Table 2. List of analyzed potassium channels.
| Name | Gene | Locus | Reference |
| Six-TMD, voltage-dependent K+ channels | |||
| Kv1.1 | KCNA1 | 12p13.32 | [44] |
| Kv1.3 | KCNA3 | 1p13.3 | [44] |
| Kv1.4 | KCNA4 | 11p14 | [44] |
| Kv1.5 | KCNA5 | 12p13 | [45] |
| Kv1.7 | KCNA7 | 19q13.3 | [45] |
| Kv2.2 | KCNB2 | 8q | [44] |
| Kv4.1 | KCND1 | Xp11.23 | [44] |
| Kv4.2 | KCND2 | 7q31 | [44] |
| Kv4.3 | KCND3 | 1p13.3 | [44] |
| Kv6.1 | KCNG1 | 20q13 | [45] |
| KvLQT1 (Kv7.1) | KCNQ1 | 11p15.5 | [46], [47] |
| Kv7.2 | KCNQ2 | 20q13.3 | [48] |
| Kv7.3 | KCNQ3 | 8q24 | [48] |
| Kv7.4 | KCNQ4 | 1p34 | [48] |
| Kv7.5 | KCNQ5 | 6q14 | [48] |
| Kv9.3 | KCNS3 | 2p24 | [44] |
| β-subunits | |||
| MiRP1 | KCNE2 | 21q22.12 | [49] |
| MiRP2 | KCNE3 | 11q13-q14 | [49] |
| MiRP3 | KCNE4 | 2q36.3 | [50] |
| KChIP2 | KCNIP2 | 10q24 | [44] |
| KChIP3 | KCNIP3 | 2q21.1 | [44] |
| Kvβ1 | KCNAB1 | 3q26.1 | [44] |
| Kvβ2 | KCNAB2 | 1q36.3 | [44] |
| Kvβ3 | KCNAB3 | 17p13.1 | [44] |
| Six-TMD, Ca2+ activated K+ channels | |||
| SK1 | KCNN1 | 19q13.1 | [51] |
| KCa3.1 | KCNN4 | 19q13.2 | [46] |
| BKca | KCNMA1 | 10q22.3 | [52], [53] |
| Two-TMD, inward-rectified K+ channels | |||
| Kir2.1 | KCNJ2 | 17q23-1q24.2 | [54] |
| Kir3.1 GIRK1 | KCNJ3 | 2q24.1 | [55] |
| Kir3.2 GIRK2 | KCNJ6 | 21q22.13-q22.2 | [55] |
| Kir3.3 GIRK3 | KCNJ9 | 1q21-q23 | [55] |
| Kir3.4 GIRK4 | KCNJ5 | 11q24 | [55] |
| Kir4.2 | KCNJ15 | 21q22.2 | [56] |
| Kir6.1 | KCNJ8 | 12p11.23 | [56] |
| Kir7.1 | KCNJ13 | 2q37 | [57] |
| Four-TMD, 2 pore K+ channels | |||
| Twik 1 | KCNK1 | 1q42-q43 | [58] |
| Twik 2 | KCNK6 | 19q13.1 | [58] |
| Trek 1 | KCNK2 | 1q41 | [58] |
| Trek 2 | KCNK10 | 14q31 | [58] |
| Task 2 | KCNK5 | 6p21 | [58] |
| Task 3 | KCNK9 | 8q24.3 | [58] |
| Task 4 | KCNK17 | 6p21.2-p21.1 | [58] |
| Thik 1 | KCNK13 | 14q31-q32 | [58] |
| KCNK7 | KCNK7 | 11q13 | [58] |
Table 3. Results of gene set-based analyses of 44 potassium channel genes.
| Caucasian | African American | |||||||||||||
| Gene | Chr | Start (bp, hg18) | End (bp, hg18) | Size (kb) | NSNP | NSIG | ISIG | P-value | Adj. P-value | NSNP | NSIG | ISIG | P-value | Adj. P-value |
| KCNA1 | 12 | 4889333 | 4897683 | 8.35 | 12 | 0 | 0 | 1 | 1 | 11 | 0 | 0 | 1 | 1 |
| KCNA3 | 1 | 111015832 | 111019178 | 3.35 | 6 | 0 | 0 | 1 | 1 | 6 | 0 | 0 | 1 | 1 |
| KCNA4 | 11 | 29988340 | 29995064 | 6.72 | 10 | 0 | 0 | 1 | 1 | 10 | 0 | 0 | 1 | 1 |
| KCNA5 | 12 | 5023345 | 5026210 | 2.87 | 33 | 0 | 0 | 1 | 1 | 31 | 0 | 0 | 1 | 1 |
| KCNA7 | 19 | 54262486 | 54268010 | 5.52 | 9 | 0 | 0 | 1 | 1 | 7 | 0 | 0 | 1 | 1 |
| KCNAB1 | 3 | 157321030 | 157739621 | 418.59 | 92 | 0 | 0 | 1 | 1 | 96 | 1 | 1 | 0.18 | 1 |
| KCNAB2 | 1 | 6008966 | 6083110 | 74.14 | 14 | 0 | 0 | 1 | 1 | 14 | 0 | 0 | 1 | 1 |
| KCNAB3 | 17 | 7766751 | 7773478 | 6.73 | 4 | 0 | 0 | 1 | 1 | 4 | 0 | 0 | 1 | 1 |
| KCNB2 | 8 | 73612179 | 74013138 | 400.96 | 120 | 0 | 0 | 1 | 1 | 121 | 1 | 1 | 0.19 | 1 |
| KCND1 | X | 48703582 | 48713195 | 9.61 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 1 |
| KCND2 | 7 | 119700957 | 120177623 | 476.67 | 59 | 2 | 2 | 0.23 | 1 | 63 | 0 | 0 | 1 | 1 |
| KCND3 | 1 | 112119976 | 112333300 | 213.32 | 112 | 1 | 1 | 0.20 | 1 | 115 | 1 | 1 | 0.56 | 1 |
