Abstract
Purpose
The aim of the present study is to assess whether the single nucleotide polymorphism in the GDF5 (+104T/C; rs143383) is associated with the symptomatic lumbar disc herniation in the Chinese Han population and the identification of the mechanisms of its action.
Methods
This study consisted of 231 patients with symptomatic lumbar disc herniation as the case group and 370 patients who had a lifetime lack of symptoms as the control group. PCR products were genotyped. Thirty-eight disc specimens derived from the cases were analyzed by immunohistochemical staining. The stain intensity of immunohistochemistry was quantified using a computerized image analysis system.
Results
Significant differences in genotypic and allelic frequencies were found between case group and control group (TT genotype P < 0.001; CC genotype P = 0.002; T allele P < 0.001). The T allele was more frequent in the case group regardless of gender (Female P = 0.018; Male P < 0.001). Significant differences were found in the genotype frequencies when stratified by gender except the comparison between the CC genotype and other genotypes combined among the female samples (P > 0.05). A semi-quantification of collagen protein in the nucleus pulposus showed that the average collagen protein content in TC group was higher than in TT group (P < 0.05).
Conclusion
Our results suggested that the GDF5 polymorphism is associated with a susceptibility to symptomatic lumbar disc herniation in the Chinese Han population and type II collagen in the nucleus pulposus may be a key factor in susceptibility to symptomatic lumbar disc herniation.
Keywords: GDF5, Polymorphism, Lumbar disc herniation, Han population
Introduction
As is known, lumbar disc degeneration (LDD) and lumbar disc herniation (LDH) are major musculoskeletal diseases causing low back pain, which is quite a common ailment all over the world affecting our work and life. Lumbar disc herniation is a serious lumbar disc disease.
The etiology of lumbar disc disease is complex. The traditional view generally attributes it to age, gender, height, smoking, physical activity and loading, which can explain a portion of lumbar disc degeneration and lumbar disc herniation [1, 2]. These environmental and constitutional risk factors played a role in the disease. Even though their effects are more important at lower lumbar levels [3], LDD and LDH may be explained to a large degree by genetic risk factors [4, 5]. In recent years, it is suggested in the twin and family studies that genetic risk factors contribute to the development of lumbar disc degeneration and lumbar disc herniation.
Growth differentiation factor 5 (GDF5; also known as cartilage-derived morphogenetic protein-1) is a member of the transforming growth factor-β (TGF-β) super-family and is closely related to the subfamily of bone morphogenetic proteins (BMPs). Previous studies found it exhibited a number of biological activities on the morphogenesis and development of tendon, ligament and bone [6–12]. In the common human intervertebral disc (IVD), the major components of the nucleus pulposus (NP) are collagen type II and proteoglycans (PGs), whereas the annulus fibrosus (AF) is composed primarily of collagen I and PGs [13]. Type I collagen can give IVD the tensile strength and type II collagen with PGs can give the tissue stiffness and resilience to compression [14]. Previous studies showed that GDF5 can promote accumulation of type II collagen during pellet culture [15].
Previous studies suggested that a functional single nucleotide polymorphism (SNP) located within the 5′-untranslated region of GDF5 (+104T/C) with a change of base from T to C is associated with susceptibility to osteoarthritis at different joint locations across different ethnic groups and it exerts allelic differences on transcriptional activity in chondrogenic cells, with the susceptibility allele showing reduced activity [16, 17]. The association of the SNP with osteoarthritis has been replicated in one European case-control study [18], while it was not confirmed in a Greek case-control study [19]. A recent study showed that there is an association between GDF5 (+104T/C) and lumbar disc degeneration in Northern European population [20]. In these studies, the carriers of the T allele have a higher risk than the carriers of C allele, so T allele can be considered as a predisposing allele in osteoarthritis and lumbar disc degeneration.
The aim of our study is to evaluate whether the SNP (+104T/C) is associated with symptomatic lumbar disc herniation in the Chinese population and how the polymorphism works.
Materials and methods
Specimens
A total of 601 subjects with or without symptomatic lumbar disc herniation were recruited from the Department of Spinal Surgery of the 150th Hospital and Xijing Hospital between March 2011 and June 2012.
