Abstract
Hematological and gastrointestinal toxicities are common among patients treated with cyclophosphamide and doxorubicin for breast cancer. To examine whether single nucleotide polymorphisms (SNPs) in key pharmacokinetic genes were associated with risk of hematological or gastrointestinal toxicity, we analyzed 78 SNPs in ABCB1, ABCC1 and ALDH1A1 in 882 breast cancer patients enrolled in the SWOG trial S0221 and treated with cyclophosphamide and doxorubicin. A two-SNP haplotype in ALDH1A1 was associated with an increased risk of grade 3 and 4 hematological toxicity (odds ratio [OR]=1.44, 95% confidence interval [CI]=1.16-1.78), which remained significant after correction for multiple comparisons. In addition, 4 SNPs in ABCC1 were associated with gastrointestinal toxicity. Our findings provide evidence that SNPs in pharmacokinetic genes may have an impact on the development of chemotherapy-related toxicities. This is a necessary first step towards building a clinical tool that will help assess risk of adverse outcomes prior to administration of chemotherapy.
Keywords: breast cancer, chemotherapy, toxicity, pharmacokinetics, pharmacogenetics, ALDH1A1
Introduction
Systematically administered breast cancer chemotherapeutic drugs, including cyclophosphamide (C), doxorubicin (A) and taxanes (T), are cytotoxic compounds which target not only rapidly dividing cancer cells, but also normal cells in other exposed organs, resulting in debilitating and sometimes life-threatening toxicities1. Hematological toxicities, such as neutropenia and leucopenia, and gastrointestinal toxicities, such as nausea, vomiting, stomatitis and diarrhea, are commonly seen in patients treated with cyclophosphamide and doxorubicin. In addition to adversely affecting patients' quality-of-life, these acute side effects can lead to delay, alteration, or discontinuation of treatment, which may subsequently lead to cancer recurrence or premature mortality2. It is thus important to assess the relative risk/benefit ratio of adjuvant chemotherapy, which may facilitate physicians' and patients' decision-making in selection of chemotherapy regimens.
In the search for genetic biomarkers that are predictive of chemotherapy toxicity in breast cancer patients, genes in drug pharmacokinetic pathways, which may have direct impact on the effective drug dose reaching target cells and the rate of drug disposal, are excellent candidates3, 4. An important gene for cyclophosphamide pharmacokinetics is aldehyde dehydrogenase 1 family, member A1 (ALDH1A1), which is a major enzyme responsible for the detoxification of active cyclophosphamide metabolites5. In a small study of cancer patients (primarily breast) treated with high dose cyclophosphamide, patients heterozygous for ALDH1A1*2 had an increased risk of liver toxicity6. For doxorubicin, two major efflux transporters, ATP-binding cassette, subfamily B, member 1 (ABCB1), also known as multidrug resistance protein 1 (MDR1), and ATP-binding cassette, subfamily C, member 1 (ABCC1), known as multidrug resistance associated protein 1 (MRP1), have been implicated in cellular transport of doxorubicin7. Although germline genetic variants in ABCB1 and ABCC1 have been examined with survival outcomes after chemotherapy for breast cancer, few studies have examined them in relation to chemotherapy-induced toxicity.
In this study, we analyzed 78 tagSNPs in ABCB1, ABCC1 and ALDH1A1 which were selected to comprehensively capture germline genetic variations in these key pharmacokinetic genes for cyclophosphamide and doxorubicin, among 882 breast cancer patients enrolled in a cooperative group clinical trial SWOG S0221. We examined potential associations of single SNPs and haplotypes with risk of high grade hematological and gastrointestinal toxicities induced by doxorubicin and cyclophosphamide.
