Abstract
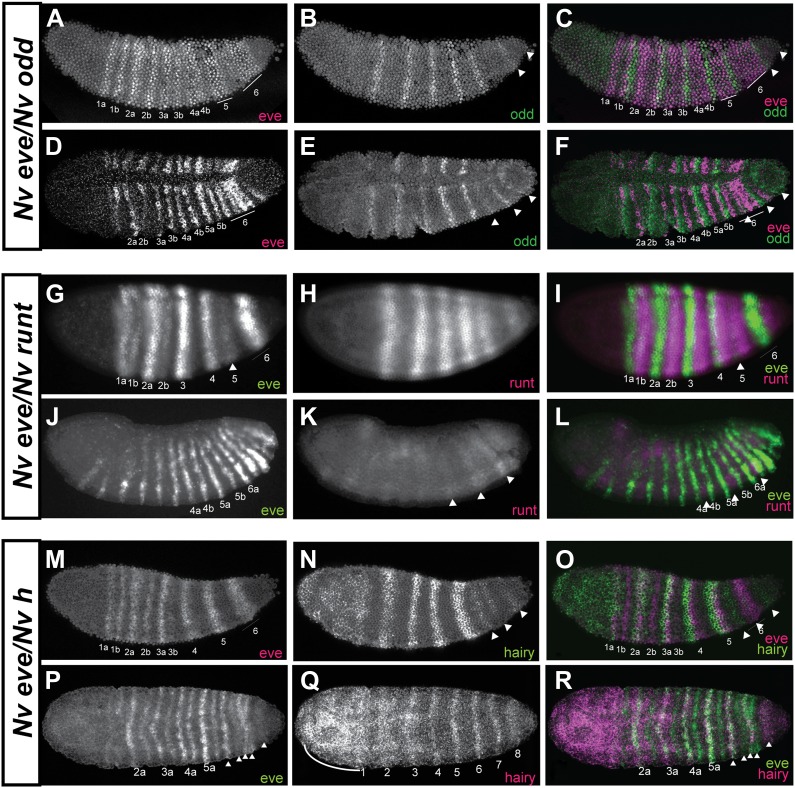
Embryonic anterior–posterior patterning is well understood in Drosophila, which uses ‘long germ’ embryogenesis, in which all segments are patterned before cellularization. In contrast, most insects use ‘short germ’ embryogenesis, wherein only head and thorax are patterned in a syncytial environment while the remainder of the embryo is generated after cellularization. We use the wasp Nasonia (Nv) to address how the transition from short to long germ embryogenesis occurred. Maternal and gap gene expression in Nasonia suggest long germ embryogenesis. However, the Nasonia pair-rule genes even-skipped, odd-skipped, runt and hairy are all expressed as early blastoderm pair-rule stripes and late-forming posterior stripes. Knockdown of Nv eve, odd or h causes loss of alternate segments at the anterior and complete loss of abdominal segments. We propose that Nasonia uses a mixed mode of segmentation wherein pair-rule genes pattern the embryo in a manner resembling Drosophila at the anterior and ancestral Tribolium at the posterior.
DOI: http://dx.doi.org/10.7554/eLife.01440.001
Research organism: D. melanogaster, other
eLife digest
Networks of genes that work together are widespread in nature. The conservation of individual genes across species and the tendency of their networks to stick together is a sign that they are working efficiently. Furthermore, it is common for existing gene networks to be adapted to perform new tasks, instead of new networks being invented every time a similar but distinct demand arises. One important question is: how can evolution use the same building blocks—such as the genes in a functioning network—in different ways to achieve new outcomes?
The gene network that sets up the ‘body plan’ of insects during development has been well studied, most deeply in the fruit fly, Drosophila. Like all insects, the body of a fruit fly is divided into three main parts—the head, the thorax and the abdomen—and each of these parts is made up of several smaller segments. There is a remarkable diversity of insect body plans in nature, and yet, these seem to arise from the same gene networks in the embryo.
When a Drosophila embryo is growing into a larva, all the different body segments develop at the same time. In most other insects, however, segments of the abdomen emerge later and sequentially during the development process. The ancestors of most insects are also thought to have developed in this way, which is known as ‘short germ embryogenesis’. So how did the so-called ‘long germ embryogenesis’, as observed in Drosophila, evolve from the short germ embryogenesis that is observed in most other insects?
The gene network that controls development includes the ‘pair-rule genes’ that are expressed in a pattern of alternating stripes that wrap around, top to bottom, along most of the length of the embryo. These stripes mark where the edges of each body segment will eventually develop. In fruit flies, this pattern extends along the entire length of the embryo and the stripes all appear at one time. However, in the abdominal region of short germ insects, the pair-rule genes are expressed in waves that pass through the posterior region as it grows, with new segments being added one behind the other.
Now, Rosenberg et al. have attempted to explain how the same genes can be used to direct the segmentation process in such different ways by studying another long germ insect species, the jewel wasp. Analysis of the expression of pair-rule genes in the jewel wasp shows that it uses a mixed strategy to control segmentation. The development of segments at the front of its body is directed in the same way as the fruit fly, with all these segments laid down together. However, the segments at the rear of the body are only patterned later, one after the other, like most other insects.
The work of Rosenberg et al. suggests that the jewel wasp represents an intermediate step between ancestral insects and Drosophila in the evolution of the gene network that patterns the ‘body plan’. Identifying and studying these intermediate forms allows us to understand the ways in which evolution can innovate by building upon what has come before.
Introduction
Control of axial patterning and embryonic development is well understood in Drosophila (reviewed in Liu and Kaufman, 2005b; Peel et al., 2005; Rosenberg et al., 2009; Pankratz and Jaj, 1993). Extensive work has elucidated the genetic basis of establishment of the anterior–posterior (A–P) and dorsal–ventral (D–V) axes of the fly embryo. For the A–P axis, maternally loaded mRNAs generate localized signaling centers at each pole of the egg to establish morphogenetic gradients. These gradients instruct, in a concentration dependent manner, broad domains of expression of early zygotic genes, the ‘gap genes’ (Chen et al., 2012). This is made possible in part by the syncytial environment of the early blastoderm embryo where nuclei are not bounded by membranes, allowing diffusion of morphogen transcription factors through a shared cytoplasm without the need for cell–cell signaling. In this environment, broad activation by maternal factors coupled with repressive activities by the gap genes leads to the expression of the pair-rule genes in two-segment periodicity, as pair-rule stripes. The overlapping registers of different pair-rule genes ultimately establish segment polarity through activation of the segment polarity genes, each expressed in stripes with single segmental register. This mode of development is termed ‘long germ’ embryogenesis because the embryo occupies all of the blastoderm apart from a dorsal region representing the extraembryonic ammnioserosa. A striking feature of long germ embryogenesis is that virtually all of segment patterning is completed synchronously in the syncytial environment. However, forays into other insect models have revealed that the Drosophila paradigm is an evolutionarily derived state, and that insects generally undergo a very different type of embryogenesis and segmentation (reviewed in Liu and Kaufman, 2005b; Peel et al., 2005; Rosenberg et al., 2009).
Unlike flies, most insect embryonic primordia occupy only a small portion of the blastoderm and only few anterior segments (head and thorax) are patterned in a syncytial environment. The remainder of the embryo is generated after cellularization via a ‘growth zone’, at the posterior region of the embryo. This mode is termed ‘short germ’ embryogenesis. Recently, the mechanisms governing posterior segment patterning and growth in the Tribolium embryo were characterized in elegant detail (Choe et al., 2006; Choe and Brown, 2009; El-Sherif et al., 2012; Sarrazin et al., 2012): Oscillations of the pair-rule gene Tc’odd-skipped (Tc’odd) in the growth zone are in turn linked to a circuit of two other pair-rule genes, Tc’runt and Tc’even-skipped (Tc’eve), such that each new pair of segments experiences a pulse of Tc’odd and requires both Tc’eve and Tc’runt expression in order to progress; the driver of these oscillations is still unknown. The waves of expression of Tc’odd-skipped pass through the growth zone rhythmically, generating segments and new stripes of stable expression with each periodic pulse (Sarrazin et al., 2012). RNAi of Tc’odd, Tc’runt, or Tc’eve results in asegmental embryos, underscoring their requirement in both growth zone-derived segments and earlier blastoderm anterior segments (Choe et al., 2006). In contrast, the pair-rule genes Tc’sloppy paired (Tc’slp) and Tc’paired (Tc’prd) appear in two-segment periodicity in head stripes and in stripes that emerge from the growth zone, and RNAi of those genes produce classical pair-rule phenotypes, in which alternating segments are lost (Choe and Brown, 2007). Live imaging revealed that formation of posterior segments results primarily from convergent extension and short-range cell movements and not strictly from cell division within the ‘growth zone’. This mechanism appears similar to both segmentation of vertebrate presomitic mesoderm (reviewed in [Dubrulle and Pourquie, 2004] and [Pourquie, 2011]) and to segment formation in more basal arthropods, including the centipede Strigamia maritima (Chipman et al., 2004; Chipman and Akam, 2008) and the spider Cupiennius salei (Stollewerk et al., 2003), suggesting it as an ancient mechanism inherited from the last common ancestor of all segmented animals (though this interpretation is still debated; reviewed in [Davis and Patel, 1999]).
As Drosophila is only one example of a derived long germ strategy, one outstanding question is how transitions from short germ to long germ embryogenesis occurred, such that the same set of segmentation genes possesses different functions. The careful study of additional long germ insects should shed light on what aspects of Drosophila development are essential facets of long germ embryogenesis and which aspects are more evolutionarily labile. Other model species have been studied, including long germ beetles (e.g., Callosobruchus order: Coleoptera; (Patel et al., 1994)), and several members of the order Hymenoptera, including the honeybee, Apis mellifera (Dearden et al., 2006; Wilson et al., 2010; Wilson and Dearden, 2011, 2012) and the jewel wasp, Nasonia vitripennis (Nv) (Pultz et al., 1999; Werren et al., 2010). However, systematic characterization of their pair-rule genes and segmentation mechanisms is still incomplete.
We use the wasp Nasonia vitripennis as a model for the study of A–P patterning, as a species that appears to have evolved, independently of Drosophila, a similar mode of long germ embryogenesis. We have previously characterized the early patterns of Nasonia segmentation genes and found that maternal and gap gene expression confirms a long germ mode of embryogenesis. This conclusion was based on the existence of two polar signaling centers, each utilizing localized maternal Nv orthodenticle (otd) mRNA that encodes a morphogen. Nv otd acts in combination with Nv hunchback (hb) and localized maternal Nv giant (gt) at the anterior, and with localized maternal Nv caudal (cad) at the posterior, to specify positional identity. The domains of zygotic expression of Nv hb, Nv gt, Nv cad, Nv Krüppel (Kr), Nv tailless (tll), and Nv knirps (kni) closely resemble their Drosophila counterparts, consistent with a similar mode of blastoderm allocation (Pultz et al., 2005; Lynch et al., 2006; Olesnicky et al., 2006; Brent et al., 2007). Although these data support Drosophila-like early regulatory interactions and a long germ mode of embryogenesis, little was known about later stages of Nasonia embryonic patterning.
We analyzed the expression and function of the pair-rule genes Nv eve, Nv odd, Nv runt and Nv h during embryogenesis. We found that each gene is expressed in both a canonical long-germ pair-rule stripe pattern at the anterior, as well as late-forming posterior stripes, indicating a dual mode of regulation. Strikingly, Nv eve is ultimately expressed in a total of 16 segmental stripes, of which six are derived from a single posterior stripe in the cellularized blastoderm. We also observe waves of Nv odd expression that resemble the waves of Tribolium odd expression, suggesting the residual activity of a segmentation clock in Nasonia. As in Tribolium, we found that mitoses do not occur exclusively at the site of late forming segments, but mitotic figures are not randomly distributed throughout the embryo. Instead, coordinated mitoses resembling the later mitotic domains of Drosophila (Foe, 1989) appear and progress in waves from anterior to posterior, and are largely excluded from stripes of eve expression, suggesting a coordination of mitoses by segmentation genes. Using morpholinos to knock down gene function, we found that Nv eve, Nv odd and Nv h phenotypes do not affect alternating segments at the posterior, unlike what is observed in Drosophila. Instead, these ‘pair-rule’ genes are required for the formation of a continuous posterior region comprising abdominal segments A5–A10. Phenotypes in the anterior of the embryo are gene-specific; each gene exhibits a partial pair-rule phenotype in the allelic series. We suggest that Nasonia uses ‘pair-rule’ genes to pattern the embryo in a manner that resembles both Drosophila and Tribolium. We present a model for how this mixed mode of segmentation is achieved.
