Abstract
The origin of plasmid DNA transfer, oriT, has been localized on RK2, a conjugative drug-resistance plasmid of the IncP group with a very broad host range in gram-negative bacteria. The transfer origin is contained in a 760-base-pair Hae II restriction fragment that maps in the same region as the single-strand nick made by the RK2 relaxation complex. The functional oriT was subcloned as a 112-base-pair Hpa II fragment, and the DNA sequence of this region was determined. The dominant structural feature of the oriT sequence is a 19-base-pair inverted repeat, with 15 of the 19 bases able to form pairs in a hairpin structure. This inverted repeat may be the recognition site for the relaxation complex proteins, which nick the plasmid DNA molecule and initiate the transfer process.
Full text
PDF
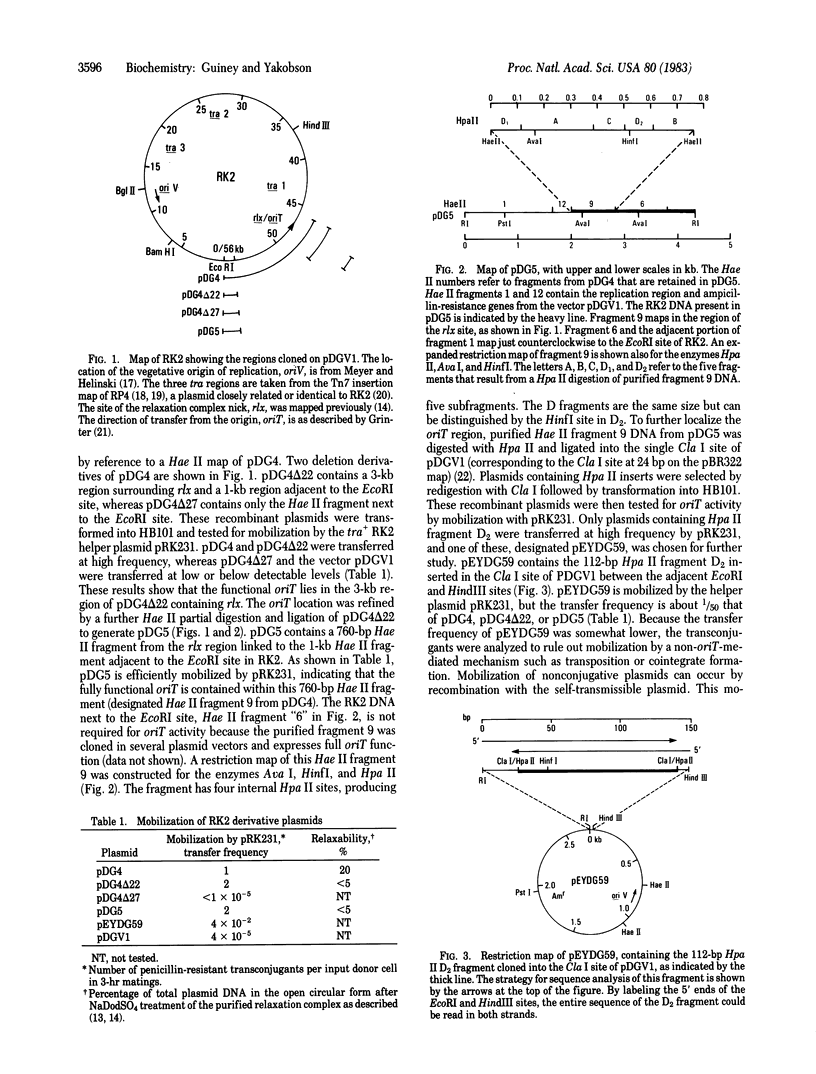
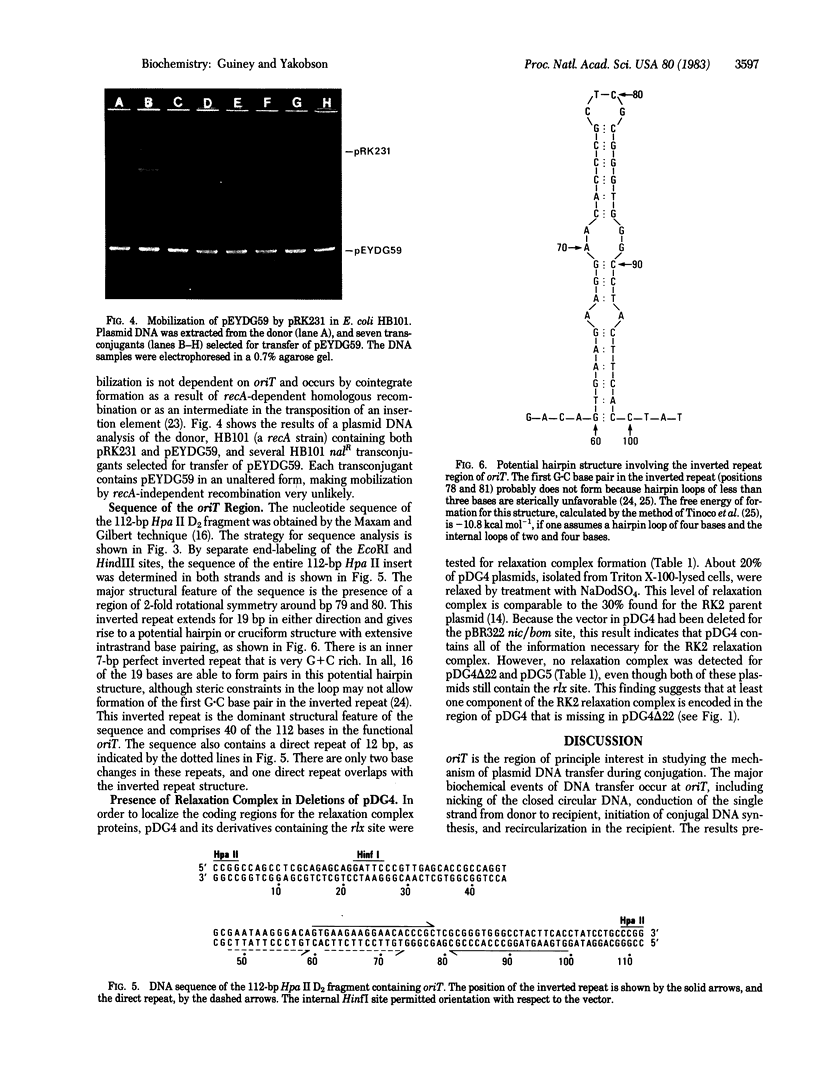

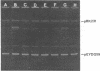
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth P. T., Grinter N. J., Bradley D. E. Conjugal transfer system of plasmid RP4: analysis by transposon 7 insertion. J Bacteriol. 1978 Jan;133(1):43–52. doi: 10.1128/jb.133.1.43-52.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D. Determination of restriction sites and the nucleotide sequence surrounding the relaxation site of ColE1. J Mol Biol. 1978 Oct 5;124(4):601–639. doi: 10.1016/0022-2836(78)90174-2. [DOI] [PubMed] [Google Scholar]
- Blair D. G., Helinski D. R. Relaxation complexes of plasmid DNA and protein. I. Strand-specific association of protein and DNA in the relaxed complexes of plasmids ColE1 and ColE2. J Biol Chem. 1975 Nov 25;250(22):8785–8789. [PubMed] [Google Scholar]
- Burkardt H. J., Riess G., Pühler A. Relationship of group P1 plasmids revealed by heteroduplex experiments: RP1, RP4, R68 and RK2 are identical. J Gen Microbiol. 1979 Oct;114(2):341–348. doi: 10.1099/00221287-114-2-341. [DOI] [PubMed] [Google Scholar]