Patients and Methods
Patient population
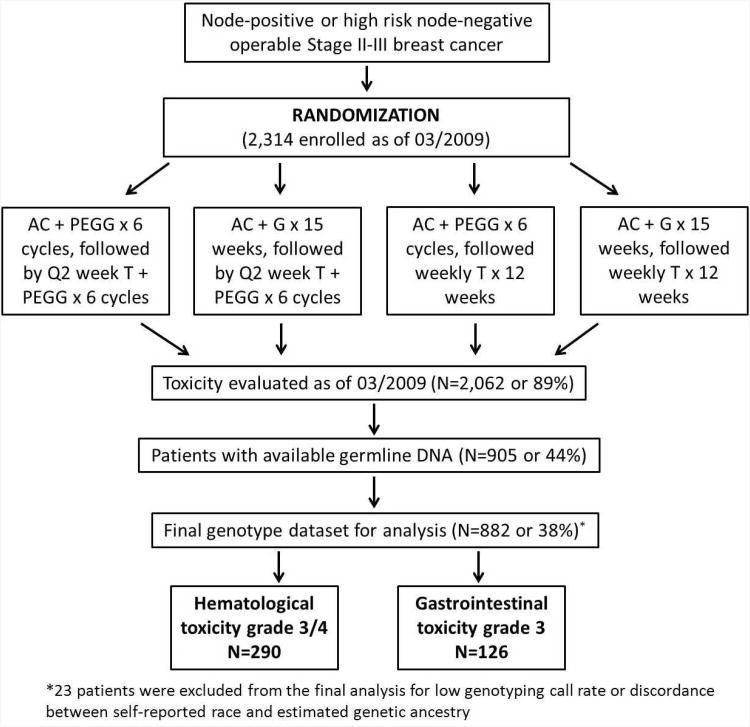
Data and DNA samples for this study were from a North American Breast Cancer Intergroup clinical trial S0221 led by SWOG. S0221 is a Phase III trial to test if a continuous “metronomic” schedule of AC (doxorubicin and cyclophosphamide) for 15 weeks was superior to a more conventional Q2 week schedule of AC for 6 cycles, followed by either weekly T (paclitaxel) for 12 weeks or Q2 week T for 6 cycles. All patients received subcutaneous filgrastim at 5 mcg/kg or pegfilgrastim at 6 mg/kg starting 24 hours after chemotherapy injection. The randomization schema is shown in Figure 1. Details of the trial design and patients included in this pharmacogenetic study have been described previously8. In brief, patients of age ≥18 with a histologically confirmed operable Stage II or III high risk invasive breast cancer with known estrogen or progesterone receptor status and no previous malignancies were eligible to participate in the trial. Blood samples were drawn at the time of registration and shipped to the Roswell Park Cancer Institute (RPCI) laboratory for processing and DNA extraction. Because blood collection was not instituted until after the trial had begun, and there were delays because of the need for the multiple institutions involved to get IRB approval for the amendment, blood specimens were not collected from the approximately 1,000 patients enrolled on the trial before approval for specimen collection. At the time of the genotyping performed in March 2009, 2,314 patients had been enrolled in the trial and 2,062 had toxicity evaluation completed. Among them, a total of 905 patients with available DNA samples were included in this study. The enrollment of new patients in S0221 was closed in January 2012 with continued follow up ongoing. Because the main findings from S0221 have not been published, we cannot disclose the treatment arm assignment, examine toxicities by treatment arm, or evaluate genotypes with survival outcomes in this study.
Figure 1. Randomization schema of SWOG S0221 clinical trial.
Abbreviations: A, doxorubicin; C, cyclophosphamide; T, paclitaxel; G, filgrastim; PEGG, pegfilgrastim
Collection of toxicity data
In S0221, toxicities were monitored and reported according to the NCI Common Toxicity Criteria for Adverse Events (CTCAE) Version 3.09. Each adverse event was graded based on the severity, where grade 3 toxicities interfere with activities of daily living, and grade 4 toxicities are life-threatening and usually require hospitalization. In this analysis, grade 3 and 4 hematological toxicities and grade 3 gastrointestinal toxicities that occurred during the AC segment of chemotherapy before the T segment treatment, two major side effects related to cyclophosphamide and doxorubicin, were used as the major analytical endpoints. Toxicity data were recoded as occurring or not during AC treatment, and timing relative to the treatment period was not collected in this study.
SNP selection and genotyping
To systematically interrogate common genetic variations in ABCB1, ABCC1 and ALDH1A1, tagSNPs were selected using the SNAGGER program10 based on the HapMap genotype data from the CEU population of European ancestry and the YRI population of African ancestry11. The minimum allele frequency (MAF) was set at 0.05 and r2 threshold of 0.80, with a consideration of position relative to functional genomic regions and Illumina design scores when prioritizing selections. A total of 16 SNPs in ABCB1, 48 SNPs in ABCC1 and 15 SNPs in ALDH1A1 were selected and genotyped using the Illumina GoldenGate platform in the Genomics Core Facility at Roswell Park Cancer Institute. To control for population admixture bias, 70 uncorrelated SNPs were selected and genotyped as ancestry informative markers to estimate individual ancestry using the STRUCTURE program12. The average call rate was 97.1%, and the concordance rate among 5% blind duplicates was 100%. A total of 23 patients were excluded from the analysis for genotyping call rates lower than 90%, or discordance between self-reported race and estimated genetic ancestry. None of the SNPs in the three selected genes were excluded due to call rate below 90%. One SNP in ABCC1 was removed for violation of Hardy-Weinberg Equilibrium in patients of European ancestry, leaving a total of 78 SNPs and 882 patients in the final analysis.
Statistical methods
Unconditional logistic regression was used to compute odds ratios (OR) and 95% confidence intervals (CI) for both single SNP and haplotype analysis, with adjustment for age, proportion of genetic ancestry, and treatment arm. Codominant genetic models, i.e. independent effects of all three genotypes, and dominant models, i.e. identical effect of the rare homozygote and heterozygote, were both tested. The R package snp.plotter13 was used to generate the linkage disequilibrium (LD) map with p-values from single SNP analysis. The criteria by Gabriel et al14 were used to determine haplotype blocks and to estimate haplotype frequency for each gene. Each haplotype was tested in comparison to all other haplotypes combined. In addition to adjusting for genetic ancestry estimates, subgroup analyses were also performed within patients of European ancestry only, the majority of the study population (83%), to control for potential population admixture bias. In addition to combining grade 3 and 4 hematological toxicity as one endpoint, grade 4 hematological toxicity versus grade 0-2 toxicity was also tested as a separate endpoint, and ordinal logistic regression model was also used, which treated hematological toxicity grade ≤2, grade 3 and grade 4 as an ordinal response variable. Multiple testing was controlled using 10,000 permutation using the PLINK program15. The results are reported in accordance with the REMARK criteria16.