| KCNE2 | 21 | 34658192 | 34665310 | 7.12 | 14 | 0 | 0 | 1 | 1 | 13 | 0 | 0 | 1 | 1 |
| KCNE3 | 11 | 73843533 | 73856248 | 12.72 | 16 | 0 | 0 | 1 | 1 | 17 | 0 | 0 | 1 | 1 |
| KCNE4 | 2 | 223625105 | 223628599 | 3.49 | 7 | 0 | 0 | 1 | 1 | 9 | 0 | 0 | 1 | 1 |
| KCNG1 | 20 | 49053599 | 49073082 | 19.48 | 8 | 0 | 0 | 1 | 1 | 8 | 0 | 0 | 1 | 1 |
| KCNIP2 | 10 | 103575720 | 103593667 | 17.95 | 5 | 0 | 0 | 1 | 1 | 5 | 0 | 0 | 1 | 1 |
| KCNIP3 | 2 | 95326798 | 95415552 | 88.75 | 8 | 0 | 0 | 1 | 1 | 8 | 0 | 0 | 1 | 1 |
| KCNJ13 | 2 | 233339103 | 233349519 | 10.42 | 8 | 0 | 0 | 1 | 1 | 6 | 0 | 0 | 1 | 1 |
| KCNJ15 | 21 | 38550533 | 38595616 | 45.08 | 17 | 0 | 0 | 1 | 1 | 19 | 0 | 0 | 1 | 1 |
| KCNJ2 | 17 | 65677270 | 65687778 | 10.51 | 7 | 0 | 0 | 1 | 1 | 8 | 0 | 0 | 1 | 1 |
| KCNJ3 | 2 | 155263338 | 155421260 | 157.92 | 47 | 0 | 0 | 1 | 1 | 49 | 0 | 0 | 1 | 1 |
| KCNJ5 | 11 | 128266522 | 128293161 | 26.64 | 22 | 0 | 0 | 1 | 1 | 22 | 0 | 0 | 1 | 1 |
| KCNJ6 | 21 | 37918656 | 38210566 | 291.91 | 107 | 0 | 0 | 1 | 1 | 109 | 0 | 0 | 1 | 1 |
| KCNJ8 | 12 | 21809155 | 21819014 | 9.86 | 1 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 1 | 1 |
| KCNJ9 | 1 | 158317983 | 158325836 | 7.85 | 10 | 0 | 0 | 1 | 1 | 10 | 0 | 0 | 1 | 1 |
| KCNK1 | 1 | 231816372 | 231874607 | 58.24 | 42 | 0 | 0 | 1 | 1 | 43 | 0 | 0 | 1 | 1 |
| KCNK10 | 14 | 87720998 | 87863004 | 142.01 | 51 | 0 | 0 | 1 | 1 | 53 | 0 | 0 | 1 | 1 |
| KCNK13 | 14 | 89597860 | 89721948 | 124.09 | 33 | 0 | 0 | 1 | 1 | 34 | 0 | 0 | 1 | 1 |
| KCNK17 | 6 | 39374754 | 39390214 | 15.46 | 23 | 0 | 0 | 1 | 1 | 22 | 0 | 0 | 1 | 1 |
| KCNK2 | 1 | 213245507 | 213477059 | 231.55 | 43 | 0 | 0 | 1 | 1 | 47 | 1 | 1 | 0.26 | 1 |
| KCNK5 | 6 | 39264724 | 39305229 | 40.51 | 22 | 1 | 1 | 0.027 | 1 | 20 | 3 | 2 | 0.027 | 1 |
| KCNK6 | 19 | 43502323 | 43511489 | 9.17 | 6 | 0 | 0 | 1 | 1 | 6 | 0 | 0 | 1 | 1 |
| KCNK7 | 11 | 65116901 | 65120043 | 3.14 | 3 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 1 | 1 |
| KCNK9 | 8 | 140693985 | 140784481 | 90.5 | 45 | 0 | 0 | 1 | 1 | 44 | 2 | 2 | 0.042 | 1 |
| KCNMA1 | 10 | 78299367 | 79067583 | 768.22 | 224 | 2 | 2 | 0.0005 | 0.022 | 227 | 1 | 1 | 0.43 | 1 |
| KCNN1 | 19 | 17923110 | 17970930 | 47.82 | 18 | 0 | 0 | 1 | 1 | 18 | 0 | 0 | 1 | 1 |
| KCNN4 | 19 | 48962524 | 48977249 | 14.73 | 11 | 0 | 0 | 1 | 1 | 11 | 0 | 0 | 1 | 1 |
| KCNQ1 | 11 | 2422796 | 2826916 | 404.12 | 123 | 2 | 2 | 0.14 | 1 | 124 | 1 | 1 | 0.49 | 1 |
| KCNQ2 | 20 | 61507985 | 61574437 | 66.45 | 14 | 0 | 0 | 1 | 1 | 14 | 0 | 0 | 1 | 1 |
| KCNQ3 | 8 | 133210437 | 133562186 | 351.75 | 140 | 1 | 1 | 0.44 | 1 | 140 | 2 | 2 | 0.5 | 1 |
| KCNQ4 | 1 | 41022270 | 41076947 | 54.68 | 23 | 0 | 0 | 1 | 1 | 23 | 0 | 0 | 1 | 1 |
| KCNQ5 | 6 | 73388555 | 73962301 | 573.75 | 135 | 2 | 2 | 0.38 | 1 | 145 | 2 | 2 | 0.0016 | 0.0704 |
| KCNS3 | 2 | 17923425 | 17977706 | 54.28 | 22 | 0 | 0 | 1 | 1 | 21 | 0 | 0 | 1 | 1 |
NSNP: number of SNPs within the gene and its upstream and downstream 20 kb genomic region; NSIG: number of significant SNPs; ISIG: number of independent significant SNPs; adj. P-value: P-value adjusted for multiple testing.