We included 231 patients, 95 women (41.1 %) and 136 men (58.9 %) in the case group only if they had a history of unilateral pain radiating from the back along the femoral or sciatic nerve to the corresponding dermatome of the nerve root for more than 1 month and positive MRI findings for LDH. The symptoms should be in accordance with compression of the lumbar disc. The mean age of the case group was 48.40 ± 11.66 years. The control group consisted of 370 other patients, 175 women (47.3 %) and 195 men (52.7 %) who had a lifetime lack of symptoms suggesting LDH. Their average age was 47.85 ± 12.30 years. OA, previous fractures of the spine, lumbar spinal stenosis, malignancies involving the spine and poliomyelitis were excluded from all the subjects. All research participants were Han Chinese and gave written informed consent to take part. This study was approved by the University of Xi’ an Ethics Committee.
MRI
Lumbar sagittal MRI was performed with a 1.5-T imaging system (Siemmens AG, Germany). A T2-weighted image with a 5-mm slice thickness, a repetition of 2,500 ms and an echo time of 90 ms of the lumbar spine was taken for all the participants. The findings of MRI were considered positive if the disk extended beyond the margins of adjacent vertebral bodies. The evaluation of the lumbar disc herniation was agreed upon by three observers.
Genotypes
We extracted genomic DNA from peripheral blood leukocytes of each patient using standard protocols. The DNA fragment containing the polymorphic sequence was amplified by PCR. Primer5 software was used to design the primers for the purpose of the PCR amplification. The primer sequences for the polymorphism were as follows: forward 5′-CAGCATTACGCCATTCTTCC-3′; reverse 5′-CGCTGAATGACACCAAAGAGA-3′. Amplification proceeded for 35 cycles, with denaturation at 95 °C for 4 min, annealing at 58 °C for 35 s and extension at 72 °C for 45 s. All samples were sequenced using ABI 3730 DNA analyzers (Applied Biosystems).
Immunohistochemistry
We collected L4–5 disc specimens derived from surgical disc procedures performed on individuals with herniated discs from 38 cases. Twenty-six were derived from the cases with TT genotype as TT group and 12 were with TC genotype as TC group. All the specimens were transported to the laboratory and fixed in 10 % neutral buffered formalin. The un-decalcified specimens were embedded in paraffin and sectioned at 4 μm. Endogenous peroxidase was blocked using 3 % H2O2. Sections were incubated with rabbit polyclonal anti-human primary antibodies against collagen II (dilution 1:100; Abcam UK), followed by incubation with goat anti-rabbit IgG secondary antibody (ZSGB-BIO, Beijing, China). Then DAB (3,3′-diaminobenzidine) was used for 10 min and the sections were counterstained with Mayer’s hematoxylin. The samples were washed, dehydrated, and mounted with resinous mounting media. Five regions of nucleus pulposus were captured randomly for each section. The yellow stain indicated the presence of type II collagen. The stain intensity in the nucleus pulposus was quantified using a computerized image analysis system (Motic Med 6.0). The average stain intensity of every section represents the semi-quantification of the collagen-II content in the nucleus pulposus.
Statistical analysis
All statistical analyses were performed with SPSS software version 19.0 for Windows. The characteristics of case group and control group were compared with χ2 test and t test. Participants were stratified according to gender. Chi squared test and Fisher’s exact test were used to determine any differences in genotype and allele frequencies between case group and control group. Cochran–Mantel–Haenszel test was used when necessary. Odds ratios (OR) and their 95 % confidence intervals (CI) were also calculated. χ2 test was performed to assess Hardy–Weinberg equilibrium using spreadsheet software (Excel) for the purpose of ensuring population representation of the case group and the control group. A t test was used to determine whether there was a significant difference about the content of collagen II between TT group and TC group. All statistical tests were two-sided and the corrected P value < 0.05 was considered statistically significant.
Results
No significant differences in age, gender and smoking status were found between the two groups (Table 1). The data on alleles and genotypes in the case group and control group are shown (Table 2). The distributions of genotypes in the case group and the control group were consistent with the Hardy–Weinberg equilibrium law (Table 2).
Table 1.
The characteristics of case group and control group
| Characteristics | Case group (n = 231) | Control group (n = 370) | P value |
|---|---|---|---|
| Age (years) | 48.40 ± 11.66 | 47.85 ± 12.30 | 0.591 |
| Smoking | 55 (23.8 %) | 79 (21.4 %) | 0.481 |
| Gender (female/male) | 95/136 | 175/195 | 0.139 |
Table 2.