Results
For purposes of this study, only the AC segment of the trial was examined for associations between genotypes and toxicity, since paclitaxel rarely causes either Grade 3/4 hematological or gastrointestinal toxicity, especially in the setting of routine growth factor administration. Descriptive characteristics of the patient population included in this study are summarized in Table 1. The average age of breast cancer diagnosis was 50 years, with 83% of patients self-reported as European American, 8% as African American, 5% as Asian American, and 4% as other race or ethnicity. Data are shown for patients included in this study as well as those for whom DNA was not available in Table 1; characteristics and outcomes did not differ between those with and without DNA. According to the CTCAE, in the AC segment of the chemotherapy, 19% and 14% of the patients developed grade 3 and grade 4 hematological toxicities, respectively; 14% of patients developed grade 3 gastrointestinal toxicity. No grade 4 gastrointestinal toxicities were observed in the AC segment for those with DNA and only four Grade 4 events were observed in those without DNA.
Table 1. Descriptive characteristics of breast cancer patients enrolled in the S0221 trial with assessed toxicity by the availability of DNA at the time of genotyping.
| Variable | Patients with DNA available (n=882) | Patients without DNA available (n=1,180) |
|---|---|---|
| Age, years, mean ± standard deviation | 50.0 ± 10.2 | 50.4 ± 9.9 |
| Self-reported race, N (%) | ||
| European American | 734 (83) | 921 (78) |
| African American | 69 (8) | 149 (13) |
| Asian | 41 (5) | 61 (5) |
| Other | 38 (4) | 49 (4) |
| AC Treatment arm, N (%) | ||
| Arm 1 | 446 (51) | 619 (52) |
| Arm 2 | 436 (49) | 561 (48) |
| CTCAE Grade of hematological toxicity, N (%) | ||
| Grade 0-2 | 592 (67) | 830 (70) |
| Grade 3 | 165 (19) | 175 (15) |
| Grade 4 | 125 (14) | 175 (15) |
| CTCAE Grade of gastrointestinal toxicity, N (%) | ||
| Grade 0-2 | 756 (86) | 1004 (85) |
| Grade 3 | 126 (14) | 172 (15) |
| Grade 4 | 0 (0) | 4 (0) |
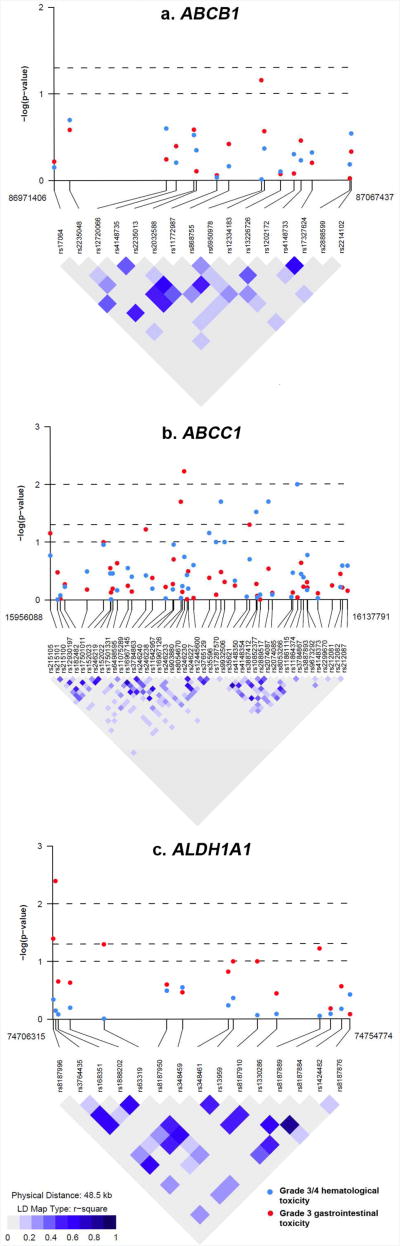
P-values for the associations of the 78 SNPs in ABCB1, ABCC1 and ALDH1A1 with hematological and gastrointestinal toxicity plotted with LD map for each gene are shown in Figure 2, and ORs and 95% CIs in Supplementary Tables S2 and S3. Using an uncorrected p<0.05 for statistical significance, we observed an association between grade 3 and 4 hematological toxicity and three SNPs in ABCC1 (rs903880, rs16967126 and rs4148350) and three SNPs in ALDH1A1 (rs8187996, rs3764435 and rs63319) (Table 2). Except for two SNPs (rs4148350 in ABCC1 and rs63319 in ALDH1A1), these associations remained significant when the analysis was restricted to European American women only. However, none of the four SNPs was significant after correction for multiple comparisons (Table 2). For the most significant SNP rs3764435 in ALDH1A1, the AA genotype was associated with a 76% increased risk of grade 3 and 4 hematological toxicity (codominant model, OR=1.76, 95% CI=1.18-2.61, uncorrected p=0.004, and corrected p=0.21). A similar increased risk was found in patients carrying the AA genotype when the analysis was restricted to European American women only (OR=1.84, 95% CI=1.22-2.78), or when grade 4 