In the smaller African American cohort, KCNQ5 demonstrated the strongest association, although it did not reach strict statistical significance (nominal p-value = 0.0016, corrected p-value = 0.0704 after multiple-testing correction).
In addition, two other genes were also associated with CRS with nominal p-value<0.05 (p = 0.027 for KCNK5 in both Caucasian cohort and African American cohort; p = 0.042 for KCNK9 in the African American cohort).
Detailed information on the independent significant SNPs for KCNMA1 and KCNQ5 are shown in Table 4. They are all located within the intron of their respective genes. Interestingly, the minor allele A of rs7900261 in KCNMA1 renders a protective effect for the development of CRS (OR = 0.82). It is estimated that 0.9% of the phenotypic variance in CRS occurrence is explained by the two independent significant SNPs in KCNMA1. Similarly, the two independent significant SNPs in KCNQ5 explained about 1% of the variance in CRS occurrence. We further investigated but found no interaction between independent significant SNPs in genes KCNMA1 and KCNQ5 respectively (Table S2). Allelic test results for all SNPs in these two genes are presented in Table S3.
Table 4. Significantly associated SNPs in genes KCNMA1 and KCNQ5.
| Cohort | Gene | SNP | Chr | bp(hg18) | Function | Minor/Major Allele | MAF (cases) | MAF (controls) | OR | SE | P-value |
| Caucasian | KCNMA1 | rs2917454 | 10 | 78562421 | intron | G/A | 0.09 | 0.05 | 1.84 | 0.12 | 1.39×10−7 |
| rs7900261 | 10 | 78419202 | intron | A/G | 0.36 | 0.41 | 0.82 | 0.06 | 2.91×10−3 | ||
| African American | KCNQ5 | rs6907229 | 6 | 73940870 | intron | C/T | 0.28 | 0.18 | 1.74 | 0.11 | 9.32×10−7 |
| rs9343015 | 6 | 73943714 | intron | C/T | 0.56 | 0.47 | 1.41 | 0.10 | 6.29×10−4 |
SNP = single nucleotide polymorphism; Chr = chromosome; bp = base pair; MAF = minor allele frequency; OR = odds ratio; SE = standard error.
We also performed imputation analysis and identified an additional 19 variants in KCNMA1 in LD with SNP rs2917454 associated with CRS (p-value<10−4) in the Caucasian cohort (Table 5). Further, we found 9 imputed variants in LD with rs6907229 in KCNQ5 were also associated with CRS (p-value<10−4) in the African American cohort (Table 6).
Table 5. Imputed variants in gene KCNMA1 with p-value<10−4 in the Caucasian cohort.
| Variant | Chr | Pos (hg19) | Effect Allele | Reference Allele | Effect Allele Frequency | OR | 95% CI | P-value |
| rs2250841 | 10 | 78910042 | T | G | 0.052 | 1.85 | 1.47, 2.34 | 1.85×10−7 |
| rs1871063 | 10 | 78899404 | T | C | 0.053 | 1.84 | 1.46, 2.31 | 2.05×10−7 |
| rs2766619 | 10 | 78897627 | T | C | 0.054 | 1.83 | 1.45, 2.31 | 2.36×10−7 |
| rs2766624 | 10 | 78893799 | C | T | 0.054 | 1.83 | 1.45, 2.31 | 2.41×10−7 |
| rs2616645 | 10 | 78889487 | G | A | 0.034 | 1.94 | 1.47, 2.57 | 1.41×10−6 |
| rs1871064 | 10 | 78899595 | A | G | 0.029 | 1.99 | 1.47, 2.69 | 4.09×10−6 |
| chr10:78889552:I | 10 | 78889552 | GA | G | 0.033 | 1.91 | 1.44, 2.55 | 5.41×10−6 |
| rs2574805 | 10 | 78905128 | A | G | 0.029 | 1.96 | 1.45, 2.65 | 6.49×10−6 |
| rs2616650 | 10 | 78901708 | A | G | 0.029 | 1.95 | 1.44, 2.64 | 7.10×10−6 |
| chr10:78894143:I | 10 | 78894143 | AC | A | 0.032 | 1.90 | 1.42, 2.53 | 7.81×10−6 |
| rs2766620 | 10 | 78899807 | A | T | 0.029 | 1.94 | 1.44, 2.63 | 7.88×10−6 |
| chr10:78894142:I | 10 | 78894142 | CAA | C | 0.033 | 1.89 | 1.42, 2.52 | 9.02×10−6 |
| rs2574799 | 10 | 78894351 | T | A | 0.033 | 1.89 | 1.42, 2.52 | 9.20×10−6 |
| rs2574797 | 10 | 78897797 | T | A | 0.030 | 1.92 | 1.43, 2.60 | 1.05×10−5 |
| rs2616647 | 10 | 78894478 | T | G | 0.030 | 1.92 | 1.42, 2.60 | 1.08×10−5 |
| rs2925826 | 10 | 78892469 | G | A | 0.030 | 1.92 | 1.42, 2.60 | 1.10×10−5 |
| chr10:78894141:I | 10 | 78894141 | TCA | T | 0.033 | 1.87 | 1.40, 2.49 | 1.31×10−5 |
| rs11002022 | 10 | 78878445 | T | C | 0.023 | 1.96 | 1.40, 2.75 | 4.79×10−5 |
| rs11002021 | 10 | 78877806 | A | G | 0.024 | 1.90 | 1.36, 2.65 | 8.45×10−5 |
Chr = chromosome; Pos = Position; OR = odds ratio; CI = confidence interval.