Results
Nasonia even-skipped exhibits character of both long and short germ patterning
The expression of eve has been studied in many insects, owing to a widely cross-reacting antibody. Its promoter has also been well studied in Drosophila and has become a classic example of modular gene control (Patel et al., 1992; Small et al., 1992, 1996). We used circular RACE from total embryo RNA (McGrath, 2011) to generate a fragment of approximately 1 kb corresponding to the coding region of Nv eve, including the highly conserved homeodomain (Genbank Accession# KC168090). Several minor transcript variants were captured and sequenced, but not studied further (see ‘Materials and methods’ for GenBank accession numbers). The homeodomain of Nv Eve shares 81.7% amino acid identity with its Drosophila counterpart.
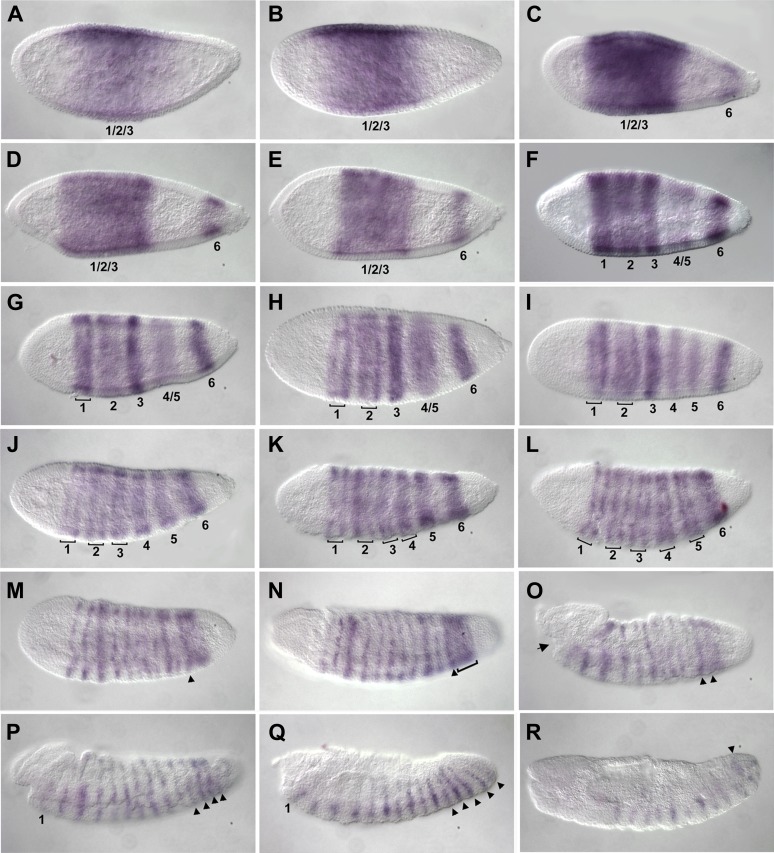
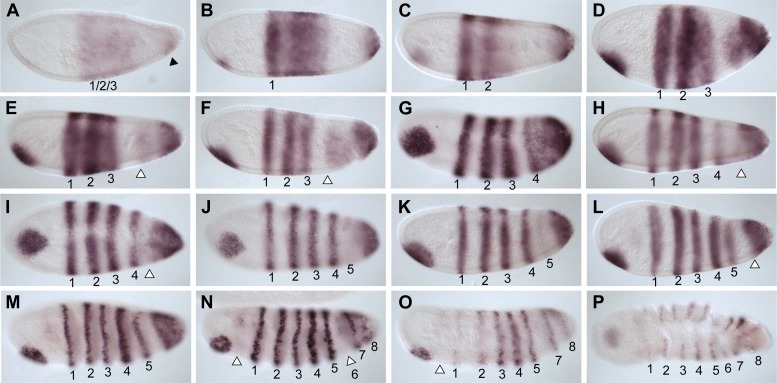
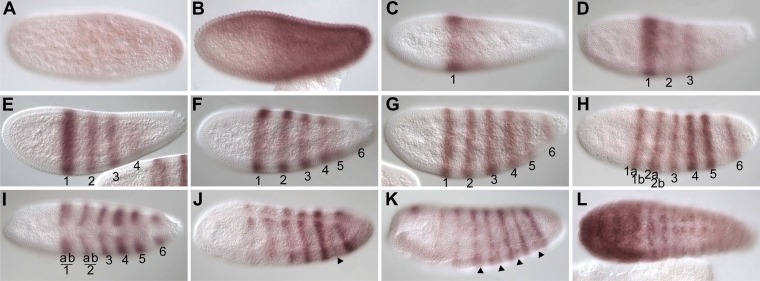
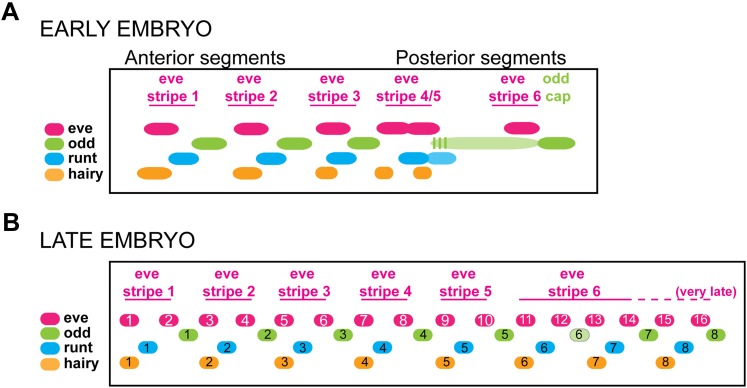
We used in situ hybridization to look at the expression pattern of Nv eve during Nasonia embryogenesis (Figure 1). Nv eve expression becomes detectable as a broad early domain in the blastoderm embryo at around 3 hr after egg laying (AEL) (Figure 1A). This domain broadens and its boundaries sharpen between 3 and 4 hr, by which time a faint posterior stripe (hereafter referred to as ‘stripe 6’, see below) becomes evident (Figure 1B,C). As embryogenesis progresses toward cellularization, the anterior domain splits into three distinct double-segment periodicity pair-rule stripes, stripes 1, 2 and 3 (Figure 1D–F). By cellularization, around 6 hr, a faint 4/5 stripe appears between the anterior stripes and stripe 6, which has become more intense (Figure 1F). Between 6 and 8 hr AEL, stripe 4/5 splits into distinct double-segment periodicity stripes 4 and 5, whereas stripes 1 and 2 split into single-segment periodicity segmental stripes (Figure 1G–I). In Drosophila, eve secondary stripes form de novo, between primary pair-rule stripes, in contrast to secondary paired stripes that later split from primary stripes, forming segmental stripes that affect all segments (Macdonald et al., 1986; Kilchherr et al., 1986). Splitting of Nasonia eve double-segment stripes into single-segment stripes may occur by a similar mechanism (see below). As gastrulation progresses between 8 and 10 hr AEL, double-segment pair-rule stripes 3–5 also split to give rise to two distinct single-segment stripes each (Figure 1J–L). This anterior to posterior progression of Nv eve stripes is consistent with the sequential appearance of the segment polarity genes Nv wg (Figure 1—figure supplement 1) and Nv en (Pultz et al., 1999), which are first detected around cellularization in a few anterior segments and then appear in stripes progressively, in an anterior to posterior manner.
Figure 1. Summary of Nasonia eve mRNA expression.
Embryos are shown with anterior left and dorsal up. Nv eve is initially expressed in a broad domain (A and B), which sharpens as a posterior stripe becomes visible at around 4 hr after embryo laying (AEL) (C and D). The broad domain retracts anteriorly and gives rise to three apparently double-segment stripes (E and F). Between stripes 3 and posterior stripe 6, an additional double stripe precursor comes up at around 6 hr AEL (stripe 4/5; panels F and G) and this splits to form two double-segment stripes, ‘4’ and ‘5’ as double-segment stripes 1–3 split into two single-segment stripes each between 6 and 8 hr AEL (F–J). Stripes 4 and 5 also split to form single-segment stripes during early gastrulation, and stripe 6 broadens (K and L), giving rise to stripes that are visibly distinct during germ band extension in non-fluorescent staining by 10–12 hr AEL (M–R, arrowheads). There are a total of 16 single-segment stripes of Nv eve.
Figure 1—figure supplement 1. Nv wingless (Nv wg) mRNA expression in the embryo.
A remarkable feature of Nv eve expression is that the posterior stripe 6 broadens significantly at about 10 hr AEL (Figure 1L–N), before generating six additional stripes with single-segment periodicity, allowing the embryo to reach 16 individual segmental stripes by the time the germ band is fully extended. This division of stripe 6 initiates with an anterior band that will give rise to four stripes (segmental stripes 11–14; Figure 1P,Q, arrowheads; see below), and two later-appearing segmental stripes 15 and 16 (Figure 1Q, arrowheads). The last stripe, Nv eve 16, appears only at full germ band extension (Figure 1R) completing the 16 stripes observed at this stage.
The Nasonia embryo has 16 segments, whereas Drosophila has only 14 (Figure 2A). In Drosophila, eve and other pair-rule genes are expressed with double-segment periodicity: seven transverse ‘pair-rule’ stripes are evident as a full complement in the blastoderm embryo at cellularization. If the Nasonia embryo were patterned using the same mechanisms as Drosophila, then eight pair-rule stripes would be predicted. However, only five truly pair-rule (double segment) stripes are apparent at cellularization, while stripe 6 gives rise later to four, then six single-segment stripes and six segments and is therefore not pair-rule (Figure 2E). This delayed sequential posterior segmentation is therefore more reminiscent of the segmentation described in short germ insects.
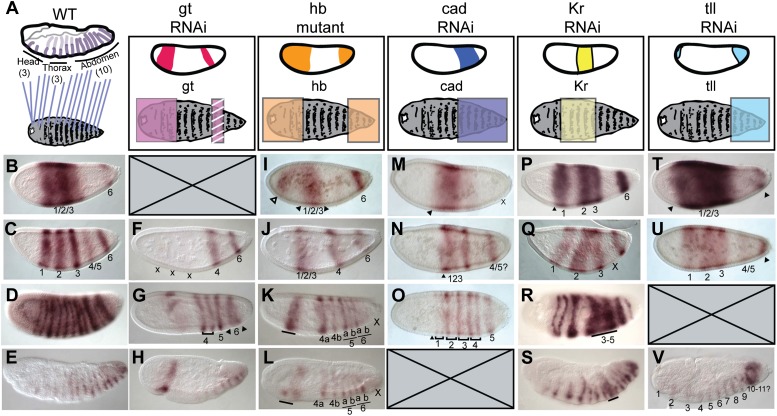
Figure 2. Nv eve epistasis with maternal and gap genes.
(A) Schematic representation of the germ-band-extended embryo, showing 16 single-segment stripes of Nv eve expression, and their segment counterparts in the patterned larval cuticle. Colored boxes cover the segments of the larval cuticle that are lost or fused in each RNAi background. All embryos are shown anterior left, dorsal up (except where indicated). Nv eve mRNA expression is shown in each embryo (B–G). Wild-type (WT) embryos are shown as staged controls for RNAi embryos. (B) WT early blastoderm embryo. (C) WT cellular blastoderm embryo. (D) WT early gastrula extension embryo. (E) WT germ-band-retracted embryo. (F–H) gt RNAi embryos stained for Nv eve mRNA expression. (F) Cellular blastoderm embryo with reduced Nv gt exhibits loss of anterior Nv eve stripes (x). (G) Nv gt RNAi embryo in early germ-band-extension exhibits loss of anterior Nv eve stripes and improper splitting of Nv eve stripe 5, as well as aberrant dorsal anterior expression of Nv eve. (H) Nv gt RNAi embryo at dorsal closure exhibits a stripe of Nv eve at the anterior, as well as a reduced number of posterior segmental Nv eve stripes. (I–L) Nv hb mutant embryos stained for Nv eve mRNA expression. (I) Early blastoderm Nv hb mutant embryos have a reduced central Nv eve domain (bounded by black arrowheads), and an ectopic anterior Nv eve stripe (white arrowhead). (J) Nv hb mutant cellular blastoderm embryo with a single anterior domain of Nv eve that has failed to resolve, and a single stripe 4 which exhibits delayed splitting. (K) Nv Hb mutant germ-band extension embryo with fused anterior domain (line) and 6 segmental stripes, representing derivatives of Nv eve stripes 4 and 5 and two derivatives of stripe 6; additional stripe 6 derivatives are absent (x). (L) hb mutant dorsal closure embryo exhibiting fused anterior domain (line) and the same number of derivatives as in (M), with more posterior segments missing (x). (M–O) Nv cad RNAi embryos stained for Nv eve mRNA expression. (M) Nv cad RNAi early blastoderm with reduced central Nv eve domain that is also posteriorly shifted (anterior boundary indicated by black arrowhead). (N) Nv cad RNAi cellular blastoderm embryo with posteriorly shifted (arrowhead), reduced Nv eve central domain, whose splitting is delayed. (O) Nv cad RNAi early gastrula embryo with posterior shift in Nv eve expression (black arrowhead). Four double-segment periodicity stripes are split into single-segment stripes and stripe 5 remains intact. (P–S) Nv Kr RNAi embryos stained for Nv eve mRNA expression. (P) Nv Kr RNAi precellular blastoderm embryo with aberrant Nv eve central domain resolution, where stripes 2–3 appear posteriorly shifted. (Q) Dorsolateral view of a Nv Kr RNAi embryo where stripes 2 and 3 are less refined than WT and 3 is posteriorly shifted. No stripe 4/5 expression is detected (X). (R) Nv Kr RNAi early gastrula embryo with aberrant stripe 2 splitting and aberrant resolution of stripes 3–5. (S) Moderately affected Nv Kr RNAi germ-band retraction embryo with fused segments in the middle of the embryo (line). (T–V) Nv tll RNAi embryos stained for Nv eve mRNA expression. (T) Nv tll RNAi early blastoderm embryo with expanded Nv eve expression domains toward both poles (arrowheads). (U) Nv tll RNAi precellular blastoderm embryo showing delayed resolution of Nv eve stripes 1–3 and Nv eve stripe 6 shifted to the extreme posterior pole of the embryo (arrowhead). (V) Nv tll RNAi dorsal closure embryo showing abnormal posterior Nv eve stripe formation.