hematological toxicity was compared to grade 0-2 toxicity (OR=2.00, 95% CI=1.11-3.58). In the ordinal logistic regression model treating grade ≤2, grade 3 and grade 4 hematological toxicity as an ordinal outcome, patients carrying the AA genotype were also at an increased risk of higher toxicity (OR=1.73, 95% CI=1.17-2.55, p=0.005). Consistent with the single SNP analysis, a two-SNP haplotype consisting of the A allele of rs3764435 and a neighboring SNP rs168351 was associated with an increased risk of grade 3 and 4 hematological toxicity (OR=1.44, 95% CI=1.16-1.78, uncorrected p=0.0008), which remained significant after correction for multiple comparisons (corrected p=0.03) (Table 4), or when the analysis was restricted to European American women only (OR=1.48, 95% CI=1.19-1.85, uncorrected p=0.0005, corrected p=0.02). None of the SNPs in ABCB1 was associated with hematological toxicity.
Figure 2. Linkage disequilibrium map and single SNP associations with grade 3 and 4 hematological toxicity and grade 3 gastrointestinal toxicity.
Table 2. SNPs in significant association with grade 3 and 4 hematological toxicity in S0221 trial.
| Gene | SNP | Genotype | # Grade 3/4 vs. grade ≤2 | Adjusted OR (95% CI)* | Praw | Ppermutation |
|---|---|---|---|---|---|---|
| ABCC1 | rs903880 | CC | 148/355 | 1.00 | 0.006 | 0.23 |
| CA | 108/191 | 1.38 (1.02-1.88) | ||||
| AA | 34/46 | 1.86 (1.10-3.15) | ||||
| AA+AC | 142/237 | 1.46 (1.09-1.95) | 0.01 | 0.39 | ||
| ABCC1 | rs16967126 | AA | 229/507 | 1.00 | 0.03 | 0.65 |
| AG | 59/79 | 1.63 (1.12-2.37) | ||||
| GG | 2/6 | 0.63 (0.12-3.22) | ||||
| AG/GG | 61/85 | 1.55 (1.08-2.25) | 0.02 | 0.52 | ||
| ABCC1 | rs4148350 | CC | 244/527 | 1.00 | 0.12 | 0.72 |
| CA | 46/64 | 1.54 (1.02-2.33) | ||||
| AA | 0/1 | - | ||||
| CA/AA | 46/65 | 1.52 (1.01-2.29) | 0.05 | 0.61 | ||
| ALDH1A1 | rs8187996 | GG | 253/543 | 1.00 | 0.10 | 0.65 |
| GA | 36/48 | 1.63 (1.02-2.58) | ||||
| AA | 1/1 | 2.05 (0.13-33.87) | ||||
| GA/AA | 49/37 | 1.64 (1.04-2.58) | 0.04 | 0.61 | ||
| ALDH1A1 | rs3764435 | CC | 63/160 | 1.00 | 0.004 | 0.21 |
| AC | 127/286 | 1.11 (0.77-1.56) | ||||
| AA | 100/145 | 1.76 (1.18-2.61) | ||||
| AC+AA | 227/431 | 1.64 (1.20-2.25) | 0.002 | 0.16 | ||
| ALDH1A1 | rs63319 | CC | 71/176 | 1.00 | 0.05 | 0.65 |
| CA | 135/277 | 1.19 (0.84-1.68) | ||||
| AA | 84/139 | 1.48 (1.00-2.18) | ||||
| CA+AA | 219/416 | 1.28 (0.93-1.77) | 0.13 | 0.78 |
Footnote:
Logistic regression model adjusted for age of diagnosis, treatment arm and genetic ancestry.
Table 4. Haplotypes in significant associations with high grade hematological and gastrointestinal toxicity in S0221 trial.
| Gene | SNPs | Haplotype | MHF in high grade toxicity | MHF in low grade toxicity | Adjusted OR (95% CI)* | Praw | Ppermutation |
|---|---|---|---|---|---|---|---|
| Grade 3 and 4 hematological toxicity | |||||||
| ALDH1A1 | rs3764435-rs168351 | A-A | 0.46 | 0.37 | 1.44 (1.16-1.78) | 0.0008 | 0.03 |
|
| |||||||
| Grade 3 gastrointestinal toxicity | |||||||
| ABCC1 | rs2889517-rs2074087 | A-G | 0.09 | 0.16 | 0.54 (0.34-0.84) | 0.006 | 0.24 |
Footnote:
Logistic regression model adjusted for age of diagnosis, treatment arm and genetic ancestry. Odds ratio (OR) and 95% confidence interval (CI) were derived by comparing the specified haplotype with all other haplotypes combined.