Table 6. Imputed variants in gene KCNQ5 with p-value<10−4 in the African American cohort.
| Variant | Chr | Pos (hg19) | Effect Allele | Reference Allele | Effect Allele Frequency | OR | 95% CI | P-value |
| rs9351980 | 6 | 73885135 | A | T | 0.806 | 0.57 | 0.46, 0.71 | 8.14×10−7 |
| chr6:73879813:D | 6 | 73879813 | A | AG | 0.033 | 2.50 | 1.64, 3.81 | 9.32×10−7 |
| rs1970549 | 6 | 73883761 | A | G | 0.804 | 0.57 | 0.46, 0.72 | 9.96×10−7 |
| rs9351979 | 6 | 73880210 | C | T | 0.781 | 0.59 | 0.48, 0.74 | 1.89×10−6 |
| rs2027545 | 6 | 73879839 | A | G | 0.781 | 0.60 | 0.48, 0.74 | 2.12×10−6 |
| rs1970547 | 6 | 73881323 | T | G | 0.799 | 0.59 | 0.47, 0.74 | 2.81×10−6 |
| rs2350386 | 6 | 73887606 | C | G | 0.878 | 0.58 | 0.45, 0.75 | 2.91×10−5 |
| rs7756501 | 6 | 73886305 | A | G | 0.643 | 0.67 | 0.55, 0.82 | 8.30×10−5 |
| rs2350385 | 6 | 73891177 | C | T | 0.646 | 0.68 | 0.55, 0.82 | 8.87×10−5 |
Chr = chromosome; Pos = Position; OR = odds ratio; CI = confidence interval.
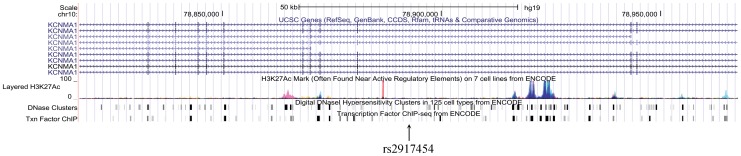
The two independent SNPs in KCNMA1, as well as those in KCNQ5 are located in introns, therefore the polymorphisms of these variants do not have a direct effect on protein coding. However, intronic sequences may have an impact on several aspects of gene transcription [32]. Recently released results from the ENCODE project revealed a huge number of functional elements in the human genome [33], including many residing within introns. The NIH Roadmap Epigenomics project has also provided an enormous resource of epigenomic data [34]. By assessing the significant loci of these genes using the Integrated Regulation from ENCODE track in the UCSC genome browser, we found clear H3K27ac modification marks in close proximity to rs2917454 in KCNMA1(Figure 1). We also performed genomic annotation with the ENCODE database and Roadmap Epigenomics data via HaploReg software [35]. The results for the Caucasian and African American cohorts are presented in Tables 7 and 8, respectively. Interestingly, the results demonstrated that SNP rs2917454 overlaps with a glucocorticoid receptor binding motif. The other identified SNP of KCNMA1 (rs7900261) overlaps with an enhancer histone mark and EWSR1-FLI1 motif suggesting a possible role in the regulation of gene expression. Additional neighboring SNPs, which are in perfect LD with rs2917454 and rs7900261, also overlap with enhancer histone marks, P300 binding sites and other transcription factor motifs (Table 7). Examination of SNPs in KCNQ5 reveals similar associations with multiple promoter and enhancer histone marks as well as transcription factor binding motifs which have overlap with neighboring SNPs in good LD with rs6907229 and rs9343015 (Table 8).
Figure 1. H3K27ac marks representing active regulatory elements found near the locus of rs2917454 in gene KCNMA1.
Table 7. Regulatory elements at the significant loci in gene KCNMA1.
| SNP | Chr | Pos(hg19) | r2 | D′ | Enhancer histone marks | DNAse hypersensitivity sites | Binding Proteins | Regulatory Motifs |
| rs2917454 | 10 | 78892415 | 1 | 1 | . | GR | ||
| rs2766624 | 10 | 78893799 | 1 | 1 | 1 cell type | Pax-8 | ||
| rs2766619 | 10 | 78897627 | 1 | 1 | 6 cell types | Nanog | ||
| rs1871063 | 10 | 78899404 | 1 | 1 | 3 cell types | AG04449,AoAF | MAZ | |
| rs2250841 | 10 | 78910042 | 0.97 | 1 | 11 cel types | P300 | EWSR1-FLI1;Pax-4;SP2 | |
| rs7900261 | 10 | 78749196 | 1 | 1 | 1 cell type | HGF | EWSR1-FLI1 | |
| rs3781153 | 10 | 78750074 | 1 | 1 | 1 cell type | HMG-IY;Hsf;STAT |
r2 and D′ are measures of LD between the indicated SNP and the significant SNPs rs2917454 or rs7900261.