Figure 2—figure supplement 1. Nv eve/Nv gt double FISH in the embryo.
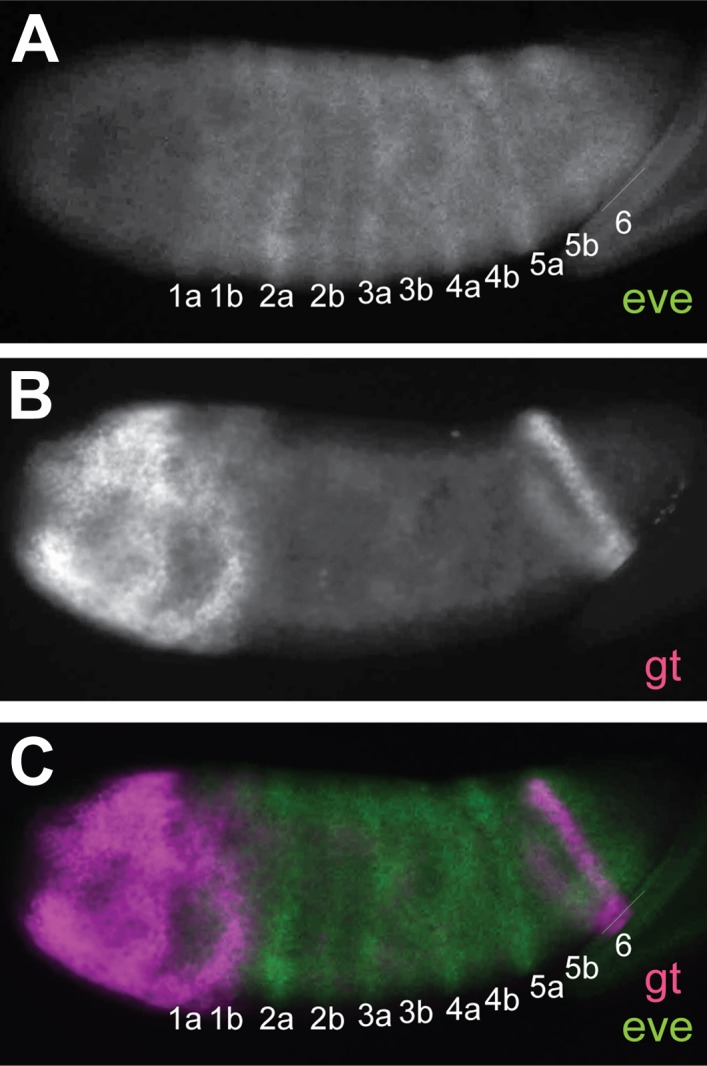
Figure 2—figure supplement 2.
Control of Nv eve by gap genes
To explore this apparent combination of short and long germ characters, we determined how Nv eve expression is controlled by upstream genes in the known Nasonia A–P patterning network. Early embryonic expression of Drosophila eve is controlled by maternal and gap genes, including bicoid (bcd), hb, Kr, gt, kni and torso (MJaJ & H, 1993; Small et al., 1992, 1996; Schroeder et al., 2004; Small and Levine, 1991), whereas later maintenance is achieved via autoregulation (Jiang et al., 1991). Some Tribolium eve pair-rule stripes are also under the control of gap genes although some of the segments themselves are born much later than gap gene expression (Sulston and Anderson, 1996; Cerny et al., 2005). For example, in Tc’Kr mutant embryos, segments anterior to the normal Kr expression domain (T1–T3) appear wild type, but expression of both Tc’eve and Tc’en is lost in posterior segments and no segments are formed posterior to A4 (Cerny et al., 2005). Therefore, Tribolium gap genes can affect the specification of segments that are not yet formed, presumably because of interactions with the growth zone. In other short germ insects, like Oncopeltus fasciatus, eve acts as a gap gene, regulating expression of hunchback and Krüppel (Liu and Kaufman, 2005a).
To determine how the known maternal and gap genes regulate Nv eve expression in the early embryo, we used parental RNAi injections in pupal Nasonia females to knockdown Nv gt, Nv Kr, Nv tll and Nv cad mRNA, as well as a null mutation in Nv hb (Pultz et al., 2000, 2005).
Nv gt
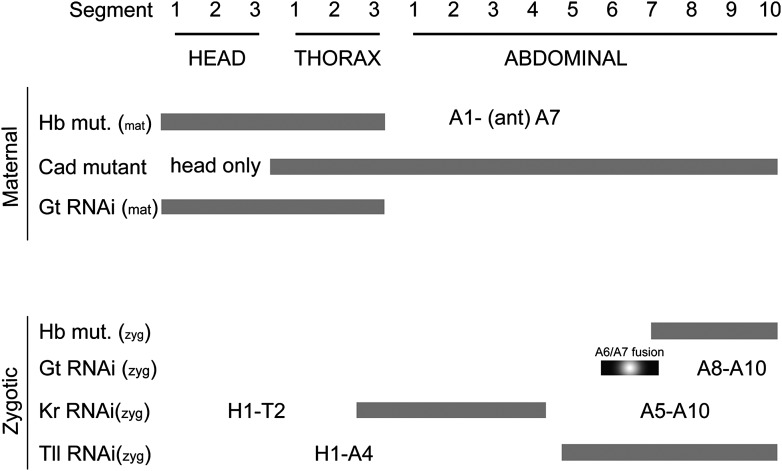
As we previously reported, Nv giant knockdown results in the loss of all segments anterior to A1 and fusion of segments A6 and A7 (Brent et al., 2007). Nv gt RNAi blastoderm embryos exhibit the loss of Nv eve double-segment stripes 1–3 (Figure 2F), as well as aberrant resolution of the first splitting events of stripe 6 (Figure 2G, arrowheads). Double in situ hybridization shows that, in the wild type, a late posterior stripe of Nv gt forms after Nv eve stripe 6 and appears to be within the Nv eve stripe 6 domain (Figure 2—figure supplement 1). These data suggest that a posterior Nv gt domain may partially affect stripe 6 splitting. Late Nv gt RNAi embryos exhibit strong anterior defects after dorsal closure, but the other posterior single-segment stripes of Nv eve appear unaffected (Figure 2H).
Nv hb
In Nv hb mutant (headless) embryos, both head and thoracic fates and abdominal fates posterior to A6 are lost (Pultz et al., 2005). Consistent with this phenotype, Nv eve double-segment stripes 1–3 (that give rise to head and thoracic fates) form but never resolve (Figure 2K, black arrowheads). An ectopic stripe of eve can be seen in the anterior of some embryos (Figure 2I, white arrowhead), consistent with the ectopic stripe of Nv cad (an activator of eve) in the head of some Nv hb mutant embryos (Olesnicky et al., 2006). Gastrula and later germ band embryos exhibit normal Nv eve double-segment stripe formation and splitting of stripes 4 and 5, which give rise to segments A1–A4. However, only the two anteriormost segments are formed from Nv eve stripe 6 (6a and 6b) in Nv hb mutant embryos (Figure 2K,L).
Nv caudal
Nv caudal (cad) is expressed maternally as a mRNA gradient with a localized posterior source (Olesnicky et al., 2006). Nv cad RNAi results in loss of all segments posterior to A1. Moderately affected Nv cad RNAi embryos exhibit a reduced early broad domain of Nv eve that is slightly shifted posteriorly (Figure 2M). This domain resolves poorly, with only weak activation of anterior Nv eve stripes and no posterior abdominal expression of Nv eve (Figure 2N–O).
Nv Kr
As in Drosophila, Nv Krüppel (Kr) is expressed in a central domain, and Nv Kr is required for formation of segments T3 to A4 (Brent et al., 2007). In Nv Kr RNAi embryos, both anterior and posterior domains of Nv hb expression expand towards the center of the embryo (Brent et al., 2007). Consistent with expansion of Nv hb, we observed that Nv eve stripe 2 and 3 exhibit aberrant resolution and Nv eve stripes 4 and 5 fail to resolve in embryos with knocked-down Nv Kr (Figure 2P–S). Posterior segments are unaffected, as reflected by normal expression of Nv eve posterior to stripe 5 (segment A4; Figure 2R,S). This phenotype is dramatically different from Tc’Kr knockdown where all posterior segments are deleted, likely because Nv Kr is expressed anterior to the growth zone while Tc’Kr abuts it.
Nv tll
tailless mRNA is expressed in both an anterior and a posterior domain, though only posterior segments are affected by Nv tll RNAi (Lynch et al., 2006). The most severely affected embryos are missing the six posterior abdominal segments. These embryos also exhibit an apparent slight anterior shift of the broad early domain of Nv eve expression and of stripe 6 (Figure 2T,U). Stripe 6 does not appear to resolve, resulting in an enduring ring of Nv eve expression and no Nv eve single-segment stripes posterior to this ring are apparent (Figure 2V).
Taken together, and consistent with previously described cuticular phenotypes for maternal and gap genes in Nasonia (Pultz et al., 2005; Lynch et al., 2006; Olesnicky et al., 2006; Brent et al., 2007; Figure 2—figure supplement 2), these data show that early Nv eve expression in blastoderm embryos involves regulatory interactions reminiscent of those underlying Drosophila long germ embryogenesis. However, since severe RNAi phenotypes of several genes, such as Nv cad and Nv tll results in total loss of posterior segments, these did not provide additional information for understanding the establishment of posterior Nv eve expression.
Nasonia embryos have mitotic domains but Nv eve posterior stripe resolution does not require localized cell division
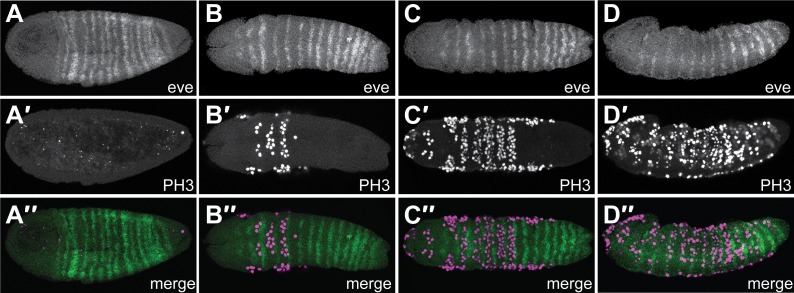
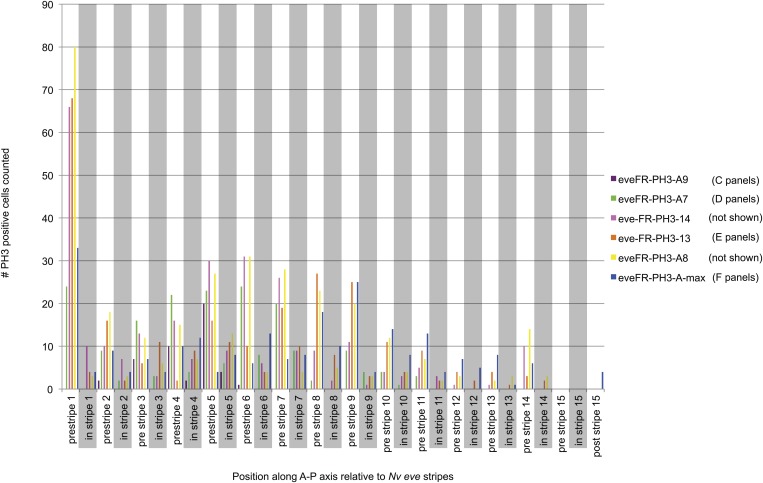
To determine whether cell division plays a role in subdivision of the Nv eve posterior domain into single-segment stripes, we used in situ hybridization to visualize Nv eve mRNA in embryos where mitotic cells were labeled with antibodies against phosphorylated histone H3. We found that there is no cell division that is consistent with a role in pair-rule stripe splitting (Figure 3A–A″), or in stripe 6 resolution (Figure 3B–B″), suggesting that the dynamics of the Nv eve mRNA pattern mostly involves transcriptional regulation. Nevertheless, later mitoses occur in restricted spatial domains, reminiscent of later Drosophila mitotic domains within segments of the expanding germ band (Figure 3B–D″). A relationship between gap gene function and regulation of mitotic domains via regulation of string has been suggested in Drosophila (Edgar et al., 1994) but not demonstrated. Nasonia mitotic domains appear in an anterior to posterior progression, allowing for progressive expansion of segments along the A–P axis via concerted cell divisions within domains. Strikingly, mitotic figures appear to be largely excluded from Nv eve stripes in these early stages of embryogenesis (Figure 3B″,C″, Figure 3—figure supplement 1). Later embryos in which anterior Nv eve stripes are beginning to fade exhibit overlap of mitotic figures with weakened Nv eve stripe expression (Figure 3D″). Ultimately, embryos exhibit widespread mitotic figures that do not correspond to any apparent concerted domains or pattern, more like the pattern of mitoses described in several short germ insects (Handel et al., 2005; Liu and Kaufman, 2009).