For grade 3 gastrointestinal toxicity, SNPs with an uncorrected p<0.05 are shown in Table 3, which included four SNPs in ABCC1 (rs35596, rs4148354, rs2889517 and rs11861115). Associations remained significant when the analysis was restricted to European American women only; however, none of the four SNPs was significant after correction for multiple comparisons. Women carrying the A allele of rs2889517 had a decreased risk of gastrointestinal toxicity as compared to those homozygous for the G allele (GA genotype: OR=0.57, 95% CI=0.37-0.86; AA genotype: OR=0.62, 95% CI=0.32-1.20). The association was confirmed in haplotype analysis, where a two-SNP haplotype consisting of the A allele of rs2889517 and the G allele of a neighboring SNP rs2074087 was associated with a decreased risk of gastrointestinal toxicity (OR=0.54, 95% CI=0.34-0.84, uncorrected p=0.006). However, the association became non-significant after correction for multiple comparisons (corrected p=0.24). None of the SNPs in ABCB1 or ALDH1A1 was associated with gastrointestinal toxicity.
Table 3. SNPs in significant association with grade 3 gastrointestinal toxicity in S0221 trial.
| Gene | SNP | Genotype | # Grade 3 vs. grade ≤2 | Adjusted OR (95% CI)* | Praw | Ppermutation |
|---|---|---|---|---|---|---|
| ABCC1 | rs35596 | AA | 58/420 | 1.00 | 0.02 | 0.52 |
| AG | 57/277 | 1.58 (1.05-2.36) | ||||
| GG | 11/56 | 1.75 (0.84-3.63) | ||||
| AG+GG | 68/333 | 1.60 (1.08-2.36) | 0.02 | 0.39 | ||
| ABCC1 | rs4148354 | AA | 49/192 | 1.00 | 0.03 | 0.59 |
| AG | 47/355 | 0.53 (0.34-0.83) | ||||
| GG | 30/208 | 0.60 (0.36-1.00) | ||||
| AG+GG | 77/563 | 0.55 (0.37-0.83) | 0.003 | 0.23 | ||
| ABCC1 | rs2889517 | GG | 76/351 | 1.00 | 0.02 | 0.52 |
| GA | 38/315 | 0.57 (0.37-0.86) | ||||
| AA | 12/90 | 0.62 (0.32-1.20) | ||||
| GA+AA | 50/405 | 0.58 (0.39-0.85) | 0.006 | 0.24 | ||
| ABCC1 | rs11861115 | GG | 87/431 | 1.00 | 0.10 | 0.95 |
| GA | 35/275 | 0.64 (0.42-0.98) | ||||
| AA | 4/48 | 0.41 (0.15-1.18) | ||||
| GA/AA | 39/323 | 0.61 (0.40-0.91) | 0.01 | 0.26 |
Footnote:
Logistic regression model adjusted for age of diagnosis, treatment arm and genetic ancestry.
Discussion
In this study, we examined tagSNPs in ABCB1, ABCC1 and ALDH1A1 in relation to risk of high grade hematological and gastrointestinal toxicities after treatment for breast cancer with doxorubicin and cyclophosphamide in a SWOG clinical trial. None of the ABCB1 SNPs were associated with either toxicity, but 3 SNPs in ABCC1 and 3 SNPs and 1 haplotype in ALDH1A1 were associated with grade 3 and 4 hematological toxicity. Four SNPs and 1 haplotype in ABCC1 were associated with grade 3 gastrointestinal toxicity. The only association that remained significant after correction for multiple comparison was a haplotype in ALDH1A1 consisting of rs3764435 and a neighboring SNP. The A-A haplotype of ALDH1A1 was associated with a 44% increased risk of grade 3 and 4 hematological toxicity as compared to the other haplotypes. Our findings suggest that common genetic variations in ALDH1A1, which plays a critical role in the detoxification of active cyclophosphamide metabolites, may affect the risk of severe hematological toxicity caused by chemotherapy, despite the prophylactic use of granulocyte colony-stimulating factors (CSFs).
Neutropenia, characterized by markedly low absolute neutrophil count (<500 cells/ul), is one of the most common hematological toxicities occurring among patients treated with myelosuppressive chemotherapy drugs17. Profound and prolonged neutropenia often leads to hospitalization due to fever or infection, which sometimes can be life-threatening. The American Society of Clinical Oncology (ASCO) recommends the use of primary prophylaxis with hematopoietic CSFs when the risk of febrile neutropenia from a specific regimen exceeds 20%18. In S0221, all patients received either filgrastim (G) or pegfilgrastim (PEGG) as prophylaxis following AC therapy. Nevertheless, 19% and 14% of patients still experienced grade 3 and 4 adverse hematological events, respectively, during the AC segment of the trial, indicating either that these patients were extremely susceptible to myelosuppressive drugs, and/or that growth factors were not adequately efficacious for them. Our findings that a haplotype in ALDH1A1 was associated with risk of grade 3 and 4 hematological toxicities, indicates that genetic variants may be useful biomarkers for identifying a subgroup of patients at high risk of neutropenia caused by myelosuppressive chemotherapy, even when given growth factors.