Table 8. Regulatory elements at the significant loci in gene KCNQ5.
| SNP | Chr | Pos(hg19) | r2 | D′ | Promoter histone marks | Enhancer histone marks | DNAse hypersensitivity sites | Regulatory Motifs |
| rs200877792 | 6 | 73873577 | 0.82 | 0.95 | Hepatocytes | Foxa;HDAC2;Homez | ||
| rs947747 | 6 | 73875824 | 0.8 | 0.93 | Irf;Pbx3;p300 | |||
| rs2350387 | 6 | 73877424 | 0.8 | 0.93 | 1 cell type | Evi-1;Hoxb13;Irf;Pbx-1;SP1 | ||
| rs7763514 | 6 | 73877796 | 0.8 | 0.93 | 2 cell types | LRH1 | ||
| rs2027545 | 6 | 73879839 | 0.81 | 1 | 1 cell type | 4 cell types | Evi-1;Foxp1 | |
| rs9351979 | 6 | 73880210 | 0.81 | 1 | 1 cell type | Pou3f2;Sox | ||
| rs1970547 | 6 | 73881323 | 0.9 | 1 | CDP;HNF1;PLZF | |||
| rs1970549 | 6 | 73883761 | 0.85 | 0.95 | COMP1;Foxj2;Foxk1;Sox | |||
| rs9351980 | 6 | 73885135 | 1 | 1 | Ik-1;Ik-2;NRSF;Sin3Ak-20 | |||
| rs9343015 | 6 | 73886993 | 1 | 1 | 1 cell type | 3 cell types | GM06990 | Pou2f2 |
| rs2882405 | 6 | 73887759 | 1 | 1 | 2 cell types | Mef2 | ||
| rs3068379 | 6 | 73887987 | 0.97 | 1 | 2 cell types | Foxp1;Hoxb9;TATA | ||
| rs2882404 | 6 | 73888146 | 1 | 1 | 1 cell type | Foxi1;Foxj2;Foxl1;Hoxa9;Nkx6-1;Pou2f2 | ||
| rs3799280 | 6 | 73888451 | 1 | 1 | 1 cell type | . | ||
| rs6937253 | 6 | 73888970 | 1 | 1 | . | E2A;Ets;Lmo2-complex;MZF1::1–4;RXRA;SIX5;TCF12;ZEB1;Znf143 | ||
| rs71696488 | 6 | 73889424 | 1 | 1 | 2 cell types | Ncx |
r2 and D′ are measures of LD between the indicated SNP and significant SNPs rs6907229 or rs9343015.
Discussion
With this study we aimed to identify whether variation harbored within genes encoding potassium channels found in airway epithelia is associated with CRS. We identified two SNPS (rs2917454 and rs790026) at the KCNMA1 locus that were associated with the development of CRS in a Caucasian pediatric population. Interestingly, in a smaller cohort of African American children, we also observed a suggestive association between variants in KCNQ5 (rs6907229 and rs9343015) and CRS. Further analysis demonstrated additional SNPs in LD with them that were associated with CRS. The primary SNP in the Caucasian group was found to be associated with a glucocorticoid receptor binding site. Multiple other intronic SNPs were predicted to have an impact on chromatin remodeling or gene expression. These findings raise the possibility that airway potassium channels physiology may contribute to the pathogenesis of CRS.
The clinical spectrum of CRS is a broad and ill-defined clinical entity. Classification of patients with CRS is difficult due to our limited understanding of the disease process which does not allow us to classify patients based on specific cellular or molecular changes. In the case of CF, ion transport dysfunction within epithelial cells, in almost all cases leads to a severe phenotype of CRS, and is evidence of the importance of precise maintenance of electrochemical gradients at the apical epithelial membrane.
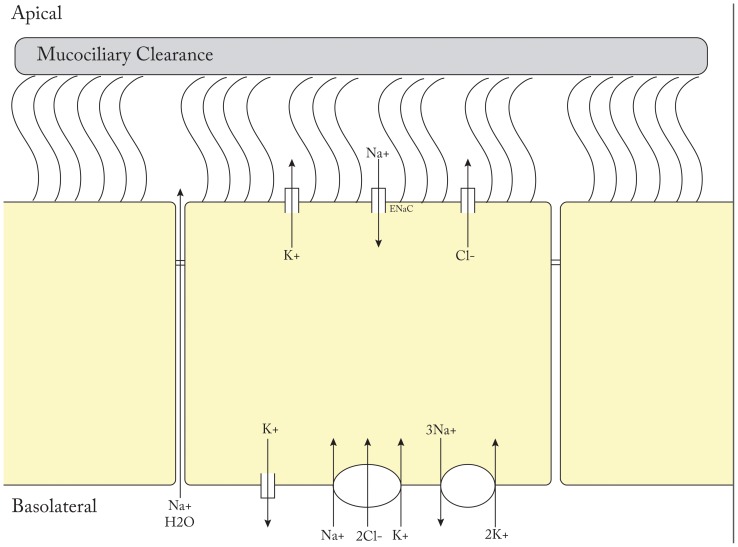
Mucociliary dysfunction is evident in essentially all patients with CRS and is a histological hallmark of the disease no matter the cause [36]. Both mucous hydration and cilia physiology are critical components of MCC [37]. Chloride anion transport is the primary ion current involved in the maintenance of air surface hydration, however, other channels contribute directly to or maintain gradients necessary for apical chloride secretion [38]. Fluid secretion models in secretory cells have supported the notion that chloride secretion is dependent on active transport mechanisms of other ions [15]. Intracellular accumulation of chloride anions is driven by the Na+K+2Cl− co-transporter present at the basolateral membrane of secretory cells where chloride ions can be concentrated up to 5-fold their normal electrochemical gradient. As a result, chloride ion movement can occur freely once chloride channels are activated at the apical membrane. Without electrical compensation for chloride efflux, this process would be inhibited, and basolateral potassium ion efflux into the interstitium plays an important role to support apical efflux of chloride ions. The apical sodium channel ENaC allows for trans-cellular transport of sodium ions and plays an important role in cell volume regulation as well as maintenance of periciliary height. Paracellular sodium movement through tight junctions balances the trans-epithelial gradient. In addition to the basolateral potassium channels discussed above, apical potassium channels have been identified and shown to contribute to chloride secretion and maintenance of ASL hydration [15]. A schematic drawing of epithelial ion transport is show in Figure 2. In the end, the regulation of ion and fluid transport at the apical surface of airway epithelium is a highly regulated system that continues to be elucidated.