Figure 3. Nv eve expression and cell division appear to be coordinated.
Embryos co-stained for Nv eve mRNA using in situ hybridization and fluorescent detection, as well as for mitotic figures, using an antibody against phospho Histone H3. Embryos are shown with anterior left and dorsal up, except columns B and C, which are ventral views. (A–A″) An early gastrula embryo exhibiting 15 stripes of Nv eve, including five derivatives of stripe 6 (A), has no evident mitotic figures in the posterior domain of Nv eve stripe 6 differentiation (A′). (A″) Merge of panels A and A′. (B–D″) Timecourse series of wild-type embryos stained for Nv eve mRNA and phospho-Histone H3. (B–D). Top panels are Nv eve in situ alone, middle panels (B′–D′) are phospho-Histone H3 antibody staining, and bottom panels (B″–D″) are merge images of upper panels, showing localization of mitotic figures relative to Nv eve stripes.
Figure 3—figure supplement 1. Quantification of PH3 positive cells relative to Nv eve stripes in the embryo.
Knockdown of Nv eve results in gap and segment polarity defects and posterior truncation
The expression of Nv eve suggests a combination of long germ and short germ character. To further explore this possibility, we knocked down Nv eve gene function. Although parental RNAi in Nasonia is effective for maternal and early zygotic genes (Lynch and Desplan, 2006; Lynch et al., 2006; Olesnicky et al., 2006; Brent et al., 2007), it often does not provide significant knockdown of later-acting genes. To overcome this limitation, we designed an Nv eve morpholino overlapping the translation start site, as well as one directed at the exon–intron junction in the homeobox. These two independent morpholinos are expected to disrupt Nv eve activity and indeed result in comparable phenotypes.
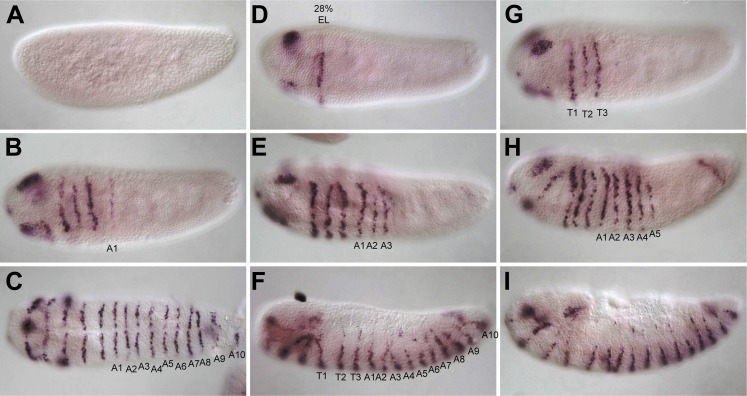
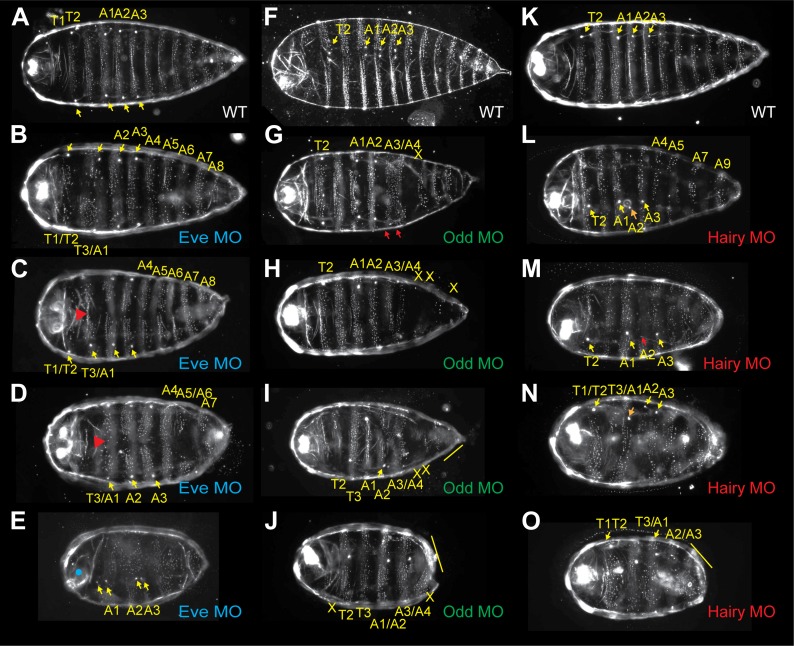
The Nasonia larval cuticle has relatively few landmarks to allow for interpretation of segmentation phenotypes. Beyond the denticle belts present on each of the three thoracic segments and ten abdominal segments, large spiracles are found on segments T2, A1, A2 and A3 (Figure 4A, yellow arrowheads). In the head, two bright structures indicate the positions of antennal papillae. Morpholino block of Nv eve causes a range of phenotypes (Figure 4B–E), resulting in severe truncation of the embryo with loss of posterior-derived segments as well as a partial pair-rule phenotype for more anterior segments. The phenotypic series includes progressive truncation at the posterior, causing fusion of segments A9–10 in the least affected cuticles, and then A8–10 (Figure 4C) with segment A6 eventually lost, whereas A7 remains virtually intact. In the most severely affected embryos, the entire posterior of the embryo is truncated with A5–A10 missing. (The approximate percentage of embryos in each phenotypic class shown in Figure 4 is indicated in the ‘Materials and methods’.)
Figure 4. Morpholino knockdown of Nv eve, Nv hairy, and Nv odd results in embryo patterning defects.
First instar larval cuticles are shown with anterior left and generally ventral denticle patterns are shown. (A, F, K) Wild-type larval cuticles. Yellow arrows indicate spiracles present on segments T2, A1, A2 and A3. Bright anterior labral appendages are apparent at the extreme anterior of the larva. (B–E) Unhatched larvae from Nv eve morpholino (MO)-injected embryos, in order of increasing phenotype severity. Red arrowheads indicate loss of midline cuticle. Blue dot indicates head open defect. Yellow arrowheads indicate position of spiracles. (G–J) Unhatched larvae from Nv odd morpholino (MO)-injected embryos, in order of increasing phenotype severity. Yellow arrows indicate position of spiracles, red arrows indicate A3/A4 fusion. X indicates naked cuticle from segment loss. Yellow line indicates multi-segment fusion. (L–O) Unhatched larvae from Nv hairy morpholino (MO)-injected embryos, in order of increasing phenotype severity. Yellow arrowheads indicate position of spiracles. Red or orange arrowheads indicate aberrantly positioned or missing spiracles. Yellow line indicates segment fusion.
At the anterior, T1 is lost with fusion of T3 and A1 (Figure 4B). Segments A2 and A3 are also fused, and there is a continuous lawn of denticles from A4 to the truncated posterior. This is accompanied by a disruption of the remaining denticle belts, leaving naked cuticle along the midline (Figure 4, red arrows). In the most severely affected embryos, segments anterior to A1 are lost and are accompanied by head closure defects. This phenotype represents a partial pair-rule phenotype, accompanied by posterior truncation of the embryo. It also does not exhibit the lawn of denticles phenotype of strong eve alleles in flies (e.g., eveR13 [Macdonald et al., 1986; Fujioka et al., 1999]), although severely affected Nasonia embryos also exhibit cuticle defects beyond a pair-rule phenotype. Hence, these results support a mixed mode of embryogenesis in Nasonia, with characteristic features resembling both long germ and short germ insects. To further examine this possibility, we then investigated the expression patterns in Nasonia of other genes acting as pair-rule in Drosophila, and undertook functional characterization of their activity during embryonic development.
Nasonia odd-skipped expression and function
In the long germ Drosophila embryo, odd is expressed with a double-segment periodicity complementary to that of eve, and its inactivation causes the absence of odd segments (Nusslein-Volhard and Wieschaus, 1980; Coulter et al., 1990). Its critical function as a mediator of the segmentation clock in the short germ beetle was recently elegantly described (Sarrazin et al., 2012). Tc’odd begins with blastoderm expression in double-segment periodicity stripes alternating with Tc’eve expression. Then, new double-segment stripes emanate from the growth zone to generate the entire complement of odd stripes. Secondary single-segment stripes arise later (Sarrazin et al., 2012).
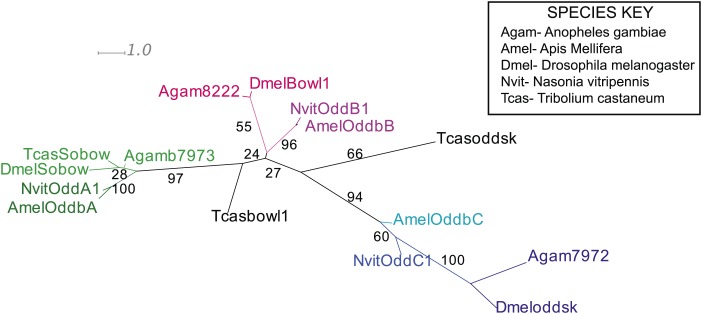
There are three odd paralogs in Nasonia, as in flies, where they are named odd, bowl, and sister of bowl (or sob; (Hart et al., 1996)). We used sequence alignment (Figure 5—figure supplement 2) and phylogenetic analysis (Figure 5—figure supplement 1) to identify the Nasonia paralog that is closest to Drosophila odd-skipped, and refer to it hereafter as Nv odd. An Nv odd cDNA fragment comprising the region encoding the conserved DNA binding domain was used as a probe for in situ hybridization (GenBank Accession # KC142194). As observed above for Nv eve, the embryonic expression of Nv odd begins as a broad early domain in syncytial blasoderm embryos (Figure 5A). As this broad domain strengthens and sharpens, a ventral head patch and a posterior cap appear (Figure 5B,C). The broad domain resolves into two clear apparent double-segment stripes (Stripes 1 and 2, Figure 5D). A third double-segment stripe arises from the second stripe, expanding posteriorly (Figure 5D–F). At the same time, a stronger posterior domain apparently advances anteriorly. Pair-rule stripe 4 (double-segment periodicity) arises at the anterior of the first advancing ‘wave’ at cellularization (Figure 5G–H) before the posterior domain recedes again (Figure 5I–J). The fifth double-segment stripe arises during a second ‘wave’ (Figure 5K–M) at the onset of gastrulation. A sixth stripe arises in an apparently similar manner, though it is much fainter and appears while more posterior stripes are already differentiated (Figure 5N, arrowhead); the posterior cap generates two thin pair-rule stripes (Figure 5O) during early germ band extension. At full germ band extension, a total of eight stripes are visible (Figure 5P) before these fade from anterior to posterior. This dynamic expression of Nv odd in the posterior of the embryo is reminiscent of the waves of growth zone expression of Tribolium odd, where blastoderm-derived stripes initially have double-segment periodicity and later single-segment periodicity (Choe et al., 2006; Sarrazin et al., 2012).
Figure 5. Summary of Nv odd-skipped mRNA expression.