The genotype and allele frequencies of the SNP in the case group and control group
| Group | Number | Genotype (frequency) | Allele (frequency) | Hardy–Weinberg equilibrium | |||
|---|---|---|---|---|---|---|---|
| TT | TC | CC | T | C | P value | ||
| Case | |||||||
| All | 231 | 144 | 79 | 8 | 367 | 95 | 0.48 |
| Female | 95 | 58 | 34 | 3 | 150 | 40 | 0.45 |
| Male | 136 | 86 | 45 | 5 | 217 | 55 | 0.77 |
| Control | |||||||
| All | 370 | 173 | 158 | 39 | 504 | 236 | 0.74 |
| Female | 175 | 84 | 75 | 16 | 243 | 107 | 0.90 |
| Male | 195 | 89 | 83 | 23 | 261 | 129 | 0.59 |
We compared the genotype frequencies (TT versus other genotypes combined and CC versus other genotypes combined) and allele frequencies, in cases versus control, for all subject and for females and males separately (Table 3). When the genotype and allele frequencies were analyzed among all the subjects, TT genotype (predisposing genotype) and CC genotype (protecting genotype) were significantly associated with the risk for LDH (P < 0.001; P = 0.002). T allele (predisposing allele) was more frequent in the case group than in the control group (P < 0.001). When stratified by gender, only CC genotype was not significantly associated with risk for LDH in the female samples (P = 0.082). Significant differences of genotype distributions were found in other comparisons when stratified by gender (P = 0.040, 0.002, 0.009). In addition, T allele was also more frequent in the case group than in the control group regardless of gender (P = 0.018; P < 0.001).
Table 3.
The association between the SNP and the symptomatic lumbar disc herniation
| Groups compared | TT versus other combined | CC versus other combined | T allele versus C allele | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | P value | 95 % CI | OR | P value | 95 % CI | OR | P value | 95 % CI | |
| All cases versus all controls | 1.89 | <0.001 | 1.35–2.63 | 0.30 | 0.002 | 0.14–0.66 | 1.81 | <0.001 | 1.38–2.38 |
| Female cases versus female controls | 1.70 | 0.040 | 1.02–2.82 | 0.32 | 0.082 | 0.09–1.14 | 1.65 | 0.018 | 1.09–2.50 |
| Male cases versus male controls | 2.05 | 0.002 | 1.31–3.21 | 0.29 | 0.009 | 0.11–0.77 | 1.95 | <0.001 | 1.36–2.81 |
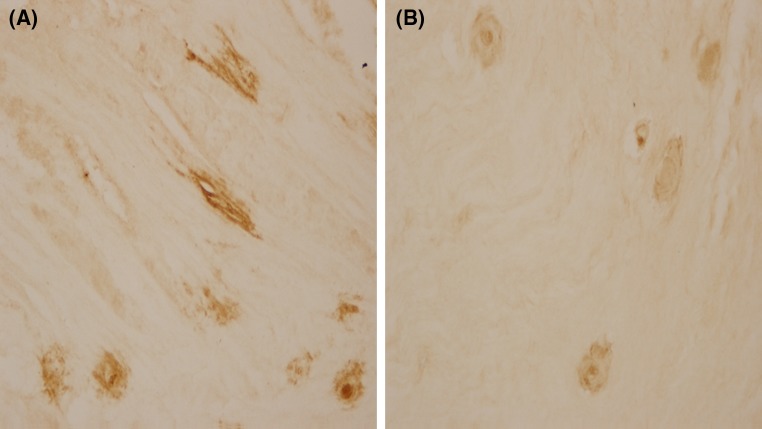
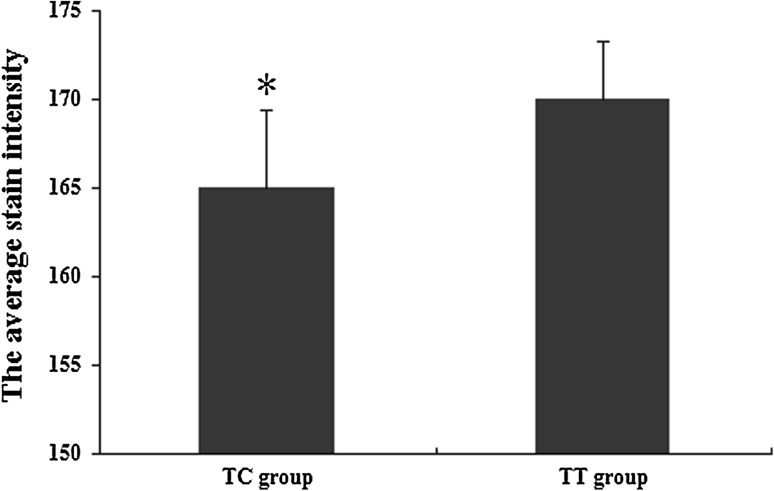
Immunohistologic staining revealed that the characteristic collagen II expression is mostly distributed in the cytoplasm and the nucleus of cells, while a relatively pale collagen II expression was seen with uneven extracellular distribution. Either intracellular or extracellular staining of collagen-II protein was darker in TC group than in TT group (Fig. 1). A semi-quantification of collagen-II protein in the nucleus pulposus (Fig. 2) showed that the average collagen-II protein content in TC group was higher than in TT group (P < 0.05).