Figure 2. Schematic drawing of ciliated epithelial airway ion transport.
The most convincing evidence that apical potassium channel physiology may directly affect MCC comes from a report by Manzanares et al. who found that BK channels were critical for the maintenance of adequate airway surface liquid volume, and inhibition of BK channels or knockdown of KCNMA1 (BK alpha subunit) led to airway dehydration in NHBE cells and, most importantly, low ciliary beat frequency [22]. Other members of the apical potassium channel family have also been shown to play important roles in the regulation of ion transport as discussed above, but they have not been directly linked to regulation of air surface hydration, ciliary beat frequency, or MCC at this time. These findings together suggest that apical potassium channels within airway epithelial cells may play an important, previously underappreciated, role in supporting normal MCC. Further work needs to be done to understand their significance in the pathogenesis of CRS.
Although our study and previous work suggests the plausible role of apical potassium channel genes in CRS pathogenesis, only a small proportion of variance in CRS occurrence is explained by the independent significant SNPs in KCNMA1 or KCNQ5. This is not surprising as CRS is a common, complex disease phenotype determined by multiple factors, including both genetic and environmental factors. A polygenic model is almost assured to be the underlying genetic mechanism.
Systemic or topical corticosteroids are an important treatment modality in patients with CRS. Corticosteroids are postulated to reduce mucosal eosinophil chemotaxis, increase eosinophil apoptosis, and prevent local histamine release. They decrease white blood cell migration, and reduce production of inflammatory mediators and antibodies [39]. Little is known about the role of glucocorticoids in ion transport. There is some evidence, however, that glucocorticoids can induce ENaC expression in lung epithelia [40]. The finding of an association between our significantly identified SNP in the Caucasian cohort with a glucocorticoid receptor site raises the possibility of a previously unrecognized therapeutic mechanism of these commonly used medications in the treatment of CRS. Further study on the role of steroid regulation on epithelial ion transport is required to test this genetic association.
Our additional identified SNPs in both cohorts were also located within introns and their biological impact may not be obvious, but they do demonstrate overlap with chromatin remodeling regions as well as gene expression enhancing and transcription factor binding sites. Intronic enhancers are not unusual. Chromatin looping and interaction between intronic enhancers and promoters have been identified for heme oxygenase-1 [41] and CFTR [42]. Additionally, alpha-fetoprotein has also been reported to harbor an enhancer and an alternative promoter in its first intron [43]. Therefore, many of the top SNPs found in our study or SNPs in good LD with them may possibly affect the risk of developing CRS through transcriptional regulation of KCNMA1 or KCNQ5.
One of the limitations of this study is how the phenotype of CRS is defined. We selected a pediatric population in an effort to minimize environmental contributions to disease and enhance a genetic signal, however, the diagnosis of CRS in a pediatric population is more challenging than adults for multiple reasons including feasibility issues with nasal endoscopy and computed tomography (CT) imaging. Every effort was made to ensure patients met strict inclusion criteria for this study with not just a history suggestive of CRS, but objective data in the form of nasal endoscopy and/or CT imaging when feasible. Further, ruling out other known contributors of pediatric CRS including CF and primary immune deficiencies were sought to the best of our ability. Another major limitation is that these analyses are based on genetic associations. As always, functional studies should be performed to elucidate the biological consequences of these findings.
In summary, this is the first report of a genetic association between potassium channel epithelial physiology and the development of CRS. Further study is needed to confirm the biological relevance of this finding in both pediatric and adult CRS populations.
Supporting Information
SNPs close to or in gene CFTR with p-value<1×10−5 before CF patients were removed and the corresponding statistics after CF patients were removed.
(DOCX)
Test of interactions between independent significant SNPs in genes KCNMA1 and KCNQ5 .
(DOCX)
Summary statistics of SNPs in the 44 potassium channel genes evaluated in this study.
(XLSX)
Acknowledgments
We thank all the children and families who participated in this research project at the Children's Hospital of Philadelphia.