Embryos are shown with anterior left and dorsal up, except where indicated. (A) Precellular blastoderm embryo showing early expression of Nv odd in a broad domain and a posterior cap with a slight clearing in between. (B) Precellular blastoderm embryo showing ventral head patch and darkened central broad domain and distinct posterior cap. (C) Precellular blastoderm embryo with sharpening pair-rule stripes and expanding posterior cap. (D) Precellular blastoderm embryo with dark ventral head patch and posterior cap, and expansion of expression between broad central domain and posterior domain. (E and F) Cellularizing blastoderm embryos with three double-segment periodicity stripes, and a continuous posterior domain of variable staining intensity. Arrowhead indicates boundary of faint expression, which prefigures position of double-segment stripe 4. (G) Ventral view of cellularizing embryo with three strong double-segment stripes, and a fourth stripe forming at the anterior boundary of a more uniformly staining posterior cap (arrowhead). (H) Cellularized blastoderm embryo with four distinct double-segment stripes and a receding posterior cap domain (arrowhead). (I) Ventral view of cellular blastoderm showing four strong double-segment stripes and receding posterior cap (arrowhead), whose anterior boundary prefigures the position of stripe 5. (J) Ventrolateral view of cellular blasoderm embryo showing early appearance of stripe 5 at the anterior boundary of receding posterior domain, whose staining intensity is now less uniform. (K) Cellular blastoderm embryo with five double-segment stripes of expression, a strong ventral head spot, and a reduced, uniform posterior cap. (L) Same as K, with five equivalently strong double-segment stripes. Arrowhead indicates slightly expanded posterior cap. (M) Early germ-band extension embryo with five double-segment periodicity stripes and two stripes becoming evident within the posterior cap. (N) Slightly later embryo than M, with 2 posterior cap stripes more clearly differentiated. (O) Slightly later embryo than N, with anterior stripes fading and posterior segments expanding. (P) Dorsal view, dorsal closure embryo exhibiting eight single-segment periodicity stripes.
Figure 5—figure supplement 1. Phylogenetic analysis of odd-skipped.
Figure 5—figure supplement 2. odd-skipped protein sequence alignment.
Using double fluorescent in situ hybridization, we confirmed that the pair-rule stripes of Nv odd and Nv eve are indeed complementary to each other, although their mode of appearance is totally different. Nv odd double-segment stripes are posterior to, and abut each posterior single-segment stripe (i.e., 1b, 2b) from each eve pair-rule doublet (Figure 6A–C), that is, the even-numbered segmental stripes. Late forming Nv eve stripe 15/16 intercalates between the two Nv odd stripes 7 and 8 that derive from the cap, with Nv odd stripe 8 remaining posterior to all Nv eve stripes (excepting the last stripe, eve 16, which is the last to appear), a relationship that may have ancestral origins (see ‘Discussion’).
Figure 6. Phasing of Nasonia pair-rule genes in embryos using double fluorescent in situ hybridization.
(A) Lateral view of Nv eve expression in early gastrula embryo. (B) Nv odd expression alone in the same embryo. (C) Merge of Nv eve and Nv odd channels, illustrating their relative phasing. Nv eve mRNA is pseudo-colored pink, Nv odd is in green. Arrowheads indicate position of a posterior doublet of odd stripes. (D) Dorsolateral view of Nv eve in later gastrula embryo. (E) Nv odd expression alone in the same embryo. Arrowheads indicate position of posterior Nv odd stripes 6, 7 and 8. (F) Merge of Nv eve and Nv odd channels, illustrating their relative phasing. (G) Lateral view of Nv eve expression in blastoderm embryo. Arrowhead indicates position of Nv eve stripe 5. (H) Nv runt expression in the same blastoderm embryo. (I) Merge of Nv eve (green) and Nv runt (pink) channels, indicating relative phasing. (J) Lateral view of Nv eve expression in germ-band-extended embryo. Numbers indicate identity of Nv eve stripe. (K) Nv runt expression alone in the same embryo. Arrowheads indicate position of posterior primary Nv runt stripes. (L) Merge of Nv eve (green) and Nv runt (pink) channels, indicating relative phasing. Note that posterior Nv runt stripes, though faint, appear to be positioned posterior to odd-numbered Nv eve segmental stripes. (M) Lateral view of Nv eve expression in early gastrula embryo. Line indicates broadening stripe 6. (N) Nv hairy expression in the same gastrula embryo. Arrowheads indicate positions of three late forming posterior double-segment stripes. (O) Merge of Nv eve (pink) and Nv hairy (green) channels, indicating relative phasing. (P) Ventral view of gastrula embryo showing Nv eve expression alone. Arrowheads indicate positions of single-segment stripes derived from eve stripe 6. (Q) Nv hairy expression alone in the same gastrula embryo. Line indicates extended anterior domain continuous with stripe 1. (R) Merge of Nv eve (green) and Nv hairy (pink) channels, illustrating relative phasing.
Figure 6—figure supplement 1. Summary of Nv runt mRNA expression.
To examine the function of Nv odd in the embryo, we used one translation blocking and one splice blocking morpholino to knock down its expression in embryos. Inactivating Nv odd function leads to loss of the most posterior germ band-derived segments A5–A10 with additional anterior defects. The most sensitive phenotypes are the fusion of segments A3 and A4, and loss of segment A5 (Figure 4G, red arrowheads and x). More severely affected embryos exhibit loss of most segments posterior to A3/A4 and naked cuticle anterior to T2, though larval head structures are still present (Figure 4I–J). In many severely affected embryos, Nv odd knockdown causes additional loss/fusion of A1, and of T2. Thus, phenotypes comprise loss of segments T2, A1, A3 and A5 and resemble a pair-rule phenotype. A small percentage of embryos are nearly asegmental, with only small patches of denticle bands of unknown identity (not shown). Therefore, as also observed for Nv eve knockdown, the segmentation defects resulting from Nv odd inactivation only corresponds to a partial Drosophila-like pair-rule phenotype, and rather, in the most severe cases, resemble the phenotype of Tc’odd loss of function. It is of note that Nv odd stripes 4, 5 and 6 appear to emerge from waves of expression at the posterior of the embryo that likely specify segments A3–A5, which are most sensitive to loss of Nv odd function (Figure 5E–L, Figure 4G).
In summary, Nv odd is expressed initially in three sequentially forming anterior double-segment periodicity stripes, which appear to have Drosophila-like pair-rule character. Three more posterior double-segment stripes (PR stripes 4–6) then form sequentially, apparently as ‘waves’ of Nv odd expression, resembling the clock-driven stripes of Tc’odd. Finally, two Nv odd stripes form from a posterior cap. Nv odd knockdown affects anterior thoracic segments with a partial pair-rule phenotype; it also leads to the loss of posterior segments A5–A10.
Nasonia runt expression
The complementarity between Nv eve and Nv odd is suggestive of cross interaction between the two genes but it is only partial and only affects half of the Nv eve segmental stripes since Nv odd does not have single-segment periodicity stripes. We sought to determine whether the remaining single segment stripes where odd is not interdigitated with Nv eve may alternate with stripes of Nv runt, as is observed in Drosophila. We studied the expression of Nv runt throughout embryogenesis (Figure 6—figure supplement 1) and then used double fluorescent in situ hybridization to visualize its register with Nv eve stripes at both early and late stages. Nv runt stripes appear cleanly in an anterior to posterior progression, with six double-segment periodicity stripes visible before cellularization; two additional double-segment stripes are added at the posterior during gastrulation. Single-segment periodicity stripes only appear much later at full germ band extension when the expression of the other pair-rule genes is already well established (Figure 6—figure supplement 1). In the early embryo, Nv runt double-segment stripes appear posterior to, and partly overlapping with, each Nv eve primary double-segment stripe (Figure 6G–I). Splitting of anterior Nv eve stripes moves the posterior of each doublet (i.e., even-numbered Nv eve single-segment stripes) more posteriorly beyond each Nv runt primary double-segment stripe (Figure 6G–I). The appearance of Nv eve stripe 5 between Nv runt double-segment stripes 4 and 5 as they split (Figure 6G–I, arrowhead; Figure 6—figure supplement 1F,G) suggests that eve may help to repress Nv runt, though this remains to be tested.
Late expression of Nv runt in the extending germ band is considerably weaker than that of other genes, making its detection more challenging. Still, at the posterior of the embryo, Nv runt double-segment stripes appear between Nv eve single-segment stripes arising from splitting of double-segment stripes (Figure 6J–L). Altogether, our data support a model in which Nv eve single-segment periodicity stripes are established through the complementary action of Nv odd for odd numbered single-segment stripes and Nv runt for even-numbered stripes, as summarized in our model below. However, these interactions are still speculative since we have been technically unable to complete the epistasis experiments needed to test this model. A predicted role for Nv h is also described below.
Expression and function of Nasonia hairy
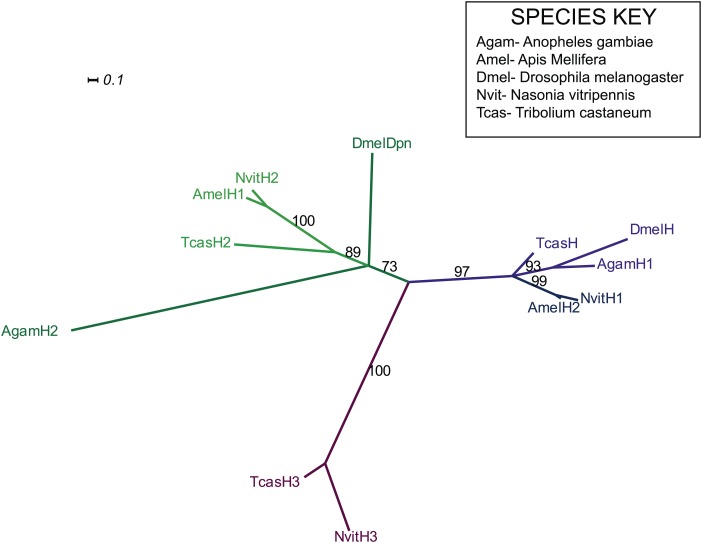
hairy is a primary pair-rule gene in Drosophila, but in Tribolium, its function is restricted to head segment differentiation (Carroll et al., 1988; Carroll and Vavra, 1989; Edgar et al., 1989; Choe et al., 2006). There are two hairy-like genes in Nasonia, and we identified the likely hairy (h) ortholog using phylogenetic analysis (Figure 7—figure supplements 1 and 2). We examined expression of Nv h using a probe directed against the full-length coding region (Genbank Accession # KC190514).
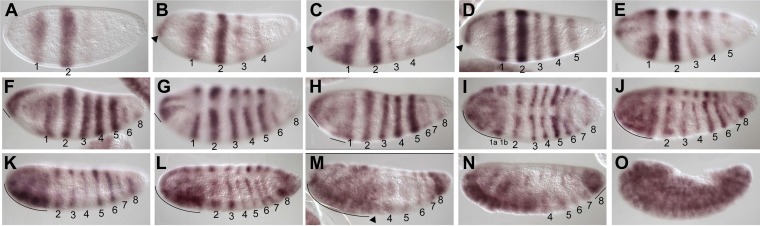
Nv h expression begins as a single broad anterior double segment stripe 1 that incompletely spans the dorso–ventral axis. It is soon followed by a second broad double-segment stripe 2 just anterior to the middle of the embryo (Figure 7A). Double-segment stripes 3, 4 and 5 are then added sequentially before cellularization (Figure 7B–E); a faint stripe at the extreme posterior of the embryo is also visible. By gastrulation, an anterior cap becomes more visible with continuous low expression between the anterior pole and the strong stripe 1 (Figure 7H–J). As gastrulation progresses, this domain becomes stronger and more uniform, whereas double-segment stripes 6 and 7 appear sequentially (Figure 7H–K). Stripe 8 broadens, becoming a posterior cap whose intensity increases during germ band extension (Figure 7J–N). The anterior of the embryo exhibits diffuse staining that expands from the anterior ventral side posteriorly, until germ band retracted embryos are faintly but uniformly stained with dark segmental stripes on top (Figure 7O). Nv h double-segment stripes appear cleanly, and the timing and presentation of expression of posterior stripes suggest that they may respond to waves of Nv odd. Like Nv odd and early Nv runt, Nv h does not have stripes with single-segment periodicity.
Figure 7. Summary of Nv hairy mRNA expression.
(A) Blastoderm embryo with two double-segment periodicity stripes of Nv hairy expression. Note that stripe 2 is broader and stronger than stripe 1. (B) Blastoderm embryo showing four double-segment periodicity stripes of expression plus an anterior accumulation of Nv hairy transcripts (arrowhead). (C) Dorsal view of embryo as in (B), illustrating the dorsal anterior expression (arrowhead) that is activated in the same pattern as the anterior domain of Nv tailless (Lynch et al., 2006). (D) Blastoderm embryo with strong anterior and dorsal anterior expression of Nv hairy and five pair-rule stripes. (E) Dorsal view of embryo as in (D) with increased dorsal anterior expression of Nv hairy, and the anterior spreading of expression from the anterior of double-segment pair-rule stripe 1. (F) Blastoderm embryo with expanding anterior domain (line), five double-segment ‘pair-rule’ stripes, and two additional stripes coming up. Note that the anterior domain between stripe 1 and the anterior pole is becoming more continuous in expression. (G) Dorsolateral view of embryo as in (F) highlighting the dorsal anterior expression. Stripe 2 is still wider than other stripes. Stripe 6 appears to be of single-segment periodicity. (H) Early gastrula embryo exhibiting a non-homogenous but largely continuous anterior cap of Nv hairy expression (that includes stripe 1). Four additional double-segment stripes and three single-segment stripes (two derived from stripe 6) are now evident. (I) Dorsal view of embryo slightly older than embryo in (H) showing the nearly continuous head domain, and the apparent splitting of stripe 1 within that domain. Double-segment stripes are thinning. (J) Dorsolateral view of extending germ-band embryo. Head domain is continuous (line). Stripes 1–7 have single-segment periodicity, are of non-uniform strength; stripe eight appears darker and broader. (K) Germ-band extending embryo with a continuous head domain (line) and eight discrete stripes. (L) Dorsolateral view of germ-band extending embryo. Stripe 8 is expanded into a wedge abutting the pole cells, and the anterior domain is expanding to include stripe 2. (M) Germ-band extension embryo with expanding anterior domain, that extends to include stripe 3 (arrowhead). Posterior domain is expanded. (N) Dorsolateral view of embryo as in (M) showing further expansion of posterior stripe 8 domain (line). (O) Germ-band-retracted embryo exhibiting ubiquitous staining with striated expression evident.