- Clark A. J., Warren G. J. Conjugal transmission of plasmids. Annu Rev Genet. 1979;13:99–125. doi: 10.1146/annurev.ge.13.120179.000531. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R., Willetts N. Characterisation of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J Mol Biol. 1980 Jan 15;136(2):129–150. doi: 10.1016/0022-2836(80)90309-5. [DOI] [PubMed] [Google Scholar]
- Everett R., Willetts N. Cloning, mutation, and location of the F origin of conjugal transfer. EMBO J. 1982;1(6):747–753. doi: 10.1002/j.1460-2075.1982.tb01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan J., Sherratt D. Plasmid ColE1 conjugal mobility: the nature of bom, a region required in cis for transfer. Mol Gen Genet. 1982;185(2):344–351. doi: 10.1007/BF00330810. [DOI] [PubMed] [Google Scholar]
- Grinter N. J. Analysis of chromosome mobilization using hybrids between plasmid RP4 and a fragment of bacteriophage lambda carrying IS1. Plasmid. 1981 May;5(3):267–276. doi: 10.1016/0147-619x(81)90004-4. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Helinski D. R. Relaxation complexes of poasmid DNA and protein. III. Association of protein with the 5' terminus of the broken DNA strand in the relaxed complex of plasmid ColE1. J Biol Chem. 1975 Nov 25;250(22):8796–8803. [PubMed] [Google Scholar]
- Guiney D. G., Helinski D. R. The DNA-protein relaxation complex of the plasmid RK2: location of the site-specific nick in the region of the proposed origin of transfer. Mol Gen Genet. 1979 Oct 3;176(2):183–189. doi: 10.1007/BF00273212. [DOI] [PubMed] [Google Scholar]
- Guyer M. S., Clark A. J. cis-Dominant, transfer-deficient mutants of the Escherichia coli K-12 F sex factor. J Bacteriol. 1976 Jan;125(1):233–247. doi: 10.1128/jb.125.1.233-247.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupersztoch-Portnoy Y. M., Miklos G. L., Helinski D. R. Properties of the relaxation complexes of supercoiled deoxyribonucleic acid and protein of the R plasmids R64, R28K, and R6K. J Bacteriol. 1974 Oct;120(1):545–548. doi: 10.1128/jb.120.1.545-548.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Relaxation complexes of plasmid DNA and protein. II. Characterization of the proteins associated with the unrelaxed and relaxed complexes of plasmid ColE1. J Biol Chem. 1975 Nov 25;250(22):8790–8795. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. J., Helinski D. R. Unidirectional replication of the P-group plasmid RK2. Biochim Biophys Acta. 1977 Sep 6;478(1):109–113. doi: 10.1016/0005-2787(77)90249-0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Hershberger C. L., Rownd R. Strand-specific nick in open circular R-factor deoxyribonucleic acid: attachment of the linear strand to a proteinaceous cellular component. J Bacteriol. 1973 Apr;114(1):300–308. doi: 10.1128/jb.114.1.300-308.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Hashimoto-Gotoh T., Timmis K. N. Location of two relaxation nick sites in R6K and single sites in pSC101 and RSF1010 close to origins of vegetative replication: implication for conjugal transfer of plasmid deoxyribonucleic acid. J Bacteriol. 1980 Dec;144(3):923–932. doi: 10.1128/jb.144.3.923-932.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki M., Tomizawa J. Asymmetric transfer of DNA strands in bacterial conjugation. Cold Spring Harb Symp Quant Biol. 1968;33:651–658. doi: 10.1101/sqb.1968.033.01.074. [DOI] [PubMed] [Google Scholar]
- Rupp W. D., Ihler G. Strand selection during bacterial mating. Cold Spring Harb Symp Quant Biol. 1968;33:647–650. doi: 10.1101/sqb.1968.033.01.073. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by d(TA) oligomers. II. Analysis of the helix-coli transitions of linear and circular oligomers. J Mol Biol. 1970 Feb 28;48(1):145–171. doi: 10.1016/0022-2836(70)90225-1. [DOI] [PubMed] [Google Scholar]
- Shafferman A., Helinski D. R. Structural properties of the beta origin of replication of plasmid R6K. J Biol Chem. 1983 Apr 10;258(7):4083–4090. [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Warren G. J., Twigg A. J., Sherratt D. J. ColE1 plasmid mobility and relaxation complex. Nature. 1978 Jul 20;274(5668):259–261. doi: 10.1038/274259a0. [DOI] [PubMed] [Google Scholar]
- Willetts N. S. Location of the origin of transfer of the sex factor F. J Bacteriol. 1972 Nov;112(2):773–778. doi: 10.1128/jb.112.2.773-778.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]