Funding Statement
This work was supported by the Institute Development Funds to The Center for Applied Genomics and an Adele S. and Daniel S. Kubert Estate gift to the Center for Applied Genomics. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kennedy DW (2004) Pathogenesis of chronic rhinosinusitis. Ann Otol Rhinol Laryngol Suppl 193: 6–9. [DOI] [PubMed] [Google Scholar]
- 2. Kennedy DW (2012) As the inflammatory nature of chronic rhinosinusitis (CRS) has become increasingly recognized, the use of steroids, both systemic and topical, as part of the disease management has significantly increased. Int Forum Allergy Rhinol 2: 93–94. [DOI] [PubMed] [Google Scholar]
- 3. Tan BK, Schleimer RP, Kern RC (2010) Perspectives on the etiology of chronic rhinosinusitis. Curr Opin Otolaryngol Head Neck Surg 18: 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soler ZM, Sauer DA, Mace J, Smith TL (2009) Relationship between clinical measures and histopathologic findings in chronic rhinosinusitis. Otolaryngol Head Neck Surg 141: 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu J, Avila PC, Kern RC, Hayes MG, Schleimer RP, et al. (2013) Genetics of chronic rhinosinusitis: State of the field and directions forward. J Allergy Clin Immunol 131: 977–93, 993.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohen NA, Widelitz JS, Chiu AG, Palmer JN, Kennedy DW (2006) Familial aggregation of sinonasal polyps correlates with severity of disease. Otolaryngol Head Neck Surg 134: 601–604. [DOI] [PubMed] [Google Scholar]
- 7. Lockey RF, Rucknagel DL, Vanselow NA (1973) Familial occurrence of asthma, nasal polyps and aspirin intolerance. Ann Intern Med 78: 57–63. [DOI] [PubMed] [Google Scholar]
- 8. Delagrand A, Gilbert-Dussardier B, Burg S, Allano G, Gohler-Desmonts C, et al. (2008) Nasal polyposis: Is there an inheritance pattern? A single family study. Rhinology 46: 125–130. [PubMed] [Google Scholar]
- 9. Greisner WA 3rd, Settipane GA (1996) Hereditary factor for nasal polyps. Allergy Asthma Proc 17: 283–286. [DOI] [PubMed] [Google Scholar]
- 10. Wang X, Kim J, McWilliams R, Cutting GR (2005) Increased prevalence of chronic rhinosinusitis in carriers of a cystic fibrosis mutation. Arch Otolaryngol Head Neck Surg 131: 237–240. [DOI] [PubMed] [Google Scholar]
- 11. Raman V, Clary R, Siegrist KL, Zehnbauer B, Chatila TA (2002) Increased prevalence of mutations in the cystic fibrosis transmembrane conductance regulator in children with chronic rhinosinusitis. Pediatrics 109: E13. [DOI] [PubMed] [Google Scholar]
- 12. Ober C, Yao TC (2011) The genetics of asthma and allergic disease: A 21st century perspective. Immunol Rev 242: 10–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oomen KP, April MM (2012) Sinonasal manifestations in cystic fibrosis. Int J Otolaryngol 2012: 789572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, et al. (1989) Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 245: 1066–1073. [DOI] [PubMed] [Google Scholar]
- 15. Hollenhorst MI, Richter K, Fronius M (2011) Ion transport by pulmonary epithelia. J Biomed Biotechnol 2011: 174306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cook DI, Young JA (1989) Effect of K+ channels in the apical plasma membrane on epithelial secretion based on secondary active cl- transport. J Membr Biol 110: 139–146. [DOI] [PubMed] [Google Scholar]
- 17. Zhao KQ, Xiong G, Wilber M, Cohen NA, Kreindler JL (2012) A role for two-pore K(+) channels in modulating na(+) absorption and cl(−) secretion in normal human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol 302: L4–L12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramsey BW, Davies J, McElvaney NG, Tullis E, Bell SC, et al. (2011) A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med 365: 1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Accurso FJ, Rowe SM, Clancy J, Boyle MP, Dunitz JM, et al. (2010) Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N Engl J Med 363: 1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Goor F, Hadida S, Grootenhuis PD, Burton B, Cao D, et al. (2009) Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci U S A 106: 18825–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bardou O, Trinh NT, Brochiero E (2009) Molecular diversity and function of K+ channels in airway and alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol 296: L145–55. [DOI] [PubMed] [Google Scholar]
- 22. Manzanares D, Gonzalez C, Ivonnet P, Chen RS, Valencia-Gattas M, et al. (2011) Functional apical large conductance, Ca2+-activated, and voltage-dependent K+ channels are required for maintenance of airway surface liquid volume. J Biol Chem 286: 19830–19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leroy C, Prive A, Bourret JC, Berthiaume Y, Ferraro P, et al. (2006) Regulation of ENaC and CFTR expression with K+ channel modulators and effect on fluid absorption across alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol 291: L1207–19. [DOI] [PubMed] [Google Scholar]
- 24. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 25. Lusk RP, Stankiewicz JA (1997) Pediatric rhinosinusitis. Otolaryngol Head Neck Surg 117: S53–7. [DOI] [PubMed] [Google Scholar]
- 26. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verschuren JJ, Trompet S, Postmus I, Sampietro ML, Heijmans BT, et al. (2012) Systematic testing of literature reported genetic variation associated with coronary restenosis: Results of the GENDER study. PLoS One 7: e42401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verschuren JJ, Trompet S, Deelen J, Stott DJ, Sattar N, et al. (2013) Non-homologous end-joining pathway associated with occurrence of myocardial infarction: Gene set analysis of genome-wide association study data. PLoS One 8: e56262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marchini J, Howie B, Myers S, McVean G, Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39: 906–913. [DOI] [PubMed] [Google Scholar]