Thus, if confirmed, these results could have clinical relevance. One might consider a number of strategies directed towards patients who are at higher risk for myelosuppression, such as increased dose of growth factor, reduction of chemotherapy dose, or administration of prophylactic antibiotics. However, each of these strategies is associated with potential risks and costs. Therefore, such strategies, following replication, need to be tested in properly designed, prospective trials.
ALDH1A1 is a major enzyme responsible for the detoxification of the cytotoxic cyclophosphamide metabolites, aldophosphamide and acrolein5. Interestingly, ALDH1A1 was proposed as a biomarker of stem cells, and in hematopoietic stem cells, deficient expression of Aldh1a1 was related to hyper-sensitivity to cyclophosphamide in mice19, consistent with its role in the detoxification of this drug. In a recent study of 513 breast cancer patients, expression of ALDH1A1 was inversely related to disease-free survival and overall survival after neoadjuvant chemotherapy containing cyclophosphamide20. To date, only a 17-bp deletion polymorphism (ALDH1A1*2 or rs615103) has been examined with pharmacokinetics and toxicity of cyclophosphamide. In 113 patients with various cancers (including breast cancer) who were treated with cyclophosphamide, patients heterozygous for ALDH1A1*2 had increased risk of hemorrhagic cystitis6. However, in another study from the same group, this variant allele was not associated with the clearance of cyclophosphamide or its 4-hydroxyl metabolite21. In S0221, we selected and genotyped 15 SNPs in ALDH1A1, which comprehensively represents common variations in this gene. We found that SNP rs3764435 was associated with grade 3 and 4, as well as grade 4 alone, hematological toxicity after AC treatment. The association was confirmed by a haplotype consisting of rs3764435 and a neighboring SNP, with the association remaining significant after correction for multiple comparison. This SNP is located in the intronic region, underneath 2 transcription factor binding sites in human ductal breast epithelial tumor cell lines (T47d), including GATA3. GATA3 yields luminal epithelial cell differentiation in the mammary gland and EP300, playing an important role in regulating cell growth and division. Furthermore, in these T47d cell lines, this SNP resides in a DNAse hypersensitivity site, and has been associated with the expression of at least 27 genes in lymphoblastoid cell lines22, indicating that rs3764435may have an impact on gene expression regulation. Because ALDH1A1 also plays a role in the conversion of acetaldehyde to acetate in the alcohol metabolism pathway23, SNPs in this gene have been previously examined with alcohol dependence, and rs3764435 was identified as the driving variant for a risk haplotype in a European population24. Although it is unclear whether this SNP has a similar impact on the metabolism of cyclophosphamide, this finding and ours call for future study to characterize the functionality of this SNP.
ABCB1 and ABCC1 belong to the ATC-binding cassette family, and are among the best characterized cellular efflux transporters implicated in resistance to multiple drugs, including doxorubicin7. The majority of the previous pharmacogenetic studies of genetic variants in ABCB1 and ABCC1 focused on survival outcomes after doxorubicin-containing chemotherapy, while studies on drug toxicity are scarce. In a small study of 62 breast cancer patients, SNPs in ABCB1 were associated with plasma concentration of doxorubicin; however, toxicity was not evaluated in that study25. A large pharmacogenetic study of 1,697 aggressive non-Hodgkin's lymphoma patients examined SNPs in a large number of genes, including ABCB1 and ABCC1, in relation to doxorubicin-induced cardiotoxicity26. A non-synonymous coding SNP rs45511401 (Gly671Val) in ABCC1 was associated with markedly increased risk of cardiotoxicity. In S0221, genetic variants in ABCC1 were associated with hematological toxicity and gastrointestinal toxicity in breast cancer patients after AC treatment, although the associations did not remain significant after controlling for multiple comparisons.
There are some limitations in the study that need to be considered when interpreting our results. First, we selected three genes that are central in the pharmacokinetic pathways of cyclophosphamide and doxorubicin to be included in this study. However, this is not a complete coverage of the important genes in these pathways. Considering the relatively moderate effect associated with each genetic variant found in our study, it may be important to consider multiple SNPs and genes and examine their combinational effects on treatment outcomes in future studies. Second, considering the sample size available in this study, we chose to focus on common variants with an MAF ≥0.05, which is likely to miss rare variants and their potential impact on chemotherapy-induced toxicity. Next-generation sequencing has now been widely used to interrogate rare variants. However, the immediate pharmacogenetic significance of rare variants is still not apparent, as screening for rare variants among a large number of patients may not be cost-effective. Third, the observed associations in our study are somewhat marginal, especially after correction for multiple comparisons by permutation. However, our study is more of exploratory nature, since pharmacogenetic studies on toxicities are still sparse, which is a clinical important research area for patients' quality-of-life and survivorship. The patient population from a cooperative group clinical trial, the vigorous and systemically evaluation and collection of toxicity data, and the consistency between single SNP analysis and haplotype analysis are all strengths making our results less likely to be spurious. Although we still cannot exclude the possibility of false positivity for the lack of replication cohort, we feel that our report will likely stimulate others' attempt to validate our findings.