Fig. 1.
Immunohistologic staining for the nucleus pulposus a TC group, b TT group. Images were captured at ×400 magnifications
Fig. 2.
Graphical representation of the immunohistologic staining in nucleus pulposus of the disc; results calculated by a computerized image analysis system (n = 38, ±SD; *P < 0.05)
Discussion
The variant GDF5 (+104T/C) is the most common single-nucleotide polymorphism (SNP) for GDF5, which increases the risk of developing musculoskeletal diseases. Recent studies showed an association between the SNP rs143383 and some joint-related diseases [17, 18, 21]. The replication studies in different ethnic populations suggested that the SNP rs143383 is associated with OA [22, 23] and congenital dislocation of the hip [24], although in the Greek populations no significant association was found [19]. One important biological function for GDF5 may be its role in the formation of diarthrodial joints [9]. Experiments on animals suggested a functional null mutation in GDF5 can lead to developmental failure of the intra-articular ligament and sutured coated with recombinant human GDF5 had an early beneficial effect in a rabbit tendon injury model [10, 25]. Because of the case-control association studies about GDF5 and its biological function, we took GDF5 as a candidate for the symptomatic LDH susceptibility gene.
In this study, the influence of other factors has been removed as much as possible to confirm the correlation between symptomatic LDH and this SNP of GDF5 gene. Although the association between OA and some SNPs was proved in the Asian cohorts, not all the effects of these SNPs can be related to the function in all the collagenous tissues. Moreover, the SNP with the susceptibility to OA may not be responsible for LDH, so we excluded the OA patients from the subjects.
We found the polymorphic T allele as a predisposing allele was more frequent in the case group than in the control group in Han Chinese cohorts. The results agreed with those of Williams et al. [20] observed in a Northern European cohort in which the SNP rs143383 correlated with lumbar disc degeneration. Our results indicated that T allele as a predisposing allele and TT genotype as a predisposing genotype were associated with the risk of symptomatic lumbar disc herniation among the female samples and male samples. However, no significant difference of genotypes distribution was found between CC genotype and other genotypes combined among the female samples when stratified by gender. This is possibly because the female sample number seemed relatively insufficient when the samples were stratified by gender. In addition, control group was short of integrated diagnosis of MRI, so the potential LDH patients without symptoms may be included into the control group. The above limitations may bias our analyses, so more studies among larger sample are necessary.
The immunohistochemistry study performed on the nucleus pulposus confirms that the type II collagen can be detected in both TT group and TC group. Moreover, we can see stronger staining for type II collagen in the samples from patients with TC genotype than those with TT genotype. The likely explanation for this result is the difference in allelic expression. A previous study demonstrated that the associated T-allele showed a reduction in expression relative to the C-allele by extracting the RNA from the cartilage of OA patient [18]. Another study suggested GDF5 resulted in an increase in type II collagen gene expression [13]. Thus, some alterations in the mechanical properties of the intervertebral disc caused by this SNP may predispose to LDH. It is reasonable to presume that there would be some other structural and biochemical constituents of the intervertebral disc mediated by GDF5, making it more vulnerable to external forces in some persons than in others.
Intervertebral disc degeneration is mainly characterized by changes in the composition of the extracellular matrix and the loss of water and disc cells. The content of GDF5 in the nucleus pulposus mostly depends on native disc cells [26], thus it seems insufficient to detect individual variances by analyzing the content of GDF5 in the nucleus pulposus. However, the interactive potential between GDF5 and type II collagen can not be neglected, and the study of the interaction is required to further elucidate the pathogenesis of lumbar disc degeneration and lumbar disc herniation.