Figure 7—figure supplement 1. Phylogenetic analysis of hairy.
Figure 7—figure supplement 2. Hairy protein sequence alignment.
Double fluorescent in situ hybridization with Nv eve reveals that early Nv h overlaps the anterior of Nv eve double-segment stripes (Figure 6M–O). Later, these stripes appear thinner, and they overlap the anterior Nv eve single-segment odd-numbered stripes in each double-segment doublet. Nv h double-segment stripe 6 anticipates the position of the most anterior derivative of the Nv eve stripe 6 quartet (segmental stripes 11–14). Nv h stripe 7 (double segment), which is thin, appears within the Nv eve early broad stripe 6 domain, coinciding with Nv eve single-segment stripe 13 (Figure 6P–R, Figure 8). A more posterior stripe, Nv h 8, anticipates, albeit more broadly, the site of Nv eve single-segment stripe 15 (Figure 6M–O,P–R, Figure 8). Thus, Nv h and Nv eve are co-expressed at the anterior of eve pair-rule stripes and in the first of each pair of eve (odd-numbered) single-segment stripes, similar to the relationship described for Drosophila eve and hairy as they initiate segment polarity (Warrior and Levine, 1990).
Figure 8. Summary model of pair-rule gene expression in the Nasonia embryo.
(A) Model of register of pair-rule gene expression in the early embryo. Nv eve and Nv odd stripes are totally complementary, whereas Nv runt stripes partly overlap each of these genes at their interface. Nv hairy stripes overlap Nv eve stripes toward the anterior of each double-segment periodicity stripe. Towards the posterior of the embryo, an extended domain of low-level Nv odd expression exhibits dynamic behavior over several nuclear cycles, and stripes 4/5 of Nv eve, Nv odd, and Nv runt each differentiate during this interval. Even more posteriorly, Nv eve stripe 6 lay anterior to a continuous Nv odd cap that extends to the posterior pole of the embryo. This region is set aside for segment specification and differentiation during germ-band extension. (B) Model of register of pair-rule gene expression in the germ-band extension (late) embryo. Single-segment periodicity stripes in the germ-band-extended embryo exhibit a variation upon early gene expression patterns. Nv eve single-segment stripes are interrupted by Nv runt and then Nv odd such that Nv runt stripes follow odd-numbered Nv eve stripes, and Nv odd stripes follow even-numbered Nv eve stripes. Each of 8 Nv hairy stripes overlaps odd-numbered Nv eve stripes that derive from the anterior of Nv eve pair-rule stripes. Additional expression of several of these genes in the ventral and head domains, which appears to rely on different regulatory logic, is not shown.
We knocked down Nv hairy function using two independent morpholinos directed at two different splice junctions. Both resulted in a range of cuticle defects that indicate that Nv h is required for the formation of all posterior-derived segments and blastoderm-derived segments in the thorax and anterior abdomen (Figure 4L–O). At the posterior, mildly affected cuticles exhibit fusion of segments A9–10 (Figure 4L,N), along with partial loss of alternating abdominal segments posterior to A4. In more affected cuticles, alternating segments posterior to A2 are fused (Figure 4L–N), resembling a pair-rule phenotype. In severely affected embryos, all segments from A4–A10 are fused with a continuous lawn of denticles that covers the posterior of a severely reduced cuticle (Figure 4O, yellow line). These phenotypes suggest a requirement for Nv hairy in specification of posterior segments. The late Nv h stripes 6–8 are positioned to affect the late forming segments as supported by double in situs (Figure 6). In spite of Nv hairy expression in the head and extreme anterior of the embryo, labral structures in Nv hairy morpholino cuticles appear to be unaffected. The expression pattern of Nv hairy is thus strikingly similar to Tc’hairy (Sommer and Tautz, 1993; Aranda et al., 2008), though functionally quite different, since Tc’h seems to act exclusively in patterning head segments (Choe et al., 2006; Aranda et al., 2008).
At the anterior, A2 is also nearly always affected, exhibiting loss of denticles and displacement or loss of the associated spiracle (Figure 4L, orange arrowhead). Segment T1 appears to be lost and fused to T2. Finally, more affected embryos show a loss of T3 (Figure 4M–O). Therefore, segments T1, T3, and A2 are missing, which resembles an anterior pair-rule phenotype.
In summary, Nv h expression is highly dynamic and proceeds in an anterior to posterior progression. It is distinct from Nv eve and does not exhibit single-segment periodicity stripes. At the end of embryogenesis, its expression becomes nearly ubiquitous (Figure 7O).
Taken together, these data support a model wherein ‘pair-rule’ genes have weak fly-type ‘pair-rule’ phenotypes in the anterior, and are required for the formation of a suite of posterior segments. Their interdigitated expression suggests extensive interactions during patterning of the posterior region after cellularization, although this has not yet been tested due to current experimental limitations. Our summary model of the phasing of ‘pair-rule’ stripes in the embryo is given in Figure 8.
Discussion
Decades of study of a variety of insects have yielded a deep understanding of the genes controlling anterior–posterior patterning of the embryo. The best-characterized model species are the extreme long germ Drosophila embryo and the short-germ Tribolium embryo. Additional insects, such as Gryllus bimaculatus (Mito et al., 2007), Bombyx mori (Xu et al., 1997), Oncopeltus fasciatus (Liu and Kaufman, 2004a, 2004b, 2005a), Schistocerca americana (Patel et al., 1992) and others, represent informative intermediates, but many of these species are on the short end of this wide spectrum, consistent with ancestral insects being short germ band. Hymenoptera, including Apis and Nasonia, have evolved a long germ embryogenesis independently from flies and therefore provide an excellent context for addressing the question of how the transition from short to long germ occurred. Other well-characterized arthropod species, such as Cupiennius salei (Damen et al., 2000; Stollewerk et al., 2003) and Strigamia maritima (Chipman et al., 2004; Chipman and Akam, 2008), provide additional models of interest for understanding more ancient evolutionary history.
Nasonia has been characterized as long germ because of the presence of two morphogenetic centers and the expression patterns and RNAi phenotypes of the gap genes (Ingham et al., 1985; Pultz et al., 2005; Lynch et al., 2006; Olesnicky et al., 2006; Brent et al., 2007). In this study, we describe the expression and loss of function phenotypes of Nasonia eve, hairy and odd, three genes that act as pair-rule genes in Drosophila. While the expression of Nv eve is controlled by maternal and gap gene factors that are largely similar to their Drosophila counterparts, we find critical deviations from the Drosophila long germ paradigm. Nv ‘pair-rule’ genes display similarity to both Drosophila and Tribolium, suggesting that Nasonia has features of both short and long germ band development. Unlike the long germ embryo of the honeybee, Apis, Nasonia pair-rule genes do not seem to be maternally expressed, and so far, regulation of Nv gap genes by pair-rule genes (as reported for Apis [Wilson and Dearden, 2012]) has yet to be studied systematically.
Expression and knockdown of Nasonia eve, hairy and odd
In contrast to Drosophila, whose pair-rule genes are expressed in the blastoderm in seven double-segment periodicity stripes to determine the formation of 14 segments, their orthologs in Nasonia are expressed in diverse and more intricate patterns. In no case do we observe simply eight precellularization double-segment stripes, confirming that pair ‘rule’ does not represent the regulatory dynamics of these genes across insects. We observe wave-like behavior of Nv odd stripe 4–6, which underscores that cycling control may remain from the ancestral segmentation clock. Nv runt and, to a large degree, Nv h, also exhibit a sequential progression of sharp stripe appearance that may be responsive to the waves of Nv odd (Figure 7, Figure 6—figure supplement 1).
It is clear that Nv eve, Nv odd, and Nv h exhibit multiple modes of regulation during embryogenesis. In each case, their anterior stripes formed in the syncytial blastoderm have double-segment periodicity and arise in a manner that could be explained by the type of enhancer logic exemplified by Drosophila eve (Small and Levine, 1991; Small et al., 1992, 1996; Schroeder et al., 2004; Schroeder et al., 2011). Anterior double-segment ‘pair-rule’ stripes of Nv eve appear to be regulated by maternal and gap genes as in Drosophila, and embryos knocked down for Nv eve, Nv odd and Nv h exhibit a pair-rule phenotype in the blastoderm-derived segments, although this phenotype is most often limited. It is worth noting that the severe Nv eve anterior defects are more regional, as observed for Oncopeltus eve that behaves as a gap gene, and is attributed to the broad early domain of expression (Liu and Kaufman, 2005a). Perhaps, as in Oncopeltus, Nv eve is required in combination with Nv hb or Nv gt for activation of their targets, which are in turn required for the proper formation of head and thoracic segments.
Another mode must control the formation of stripes of Nv eve, Nv odd, and Nv h that arise later, in a cellular environment, from a posterior domain (whether at the posterior pole of the embryo, as in the case of Nv odd and Nv hairy, or from a broad posterior stripe, as in the case of Nv eve). Knockdown of each of the three genes produces a severe posterior truncation of the embryo, deleting all six posterior segments, indicating that each gene is required for the formation of posterior-derived segments. This phenotype is unlike flies, and resembles more the short germ pair-rule gene circuit of Tribolium (Choe et al., 2006; Choe and Brown, 2009; Sarrazin et al., 2012). Together with co-expression data, their phenotypes suggest that each of these genes is required for refinement or maintenance of each other’s activity or expression (Figure 6, Figure 6—figure supplement 1).
We propose a model in which interactions among ‘pair-rule’ genes dominate in patterning the long germ Nasonia embryo. Unlike flies, posterior stripes of ‘pair-rule’ genes like Nv hairy and Nv runt appear sequentially. Indeed, the gene circuit involving interactions among Tc’odd, Tc’eve and Tc’runt in each round of posterior segment formation underscores the likely ancestral nature of this network, which might have been brought under the control of the gap and maternal genes in flies, and in the anterior segments of Nasonia. The ‘waves’ of Nv odd pair-rule stripe expression that give rise to blastoderm stripes 4, 5, and 6 suggest residual activity of a segmentation clock. The presumptive domain of six posterior segments indicated by early posterior expression of Nv eve and Nv odd may be similar to the ‘growth zone’ of short germ insects.
Waves of Nv odd 4, 5, and 6, and the sequential formation of Nv runt stripes both interrupt and likely pattern the continuous Nv eve stripe 6 domain (Figure 6A–L; Figure 8). Nv h expression anticipates the final position of several late forming Nv eve stripes, and in combination with the phenotype of Nv h knockdown and co-expression data, suggests that it is required for the formation of the posterior Nv eve stripes.
Thus, Nasonia represents a variation on embryo allocation and patterning, but the contribution of ‘pair-rule’ gene function is enduring. The use of a clock-like mechanism is not incompatible with long germ embryogenesis, and rather, retaining this character might allow for sampling transitional states between the short- and long germ strategies, which likely occurred multiple times within holometabola. Further, it may be that the absence of significant posterior elongation is the transition state that tips the balance toward elaboration of anterior segmentation control mechanisms and loss of late forming segments. Our characterization of the Nasonia pair-rule genes illustrates one way that these two strategies can co-exist.
It is also of note that although hairy-related genes are the oscillating components of vertebrate segmentation clocks (Palmeirim et al., 1997), it is generally odd-skipped-related genes that oscillate in arthropods (Chipman et al., 2004; Chipman and Akam, 2008; El-Sherif et al., 2012; Sarrazin et al., 2012). Notch-signaling has been described for its involvement in regulating hairy-related oscillations in the vertebrate clock (e.g., Jouve et al., 2000), and it may also be involved in the context of the arthropod segmentation clock (Stollewerk et al., 2003; Eriksson et al., 2013; Kainz et al., 2011); in all cases, the driver of the clock is yet to be elucidated (reviewed in Pourquie, 2003).