- 30. Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chorev M, Carmel L (2012) The function of introns. Front Genet 3: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. ENCODE Project Consortium (2012) Bernstein BE, Birney E, Dunham I, Green ED, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chadwick LH (2012) The NIH roadmap epigenomics program data resource. Epigenomics 4: 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward LD, Kellis M (2012) HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakakura Y, Majima Y, Saida S, Ukai K, Miyoshi Y (1985) Reversibility of reduced mucociliary clearance in chronic sinusitis. Clin Otolaryngol Allied Sci 10: 79–83. [DOI] [PubMed] [Google Scholar]
- 37. Ooi EH, Psaltis AJ, Witterick IJ, Wormald PJ (2010) Innate immunity. Otolaryngol Clin North Am 43: 473–87, vii. [DOI] [PubMed] [Google Scholar]
- 38. Harvey PR, Tarran R, Garoff S, Myerburg MM (2011) Measurement of the airway surface liquid volume with simple light refraction microscopy. Am J Respir Cell Mol Biol 45: 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Derendorf H, Meltzer EO (2008) Molecular and clinical pharmacology of intranasal corticosteroids: Clinical and therapeutic implications. Allergy 63: 1292–1300. [DOI] [PubMed] [Google Scholar]
- 40. Sayegh R, Auerbach SD, Li X, Loftus RW, Husted RF, et al. (1999) Glucocorticoid induction of epithelial sodium channel expression in lung and renal epithelia occurs via trans-activation of a hormone response element in the 5′-flanking region of the human epithelial sodium channel alpha subunit gene. J Biol Chem 274: 12431–12437. [DOI] [PubMed] [Google Scholar]
- 41. Deshane J, Kim J, Bolisetty S, Hock TD, Hill-Kapturczak N, et al. (2010) Sp1 regulates chromatin looping between an intronic enhancer and distal promoter of the human heme oxygenase-1 gene in renal cells. J Biol Chem 285: 16476–16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, et al. (2009) A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med 13: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scohy S, Gabant P, Szpirer C, Szpirer J (2000) Identification of an enhancer and an alternative promoter in the first intron of the alpha-fetoprotein gene. Nucleic Acids Res 28: 3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Grady SM, Lee SY (2003) Chloride and potassium channel function in alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol 284: L689–700. [DOI] [PubMed] [Google Scholar]
- 45. Coetzee WA, Amarillo Y, Chiu J, Chow A, Lau D, et al. (1999) Molecular diversity of K+ channels. Ann N Y Acad Sci 868: 233–285. [DOI] [PubMed] [Google Scholar]
- 46. Mall M, Gonska T, Thomas J, Schreiber R, Seydewitz HH, et al. (2003) Modulation of Ca2+-activated cl- secretion by basolateral K+ channels in human normal and cystic fibrosis airway epithelia. Pediatr Res 53: 608–618. [DOI] [PubMed] [Google Scholar]
- 47. Mall M, Wissner A, Schreiber R, Kuehr J, Seydewitz HH, et al. (2000) Role of K(V)LQT1 in cyclic adenosine monophosphate-mediated cl(−) secretion in human airway epithelia. Am J Respir Cell Mol Biol 23: 283–289. [DOI] [PubMed] [Google Scholar]
- 48. Moser SL, Harron SA, Crack J, Fawcett JP, Cowley EA (2008) Multiple KCNQ potassium channel subtypes mediate basal anion secretion from the human airway epithelial cell line calu-3. J Membr Biol 221: 153–163. [DOI] [PubMed] [Google Scholar]
- 49. Cowley EA, Linsdell P (2002) Characterization of basolateral K+ channels underlying anion secretion in the human airway cell line calu-3. J Physiol 538: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Teng S, Ma L, Zhen Y, Lin C, Bahring R, et al. (2003) Novel gene hKCNE4 slows the activation of the KCNQ1 channel. Biochem Biophys Res Commun 303: 808–813. [DOI] [PubMed] [Google Scholar]
- 51. Bernard K, Bogliolo S, Soriani O, Ehrenfeld J (2003) Modulation of calcium-dependent chloride secretion by basolateral SK4-like channels in a human bronchial cell line. J Membr Biol 196: 15–31. [DOI] [PubMed] [Google Scholar]
- 52. Jovanovic S, Crawford RM, Ranki HJ, Jovanovic A (2003) Large conductance Ca2+-activated K+ channels sense acute changes in oxygen tension in alveolar epithelial cells. Am J Respir Cell Mol Biol 28: 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ridge FP, Duszyk M, French AS (1997) A large conductance, Ca2+-activated K+ channel in a human lung epithelial cell line (A549). Biochim Biophys Acta 1327: 249–258. [DOI] [PubMed] [Google Scholar]
- 54. Monaghan AS, Baines DL, Kemp PJ, Olver RE (1997) Inwardly rectifying K+ currents of alveolar type II cells isolated from fetal guinea-pig lung: Regulation by G protein- and Mg2+-dependent pathways. Pflugers Arch 433: 294–303. [DOI] [PubMed] [Google Scholar]
- 55. Plummer HK 3rd, Dhar MS, Cekanova M, Schuller HM (2005) Expression of G-protein inwardly rectifying potassium channels (GIRKs) in lung cancer cell lines. BMC Cancer 5: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sakuma T, Takahashi K, Ohya N, Nakada T, Matthay MA (1998) Effects of ATP-sensitive potassium channel opener on potassium transport and alveolar fluid clearance in the resected human lung. Pharmacol Toxicol 83: 16–22. [DOI] [PubMed] [Google Scholar]
- 57. Doring F, Derst C, Wischmeyer E, Karschin C, Schneggenburger R, et al. (1998) The epithelial inward rectifier channel Kir7.1 displays unusual K+ permeation properties. J Neurosci 18: 8625–8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Inglis SK, Brown SG, Constable MJ, McTavish N, Olver RE, et al. (2007) A Ba2+-resistant, acid-sensitive K+ conductance in na+-absorbing H441 human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 292: L1304–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNPs close to or in gene CFTR with p-value<1×10−5 before CF patients were removed and the corresponding statistics after CF patients were removed.
(DOCX)
Test of interactions between independent significant SNPs in genes KCNMA1 and KCNQ5 .
(DOCX)
Summary statistics of SNPs in the 44 potassium channel genes evaluated in this study.
(XLSX)