Randomized clinical trials with archived biospecimens have unique strengths in pharmacogenetic research. Although trials are typically designed to randomize patients on treatment but not genotypes, the randomization and the prospective nature minimizes most other potential biases, such as heterogeneous disease characteristics, treatment modality, incomplete report of clinical outcomes and follow-up. It has been proposed that findings of retrospective analysis of biospecimens from prospective trials can reach a high level of evidence in biomarker studies, provided that a large number of representative samples are included, there is a well-established assay for biomarker measurement, a pre-specified analytical plan of the biomarker of interest, and validation of the initial findings in a similar but independent cohort27. As far as the above criteria are concerned, our study included a large proportion of patients enrolled in S0221 who also had banked blood samples, we selected tagSNPs to comprehensively capture common genetic variations, and the genotyping quality was high as demonstrated by very high call rate and concordance rate. In addition, the genotyped population was representative of those not genotyped (Table 1). However, our study still lacks a validation cohort to test whether the findings can be repeatedly observed in a different patient population, and these findings need to be followed up in other similar cohorts.
In conclusion, among 882 breast cancer patients from a cooperative group randomized trial S0221, we found evidence of associations between genetic variants in ABCC1 and ALDH1A1 with risk of hematological and gastrointestinal toxicities induced by AC treatment. The finding that patients carrying the risk allele of the ALDH1A1 SNP were more likely to have hematological toxicity, despite the use of prophylactic growth factors, indicates that pharmacogenetic markers may be useful in identifying a subgroup of patients at high risk of neutropenia induced by myelosuppressive chemotherapy. Future studies are warranted to replicate our findings and to examine other genes in relation to chemotherapy-induced toxicities.
Supplementary Material
Supplementary Table S1. TagSNPs selected in ABCB1, ABCC1, and ALDH1A1
Supplementary Table S2. Associations of SNPs in ABCB1, ABCC1, and ALDH1A1 with grade 3 and 4 hematological toxicity in S0221 trial
Supplementary Table S3. Associations of SNPs in ABCB1, ABCC1, and ALDH1A1 with grade 3 gastrointestinal toxicity in S0221 trial
Acknowledgments
We wish to acknowledge and thank Dr. Frank L. Meyskens, Jr., M.D., Associate Chair for the Cancer Control & Prevention Committee of the Southwest Oncology Group, for his administrative support.
Funding Support: This study was supported by NIH R01 CA116395, and the following PHS Cooperative Agreement grant numbers awarded by the National Cancer Institute, DHHS: CA32102, CA38926, CA63844, CA63845, CA 20319, CA46282, CA46441, CA35261, CA63848, CA67575, CA14028, CA35281, CA128567, CA45560, CA58882, CA13612; CA46368, CA45808, CA58658, CA76447, CA37981, CA04919; CA95860; CA27057, CA42777, CA22433, CA74647, CA86780, CA68183, CA58861, CA45807, CA35192, CA35178, CA58416, CA35176, CA67663, CA35431, CA12644, CA16385, CA11083, CA45377, CA35128, CA35262, CA52654, CA76429, CA58723, CA46113, CA76132, CA45450, CA35119, CA45461, CA21115, CA21076, CA77597, CA25224, CA77202, CCSRI15469, and in part by Amgen, Inc. Support was also provided by the Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale ™ (DFH), and Drs. Ambrosone, Gralow, Hayes, Hershman and Hortobagyi are recipients of funding from the Breast Cancer Research Foundation. Funding agencies had no involvement in the study design, data collection, analysis and interpretation, or in the writing of the report and submission.