An ancestral role for eve in specifying posterior identity may be linked to growth zone behavior in short germ insects
eve has been suggested to have its most ancestral function as a specifier of posteriorness (Ahringer, 1996; Brown et al., 1997). Indeed, the two mammalian eve genes are located at the most ‘posterior’ end of two of the Hox clusters (Bastian et al., 1992), although eve is not a part of the Hox cluster in Nasonia or Tribolium or any other insects that have been studied (Shippy et al., 2008; Werren et al., 2010; Munoz-Torres, 2009; Suen et al., 2011). Yet, even in Drosophila, where there is no apparent sequential segmentation, a delayed pair-rule stripe (stripe 8) appears early in gastrulation (Macdonald et al., 1986; Frasch et al., 1987; Kim et al., 2000). In Schistocerca, eve is expressed in a posterior mesodermal domain and no pair-rule stripes arise from this region, indicating that eve plays a role in posteriorness and not segmentation in basal insects (Patel et al., 1992). Nasonia eve sets aside stripe 6 relatively early, at about the same time as Nv odd that is expressed even more posteriorly. This feature of Nv eve in posterior segmentation is not shared by the other pair-rule genes we studied, therefore supporting the notion of an ancestral role for eve in posteriorness in Nasonia. That its expression is complementary to that of odd in both Tribolium and Nasonia in a late-differentiating posterior region may hint at how this role in posteriorness evolved into a role in posterior growth. In non-insect arthropods, there is evidence for a role for eve in both posterior identity and segmentation. The centipede Lithobius atkinsoni expresses eve in a posterior domain and between segments (Hughes and Kaufman, 2002), and the crustacean Artemia franciscana exhibits growth zone eve expression that precedes expression in stripes in emerging segments (Copf et al., 2003). In other basal arthropods, like spiders (Damen et al., 2000) and the centipede Strigamia maritima (Chipman and Akam, 2008), eve expression in stripes suggests that its ancestral role is segmental.
Mitotic domains are coordinated with pair-rule gene expression
The broad stripe 6 domain of eve appears to be subdivided by transcription control, likely through interactions with Nv odd and Nv runt (Figure 6, Figure 6—figure supplement 1). Although we observed apparent mitotic domains in the early gastrula that proceed from anterior to posterior, they do not match the pattern of initial differentiation of germ band-derived segments or the splitting of anterior pair-rule stripes. Cell division patterns in mitotic domains have not been described in most insects, apart from Drosophila and the precellular blastoderm of Bombyx (order: Lepidoptera; Nagy et al., 1994). In the short germ Tribolium and Oncopeltus embryos, cell divisions during gastrulation and elongation occur throughout the germ band, with no evidence for mitotic domains (Brown et al., 1994; Liu and Kaufman, 2009).
The relationship between Nv eve and cell division suggests coordination of cell divisions by segmentation genes, a phenomenon that has been suggested for Drosophila (Foe, 1989; Edgar and O’Farrell, 1989; Bianchi-Frias et al., 2004). Use of coordinated mitotic domains is a strategy that seems to have evolved multiple times (e.g., in flies and wasps). We propose that the apparent coordination of mitotic domains and segmentation gene expression of both Nasonia and Drosophila development may constitute a step in the transition to long germ embryogenesis.
Conclusion
In summary, despite obvious differences in their expression patterns, Nasonia eve, odd, and hairy function in both blastoderm- and germ band-derived segment formation. While Nasonia exhibits fly-type expression of maternal and gap genes in the precellular blastoderm, dynamic expression patterns and extensive interactions among ‘pair-rule’ genes appear to pattern a suite of late forming posterior segments. Indeed, their relative expression patterns suggest that the regulation of posterior segments may be through the type of mutual regulation described for the pair-rule gene circuit of Tribolium. This is unlike the long germ embryo of Drosophila, whose segmentation utilizes pair-rule interactions only during the late blastoderm stage. We propose that late-forming segments are set aside using remnants of ancestral control of posteriorness and the segmentation clock. Thus, Nasonia relies on a dynamic, dual mode of segmentation that has characteristics of both ancestral short germ and derived long germ embryogenesis.
Materials and methods
Embryo collection, non-fluorescent in situ hybridization, two-color FISH, and immunohisochemistry
Nasonia embryos were collected and fixed in 5% formaldehyde/1X PBS/Heptane for 28 min, affixed to double-sided tape, and hand peeled under 1X PBS +0.1% tween, as described previously (Pultz et al., 2005), except that the embryos were collected from host-fed, mated females. The embryos were stored under methanol at −20°C between fixation and hybridization.
In situ hybridizations were carried out as described previously (Pultz et al., 2005). Briefly, the embryos stored under methanol were gradually brought up to 1X PBT and washed three times in 1x PBS +0.1% tween-20 (PBT) before a 30-min post-fix step in 5% formaldehyde/1XPBT. The embryos were then washed three times and subjected to proteinase K treatment (final concentration of 4 μg/ml) before three PBT washes. The embryos were blocked for 1 hr in hybridization buffer before probe preparation and addition for overnight incubation at 65°C. The next day, the embryos were washed in formamide wash buffer and then 1X MABT buffer before blocking in 2% Blocking Reagent (BBR; Roche Applied Science, Germany) in 1X MABT for 1 hr, then in 10% horse serum/2% BBR/1XMABT for 2 hr. The embryos were incubated overnight with primary antibody in the second blocking solution at 4°C. Anti-DIG-AP Fab fragments (Roche Diagnostics) were used at 1:2000 for non-fluorescent in situs. On the third day, the embryos were washed in 1X MABT for ten, 20 min washes before equilibrating the embryos in AP staining buffer and developing in AP buffer with NBT/BCIP solution (Roche Diagnostics). After staining, the embryos were washed in 1× PBT three times for 5 min each before a single 25 min post-fix step in 5% formaldehyde/1XPBT. The embryos were then washed briefly and allowed to sink in 50% and then 70% glycerol/1XPBS, which were subsequently used for mounting.
For fluorescent in situs, DIG probes were detected using anti-DIG-POD Fab fragments (Roche Diagnostics) at 1:50 dilution, followed by FastRed HNPP detection system (Roche Diagnostics), according to manufacturer’s instructions. Fluorescein probes were detected using anti-Fluorescein-AP Fab fragments (Roche Diagnostics) at 1:500 dilution.
For antibody staining of mitotic cells, we used a rabbit anti-phosphorylated histone H3 serine 10 antibody (Millipore, Billerica, Massachusetts) at 1:200, and then a donkey anti-rabbit secondary conjugated to Alexa-647 (Invitrogen, Carlsbad, California) at 1:200. In combination with FastRed in situ detection using anti-DIG-AP Fab fragments (Roche Diagnostics) at 1:500, primary antibodies were added to blocking buffer together and incubated according to the in situ protocol, and secondary antibody detection was carried out after the FastRed staining was completed and following three 1X PBT washes.
Cloning of Nasonia pair-rule gene cDNA fragments
Nasonia pair-rule gene cDNAs were cloned from embryo cDNA pools generated from reverse transcription of total embryo RNA from mixed age embryos using Superscript First Strand Synthesis kit (with Superscript II; Invitrogen) according to manufacturer’s specifications. For cases in which long cDNA sequences could not be amplified with oligos designed according to automated genome annotation and prediction models, we used circular RACE to simultaneously amplify sequences 5′ and 3′ to smaller cloned cDNA fragments, as previously described (McGrath, 2011).
Phylogenetic analysis of Nasonia pair-rule gene paralogs
Nasonia paralogs of fly pair-rule genes were identified by TBLASTN and aligned against predicted or experimentally validated (virtually translated) protein sequences of the same proteins from Tribolium castaneum (Tcas), Anopheles gambiae (Agam), Apis mellifera (Amel). Protein sequences were aligned using CLUSTALW (Larkin et al., 2007) and rendered using Dendroscope (Huson et al., 2007). Evolutionary relationships were inferred using a maximum likelihood analysis with 1000-fold bootstrap support, via RaXML hosted online at CIPRES science gateway (http://www.phylo.org/index.php/portal/) (Stamatakis et al., 2008; Stamatakis, 2006; Miller et al., 2010).
Morpholino injection and larval cuticle preparations
Antisense morpholinos targeting splice junctions or transcription initiation sites were designed and ordered from GeneTools, LLC (www.gene-tools.com, Philomath, Oregon). Lyophilized morpholinos were resuspended in sterile nuclease-free water to a final concentration of 5 mM. For Nv odd splice block morpholino, which yielded a high percentage of dead embryos with no cuticle, injections were also carried out at 1 mM, 0.5 mM, and 0.05 mM dilutions. Nasonia embryos were collected for 35 min at 28°C and dehydrated for 30 min before injection with morpholinos (approximate volume injected = 0.001 μl per embryo). The embryos were allowed to develop on the injection membrane at 28°C on a 1X PBS/1% agarose plate for approximately 30 hr, to ensure complete development. Unhatched larvae were peeled and transferred to a slide for cuticle preparations in Lacto–Hoyer’s media.
Morpholino sequences used are as follows:
Eve translation block 5′ CAAAGCTCCTCTGGAATCCTTGCAT 3′
Eve E2I2 splice block: 5′ AAACGATAGTTACCTTGATGGTCGA 3′
Hairy E2I2 splice block: 5′ CTGAATCTGTCAAGATACTTACGTC 3′
Hairy E1I1 splice block: 5′ GAGCAAGTCGAGATACTAACCCGTC
Odd splice block: 5′ AGAGAGTGTACTAAC TTGTGGTCCC 3′
Odd translation block: 5′ GCTCCATCGCAAGCTGGGTAAACGT 3′
cDNA sequence accession numbers
Nv odd cDNA GenBank Accession # KC142194
Nv eve cDNA, isoform 1 GenBank Accession# KC168090
Nv eve cDNA, isoform 2 GenBank Accession# KC168091
Nv eve cDNA, isoform 3 GenBank Accession# KC168092
Nv hairy cDNA GenBank Accession # KC190514
Accession numbers for sequences used in sequence alignments and trees:
NvitH1: NP_001267498 XP_001601817 (Nvit Hairy)
NvitH2: Uniprot K7J0X7_NASVI (hairy-like/Nvit Dpn)
NvitH3: XP_001601600.2 GI:345484850 (hairy-like/HES like?)
DmelH: NP_523977.2 GI:24661088 (Dmel hairy)
DmelDpn: NP_476923.1 GI:17136808 (Dmel deadpan)
AmelH1: XP_001120814.2 GI:328784100 (hypothetical protein)
AmelH2: XP_393948.3 GI:110762302 (hairy-like)
AgambH1: XP_316733.3 GI:158296333 (corrected seq; Agam hairy)
AgambH2: XP_320206.4 GI:158300226
TcasH1: NP_001107765.1 GI:166796106 (Tcas Hairy)
TcasH2: XP_967694.1 GI:91092620 (Tcas similar to GA21268-PA)
TcasH3: XP_975187.1 GI:91083981 (Tcas HES1)
DmelOddsk: NP_722922.1 GI:24581484 (Dmel Odd skipped)
DmelSobow: NP_476882.1 GI:17136746 (Dmel Sister of odd and bowl)
DmelBowl: NP_476883.1 GI:17136748 (Dmel Brother of odd with entrails limited)
NvitOddbowlA: XP_001603713.1 GI:156545195 (predicted protein)
NvitOddbowlB: XP_001603827.2 GI:345481739(Nv bowel-like)
NvitOddbowlC: XP_001603660.1 GI:156545193(Nv odd-skipped like)
AmelOddbowlA: XP_001120949.1 GI:110762343(Amel odd-skipped like)
AmelOddbowlB: XP_393879.3 GI:110762378(Amel bowel-like)
AmelOddbowlC: XP_001120905.1 GI:110762341
TcasBowl: XP_972138.2 GI:189240088(Tcas bowl-like)
TcasOdd: XP_972086.2 GI:189240086 (Tcas odd-skipped)
TcasSob: XP_972035.1 GI:91088523(Tcas: predicted sister of odd and bowl)
Agam7972_PA: XP_306979.3 GI:118776890
Agam7973_PA: XP_317495.3 GI:118789549
Agam8222: XP_555242.1 GI:57914799
For Figure 4, the phenotypic classes are approximately as follows:
Eve: B 20.4% C 24% D 30.1% E 25%.
Odd: G 27.7% H 22.3% I 29.2% J 20.8%.
Hairy: L 13.4% M 25.0% N 36.6% O 25.0%.