To the authors’ knowledge, our study is the first report on the association between functional GDF5 SNP rs143383 and symptomatic LDH in the Han Chinese population. We also found type II collagen in the nucleus pulposus influenced by this SNP, which may be a key factor in susceptibility to symptomatic LDH. Nevertheless, it is still not well known how the polymorphism works. Further studies should be conducted to explore how it affects the structure of the lumbar disc and its surrounding tissues. Our results are expected to lead to a better understanding of the pathogenic mechanisms of LDH and to achieve a novel treatment strategy for LDH.
Acknowledgments
This study was supported by the Ministry of Science and Technology of the People’s Liberation Army (01L005).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Kuh DJ, Coggan D, Mann S, Cooper C, Yusuf E. Height, occupation and back pain in a national prospective study. Br J Rheumatol. 1993;32:911–916. doi: 10.1093/rheumatology/32.10.911. [DOI] [PubMed] [Google Scholar]
- 2.Battie MC, Videman T, Gibbons LE, Fisher LD, Manninen H, Gill K, 1995 Volvo Award in clinical sciences Determinants of lumbar disc degeneration. A study relating lifetime exposures and magnetic resonance imaging findings in identical twins. Spine (Phila Pa 1976) 1995;20:2601–2612. doi: 10.1097/00007632-199512150-00001. [DOI] [PubMed] [Google Scholar]
- 3.Battie MC, Videman T, Levalahti E, Gill K, Kaprio J. Genetic and environmental effects on disc degeneration by phenotype and spinal level: a multivariate twin study. Spine (Phila Pa 1976) 2008;33:2801–2808. doi: 10.1097/BRS.0b013e31818043b7. [DOI] [PubMed] [Google Scholar]
- 4.Ala-Kokko L. Genetic risk factors for lumbar disc disease. Ann Med. 2002;34:42–47. doi: 10.1080/078538902317338634. [DOI] [PubMed] [Google Scholar]
- 5.Paz Aparicio J, Fernandez Bances I, Lopez-Anglada Fernandez E, Montes AH, Paz Aparicio A, Pena Vazquez J, Ramos Garcia S, Anton Garcia S, Lopez Fernandez P, Valle-Garay E, Asensi V. The IL-1beta (+3953 T/C) gene polymorphism associates to symptomatic lumbar disc herniation. Eur Spine J. 2011;20(Suppl 3):383–389. doi: 10.1007/s00586-011-1915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikic B. Multiple effects of GDF-5 deficiency on skeletal tissues: implications for therapeutic bioengineering. Ann Biomed Eng. 2004;32:466–476. doi: 10.1023/B:ABME.0000017549.57126.51. [DOI] [PubMed] [Google Scholar]
- 7.Francis-West PH, Abdelfattah A, Chen P, Allen C, Parish J, Ladher R, Allen S, MacPherson S, Luyten FP, Archer CW. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–1315. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- 8.Edwards CJ, Francis-West PH. Bone morphogenetic proteins in the development and healing of synovial joints. Semin Arth Rheum. 2001;31:33–42. doi: 10.1053/sarh.2001.24875. [DOI] [PubMed] [Google Scholar]
- 9.Luyten FP. Cartilage-derived morphogenetic protein-1. Int J Biochem Cell Biol. 1997;29:1241–1244. doi: 10.1016/S1357-2725(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 10.Harada M, Takahara M, Zhe P, Otsuji M, Iuchi Y, Takagi M, Ogino T. Developmental failure of the intra-articular ligaments in mice with absence of growth differentiation factor 5. Osteoarth Cartil. 2007;15:468–474. doi: 10.1016/j.joca.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Vaes RB, Rivadeneira F, Kerkhof JM, Hofman A, Pols HA, Uitterlinden AG, van Meurs JB. Genetic variation in the GDF5 region is associated with osteoarthritis, height, hip axis length and fracture risk: the Rotterdam study. Ann Rheum Dis. 2009;68:1754–1760. doi: 10.1136/ard.2008.099655. [DOI] [PubMed] [Google Scholar]
- 12.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, Shen H, Timpson N, Lettre G, Usala G, Chines PS, Stringham HM, Scott LJ, Dei M, Lai S, Albai G, Crisponi L, Naitza S, Doheny KF, Pugh EW, Ben-Shlomo Y, Ebrahim S, Lawlor DA, Bergman RN, Watanabe RM, Uda M, Tuomilehto J, Coresh J, Hirschhorn JN, Shuldiner AR, Schlessinger D, Collins FS, Davey Smith G, Boerwinkle E, Cao A, Boehnke M, Abecasis GR, Mohlke KL. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Gen. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35:652–655. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 14.Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976) 1995;20:1307–1314. doi: 10.1097/00007632-199506000-00022. [DOI] [PubMed] [Google Scholar]