Footnotes
Conflict of interest: The authors declare no conflict of interest
Supplementary information is available at The Pharmacogenomics Journal's website
References
- 1.Hassett MJ, O'Malley AJ, Pakes JR, Newhouse JP, Earle CC. Frequency and cost of chemotherapy-related serious adverse effects in a population sample of women with breast cancer. J Natl Cancer Inst. 2006;98(16):1108–1117. doi: 10.1093/jnci/djj305. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro CL, Recht A. Side effects of adjuvant treatment of breast cancer. N Engl J Med. 2001;344(26):1997–2008. doi: 10.1056/NEJM200106283442607. [DOI] [PubMed] [Google Scholar]
- 3.Choi JY, Nowell SA, Blanco JG, Ambrosone CB. The role of genetic variability in drug metabolism pathways in breast cancer prognosis. Pharmacogenomics. 2006;7(4):613–624. doi: 10.2217/14622416.7.4.613. [DOI] [PubMed] [Google Scholar]
- 4.Yao S, Maghsoudlou D, Ambrosone CB. Breast cancer pharmacogenetics in the era of personalized medicine. Current Breast Cancer Reports. 2012 [Google Scholar]
- 5.Sladek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother Pharmacol. 2002;49(4):309–321. doi: 10.1007/s00280-001-0412-4. [DOI] [PubMed] [Google Scholar]
- 6.Ekhart C, Rodenhuis S, Smits PH, Beijnen JH, Huitema AD. Relations between polymorphisms in drug-metabolising enzymes and toxicity of chemotherapy with cyclophosphamide, thiotepa and carboplatin. Pharmacogenet Genomics. 2008;18(11):1009–1015. doi: 10.1097/FPC.0b013e328313aaa4. [DOI] [PubMed] [Google Scholar]
- 7.Lal S, Mahajan A, Chen WN, Chowbay B. Pharmacogenetics of target genes across doxorubicin disposition pathway: a review. Curr Drug Metab. 2010;11(1):115–128. doi: 10.2174/138920010791110890. [DOI] [PubMed] [Google Scholar]
- 8.Sucheston LE, Zhao H, Yao S, Zirpoli G, Liu S, Barlow WE, et al. Genetic predictors of taxane-induced neurotoxicity in a SWOG phase III intergroup adjuvant breast cancer treatment trial (S0221) Breast Cancer Res Treat. 2011;130(3):993–1002. doi: 10.1007/s10549-011-1671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Therapy Evaluation Program, Common Terminology Criteria for Adverse Events. Version 3.0, DCTD, NCI, NIH, DHHS 2006 [Google Scholar]
- 10.Edlund CK, Lee WH, Li D, Van Den Berg DJ, Conti DV. Snagger: a user-friendly program for incorporating additional information for tagSNP selection. BMC Bioinformatics. 2008;9:174. doi: 10.1186/1471-2105-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 12.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luna A, Nicodemus KK. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;23(6):774–776. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
- 14.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) British journal of cancer. 2005;93(4):387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caggiano V, Weiss RV, Rickert TS, Linde-Zwirble WT. Incidence, cost, and mortality of neutropenia hospitalization associated with chemotherapy. Cancer. 2005;103(9):1916–1924. doi: 10.1002/cncr.20983. [DOI] [PubMed] [Google Scholar]
- 18.Smith TJ, Khatcheressian J, Lyman GH, Ozer H, Armitage JO, Balducci L, et al. 2006 update of recommendations for the use of white blood cell growth factors: an evidence-based clinical practice guideline. J Clin Oncol. 2006;24(19):3187–3205. doi: 10.1200/JCO.2006.06.4451. [DOI] [PubMed] [Google Scholar]
- 19.Levi BP, Yilmaz OH, Duester G, Morrison SJ. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood. 2009;113(8):1670–1680. doi: 10.1182/blood-2008-05-156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoury T, Ademuyiwa FO, Chandraseekhar R, Jabbour M, Deleo A, Ferrone S, et al. Aldehyde dehydrogenase 1A1 expression in breast cancer is associated with stage, triple negativity, and outcome to neoadjuvant chemotherapy. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25(3):388–397. doi: 10.1038/modpathol.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekhart C, Doodeman VD, Rodenhuis S, Smits PH, Beijnen JH, Huitema AD. Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4-hydroxycyclophosphamide. Pharmacogenet Genomics. 2008;18(6):515–523. doi: 10.1097/FPC.0b013e3282fc9766. [DOI] [PubMed] [Google Scholar]
- 22.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26(2):259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crabb DW, Matsumoto M, Chang D, You M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc Nutr Soc. 2004;63(1):49–63. doi: 10.1079/pns2003327. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Zhou Z, Hodgkinson CA, Yuan Q, Shen PH, Mulligan CJ, et al. Haplotype-based study of the association of alcohol-metabolizing genes with alcohol dependence in four independent populations. Alcohol Clin Exp Res. 2011;35(2):304–316. doi: 10.1111/j.1530-0277.2010.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal S, Wong ZW, Sandanaraj E, Xiang X, Ang PC, Lee EJ, et al. Influence of ABCB1 and ABCG2 polymorphisms on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99(4):816–823. doi: 10.1111/j.1349-7006.2008.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wojnowski L, Kulle B, Schirmer M, Schluter G, Schmidt A, Rosenberger A, et al. NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation. 2005;112(24):3754–3762. doi: 10.1161/CIRCULATIONAHA.105.576850. [DOI] [PubMed] [Google Scholar]
- 27.Simon RM, Paik S, Hayes DF. Ues of archived specimens in evaluation of prognostic and predictive biomarkers. Journal of the National Cancer Institute. 2009;101(1):1–7. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. TagSNPs selected in ABCB1, ABCC1, and ALDH1A1
Supplementary Table S2. Associations of SNPs in ABCB1, ABCC1, and ALDH1A1 with grade 3 and 4 hematological toxicity in S0221 trial
Supplementary Table S3. Associations of SNPs in ABCB1, ABCC1, and ALDH1A1 with grade 3 gastrointestinal toxicity in S0221 trial