- 15.Bai X, Xiao Z, Pan Y, Hu J, Pohl J, Wen J, Li L. Cartilage-derived morphogenetic protein-1 promotes the differentiation of mesenchymal stem cells into chondrocytes. Biochem Biophys Res Comm. 2004;325:453–460. doi: 10.1016/j.bbrc.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 16.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, Fujioka M, Sudo A, Uchida A, Yamamoto S, Ozaki K, Takigawa M, Tanaka T, Nakamura Y, Jiang Q, Ikegawa S. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Gen. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 17.Chapman K, Takahashi A, Meulenbelt I, Watson C, Rodriguez-Lopez J, Egli R, Tsezou A, Malizos KN, Kloppenburg M, Shi D, Southam L, van der Breggen R, Donn R, Qin J, Doherty M, Slagboom PE, Wallis G, Kamatani N, Jiang Q, Gonzalez A, Loughlin J, Ikegawa S. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5′ UTR of GDF5 with osteoarthritis susceptibility. Hum Mol Gen. 2008;17:1497–1504. doi: 10.1093/hmg/ddn038. [DOI] [PubMed] [Google Scholar]
- 18.Southam L, Rodriguez-Lopez J, Wilkins JM, Pombo-Suarez M, Snelling S, Gomez-Reino JJ, Chapman K, Gonzalez A, Loughlin J. An SNP in the 5′-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Gen. 2007;16:2226–2232. doi: 10.1093/hmg/ddm174. [DOI] [PubMed] [Google Scholar]
- 19.Tsezou A, Satra M, Oikonomou P, Bargiotas K, Malizos KN. The growth differentiation factor 5 (GDF5) core promoter polymorphism is not associated with knee osteoarthritis in the Greek population. J Orthop Res. 2008;26:136–140. doi: 10.1002/jor.20464. [DOI] [PubMed] [Google Scholar]
- 20.Williams FM, Popham M, Hart DJ, de Schepper E, Bierma-Zeinstra S, Hofman A, Uitterlinden AG, Arden NK, Cooper C, Spector TD, Valdes AM, van Meurs J. GDF5 single-nucleotide polymorphism rs143383 is associated with lumbar disc degeneration in Northern European women. Arth Rheum. 2011;63:708–712. doi: 10.1002/art.30169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai J, Shi D, Zhu P, Qin J, Ni H, Xu Y, Yao C, Zhu L, Zhu H, Zhao B, Wei J, Liu B, Ikegawa S, Jiang Q, Ding Y. Association of a single nucleotide polymorphism in growth differentiate factor 5 with congenital dysplasia of the hip: a case-control study. Arth Res Ther. 2008;10:R126. doi: 10.1186/ar2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egli RJ, Southam L, Wilkins JM, Lorenzen I, Pombo-Suarez M, Gonzalez A, Carr A, Chapman K, Loughlin J. Functional analysis of the osteoarthritis susceptibility-associated GDF5 regulatory polymorphism. Arth Rheum. 2009;60:2055–2064. doi: 10.1002/art.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdes AM, Spector TD, Doherty S, Wheeler M, Hart DJ, Doherty M. Association of the DVWA and GDF5 polymorphisms with osteoarthritis in UK populations. Ann Rheum Dis. 2009;68:1916–1920. doi: 10.1136/ard.2008.102236. [DOI] [PubMed] [Google Scholar]
- 24.Rouault K, Scotet V, Autret S, Gaucher F, Dubrana F, Tanguy D, El Rassi CY, Fenoll B, Ferec C. Evidence of association between GDF5 polymorphisms and congenital dislocation of the hip in a caucasian population. Osteoarth Cartil. 2010;18:1144–1149. doi: 10.1016/j.joca.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Henn RF, 3rd, Kuo CE, Kessler MW, Razzano P, Grande DP, Wolfe SW. Augmentation of zone II flexor tendon repair using growth differentiation factor 5 in a rabbit model. J Hand Surg Am. 2010;35:1825–1832. doi: 10.1016/j.jhsa.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 26.Le Maitre CL, Freemont AJ, Hoyland JA. Expression of cartilage-derived morphogenetic protein in human intervertebral discs and its effect on matrix synthesis in degenerate human nucleus pulposus cells. Arth Res Ther. 2009;11:R137. doi: 10.1186/ar2808. [DOI] [PMC free article] [PubMed] [Google Scholar]