Acknowledgements
The authors would like to acknowledge Karin Kiontke and David Fitch for help with phylogenetic analyses, Jeremy Lynch for technical advice about multiple FISH, and Bruce Edgar and Pat O’Farrell for thoughtful comments and helpful advice on cell cycle data. Filipe Pinto Teixeira Sousa and Claire Bertet provided invaluable expertise and assistance with confocal imaging, and Jeremy Lynch and David Loehlin provided helpful comments on the manuscript. Terry Blackman and Cleo Tsanis provided immeasurable support, carrying out all of the injections described in the paper. The authors would also like to thank Bob Johnston, Leatt Gilboa, Zhenqing Chen, Michael Perry and Brent Wells for support and discussion during the course of this project. MIR would like to dedicate this paper in loving memory of Allen Rosenberg (1931-2013).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
National Institutes of Health F32GM084563 to Miriam I Rosenberg.
Damon Runyon Cancer Research Foundation DRG-1870-05 to Ava E Brent.
American Cancer Society 120323-PF-11-242-01-DDC to Miriam I Rosenberg.
National Institutes of Health 5R01GM064864 to Claude Desplan.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
MIR, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
AEB, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
FP, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
CD, Conception and design, Analysis and interpretation of data, Drafting or revising the article.
Additional files
Major datasets
The following previously published datasets were used:
2013, UniprotKB data set for Nasonia vitripennis, http://www.uniprot.org/uniprot/?query=author:%22EnsemblMetazoa%22, Publicly available at UniProt (http://www.uniprot.org/).
JH Werren, , 2010, Nasonia vitripennis Official Gene set, http://www.hymenopteragenome.org/nasonia/?q=sequencing_and_analysis_consortium_datasets, Publicly available at NasoniaBase (http://www.hymenopteragenome.org/nasonia/).
References
- Ahringer J. 1996. Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes & Development 10:1120–1130. 10.1101/gad.10.9.1120 [DOI] [PubMed] [Google Scholar]
- Aranda M, Marques-Souza H, Bayer T, Tautz D. 2008. The role of the segmentation gene hairy in Tribolium. Development Genes and Evolution 218:465–477. 10.1007/s00427-008-0240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian H, Gruss P, Duboule D, Izpisua-Belmonte JC. 1992. The murine even-skipped-like gene Evx-2 is closely linked to the Hox-4 complex, but is transcribed in the opposite direction. Mammalian Genome 3:241–243. 10.1007/BF00355726 [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. 2004. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLOS Biology 2:E178. 10.1371/journal.pbio.0020178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent AE, Yucel G, Small S, Desplan C. 2007. Permissive and instructive anterior patterning rely on mRNA localization in the wasp embryo. Science 315:1841–1843. 10.1126/science.1137528 [DOI] [PubMed] [Google Scholar]
- Brown SJ, Parrish JK, Denell RE, Beeman RW. 1994. Genetic control of early embryogenesis in the red flour beetle, Tribolium castaneum. American Zoologist 34:343–352 [DOI] [PubMed] [Google Scholar]
- Brown SJ, Parrish JK, Beeman RW, Denell RE. 1997. Molecular characterization and embryonic expression of the even-skipped ortholog of Tribolium castaneum. Mechanisms of Development 61:165–173. 10.1016/S0925-4773(96)00642-9 [DOI] [PubMed] [Google Scholar]
- Carroll SB, Vavra SH. 1989. The zygotic control of Drosophila pair-rule gene expression. II. Spatial repression by gap and pair-rule gene products. Development 107:673–683 [DOI] [PubMed] [Google Scholar]
- Carroll SB, Laughon A, Thalley BS. 1988. Expression, function, and regulation of the hairy segmentation protein in the Drosophila embryo. Genes & Development 2:883–890. 10.1101/gad.2.7.883 [DOI] [PubMed] [Google Scholar]
- Cerny AC, Bucher G, Schroder R, Klingler M. 2005. Breakdown of abdominal patterning in the Tribolium Kruppel mutant jaws. Development 132:5353–5363. 10.1242/dev.02154 [DOI] [PubMed] [Google Scholar]
- Chen H, Xu Z, Mei C, Yu D, Small S. 2012. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149:618–629. 10.1016/j.cell.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman AD, Akam M. 2008. The segmentation cascade in the centipede Strigamia maritima: involvement of the Notch pathway and pair-rule gene homologues. Developmental Biology 319:160–169. 10.1016/j.ydbio.2008.02.038 [DOI] [PubMed] [Google Scholar]
- Chipman AD, Arthur W, Akam M. 2004. A double segment periodicity underlies segment generation in centipede development. Current Biology 14:1250–1255. 10.1016/j.cub.2004.07.026 [DOI] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. 2007. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Developmental Biology 302:281–294. 10.1016/j.ydbio.2006.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. 2009. Genetic regulation of engrailed and wingless in Tribolium segmentation and the evolution of pair-rule segmentation. Developmental Biology 325:482–491. 10.1016/j.ydbio.2008.10.037 [DOI] [PubMed] [Google Scholar]
- Choe CP, Miller SC, Brown SJ. 2006. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proceedings of the National Academy of Sciences of the United States of America 103:6560–6564. 10.1073/pnas.0510440103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copf T, Rabet N, Celniker SE, Averof M. 2003. Posterior patterning genes and the identification of a unique body region in the brine shrimp Artemia franciscana. Development 130:5915–5927. 10.1242/dev.00835 [DOI] [PubMed] [Google Scholar]
- Coulter DE, Swaykus EA, Beran-Koehn MA, Goldberg D, Wieschaus E, Schedl P. 1990. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. The EMBO Journal 9:3795–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen WG, Weller M, Tautz D. 2000. Expression patterns of hairy, even-skipped, and runt in the spider Cupiennius salei imply that these genes were segmentation genes in a basal arthropod. Proceedings of the National Academy of Sciences of the United States of America 97:4515–4519. 10.1073/pnas.97.9.4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GK, Patel NH. 1999. The origin and evolution of segmentation. Trends in Cell Biology 9:M68–M72. 10.1016/S0962-8924(99)01663-3 [DOI] [PubMed] [Google Scholar]
- Dearden PK, Wilson MJ, Sablan L, Osborne PW, Havler M, McNaughton E, Kimura K, Milshina NV, Hasselmann M, Gempe T, Schioett M, Brown SJ, Elsik CG, Holland PW, Kadowaki T, Beye M. 2006. Patterns of conservation and change in honey bee developmental genes. Genome Research 16:1376–1384. 10.1101/gr.5108606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle J, Pourquie O. 2004. Coupling segmentation to axis formation. Development 131:5783–5793. 10.1242/dev.01519 [DOI] [PubMed] [Google Scholar]
- Edgar BA, O’Farrell PH. 1989. Genetic control of cell division patterns in the Drosophila embryo. Cell 57:177–187. 10.1016/0092-8674(89)90183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Odell GM, Schubiger G. 1989. A genetic switch, based on negative regulation, sharpens stripes in Drosophila embryos. Developmental Genetics 10:124–142. 10.1002/dvg.1020100303 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Lehman DA, O’Farrell PH. 1994. Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development 120:3131–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif E, Averof M, Brown SJ. 2012. A segmentation clock operating in blastoderm and germband stages of Tribolium development. Development 139:4341–4346. 10.1242/dev.085126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson BJ, Ungerer P, Stollewerk A. 2013. The function of Notch signalling in segment formation in the crustacean Daphnia magna (Branchiopoda). Developmental Biology 383:321–330. 10.1016/j.ydbio.2013.09.021 [DOI] [PubMed] [Google Scholar]
- Foe VE. 1989. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development 107:1–22 [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. 1987. Characterization and localization of the even-skipped protein of Drosophila. The EMBO Journal 6:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. 1999. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development 126:2527–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel K, Basal A, Fan X, Roth S. 2005. Tribolium castaneum twist: gastrulation and mesoderm formation in a short-germ beetle. Development Genes and Evolution 215:13–31. 10.1007/s00427-004-0446-9 [DOI] [PubMed] [Google Scholar]
- Hart MC, Wang L, Coulter DE. 1996. Comparison of the structure and expression of odd-skipped and two related genes that encode a new family of zinc finger proteins in Drosophila. Genetics 144:171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. 2002. Exploring myriapod segmentation: the expression patterns of even-skipped, engrailed, and wingless in a centipede. Developmental Biology 247:47–61. 10.1006/dbio.2002.0683 [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. 2007. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 22:460. 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Pinchin SM, Howard KR, Ish-Horowicz D. 1985. Genetic analysis of the hairy locus in Drosophila melanogaster. Genetics 111:463–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Hoey T, Levine M. 1991. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes & Development 5:265–277. 10.1101/gad.5.2.265 [DOI] [PubMed] [Google Scholar]
- Jouve C, Palmeirim I, Henrique D, Beckers J, Gossler A, Ish-Horowicz D, Pourquié O. 2000. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127:1421–1429 [DOI] [PubMed] [Google Scholar]
- Kainz F, Ewen-Campen B, Akam M, Extavour CG. 2011. Notch/Delta signalling is not required for segment generation in the basally branching insect Gryllus bimaculatus. Development 1:5015–5026. 10.1242/dev.073395 [DOI] [PubMed] [Google Scholar]
- Kilchherr F, Baumgartner S, Bopp D, Frei E, Noll M. 1986. Isolation of the paired gene of Drosophila melanogaster and its spatial expression during early embryogenesis. Nature 3:493–499. 10.1038/321493a0 [DOI] [Google Scholar]
- Kim J, Kerr JQ, Min GS. 2000. Molecular heterochrony in the early development of Drosophila. Proceedings of the National Academy of Sciences of the United States of America 97:212–216. 10.1073/pnas.97.1.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Liu PZ, Kaufman TC. 2004a. hunchback is required for suppression of abdominal identity, and for proper germband growth and segmentation in the intermediate germband insect Oncopeltus fasciatus. Development 131:1515–1527. 10.1242/dev.01046 [DOI] [PubMed] [Google Scholar]
- Liu PZ, Kaufman TC. 2004b. Kruppel is a gap gene in the intermediate germband insect Oncopeltus fasciatus and is required for development of both blastoderm and germband-derived segments. Development 131:4567–4579. 10.1242/dev.01311 [DOI] [PubMed] [Google Scholar]
- Liu PZ, Kaufman TC. 2005a. even-skipped is not a pair-rule gene but has segmental and gap-like functions in Oncopeltus fasciatus, an intermediate germband insect. Development 132:2081–2092. 10.1242/dev.01807 [DOI] [PubMed] [Google Scholar]
- Liu PZ, Kaufman TC. 2005b. Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evoluion & Development 7:629–646. 10.1111/j.1525-142X.2005.05066.x [DOI] [PubMed] [Google Scholar]
- Liu P, Kaufman TC. 2009. Morphology and husbandry of the large milkweed bug, Oncopeltus fasciatus. Cold Spring Harbour Protocols 2009:pdb.emo127. 10.1101/pdb.emo127 [DOI] [PubMed] [Google Scholar]
- Lynch JA, Desplan C. 2006. A method for parental RNA interference in the wasp Nasonia vitripennis. Nature Protocols 1:486–494. 10.1038/nprot.2006.70 [DOI] [PubMed] [Google Scholar]
- Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. 2006. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature 439:728–732. 10.1038/nature04445 [DOI] [PubMed] [Google Scholar]
- Lynch JA, Olesnicky EC, Desplan C. 2006. Regulation and function of tailless in the long germ wasp Nasonia vitripennis. Development Genes and Evolution 216:493–498. 10.1007/s00427-006-0076-5 [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. 1986. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell 47:721–734. 10.1016/0092-8674(86)90515-5 [DOI] [PubMed] [Google Scholar]
- McGrath PT. 2011. Characterizing cDNA ends by circular RACE. Methods in Molecular Biology 772:257–265. 10.1007/978-1-61779-228-1_15 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010 New Orleans, LA: p.1–8. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Mito T, Kobayashi C, Sarashina I, Zhang H, Shinahara W, Miyawaki K, Shinmyo Y, Ohuchi H. 2007. even-skipped has gap-like, pair-rule-like, and segmental functions in the cricket Gryllus bimaculatus, a basal, intermediate germ insect (Orthoptera). Developmental Biology 303:202–213. 10.1016/j.ydbio.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Munoz-Torres MC. 2009. Comparative genomics and molecular evolution: new genomic resources for the hymenoptera and evolutionary studies on the genes of the Nasonia Vitripennis hox complex. PhD Dissertation, Clemson University. Ann Arbor: ProQuest/UMI, 2011 (Publication No. 9781244008007) [Google Scholar]
- Pankratz M, Jäckle H. 1993. Blastoderm segmentation. In: Bate M, Martinez-Arias A, editor. The development of Drosophila melanogaster. Long Island, New York: Cold Spring Harbor Press; p. 467–516 [Google Scholar]
- Nagy L, Riddiford L, Kiguchi K. 1994. Morphogenesis in the early embryo of the lepidopteran Bombyx mori. Developmental Biology 165:137–151. 10.1006/dbio.1994.1241 [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287:795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Olesnicky EC, Brent AE, Tonnes L, Walker M, Pultz MA, Leaf D, Desplan C. 2006. A caudal mRNA gradient controls posterior development in the wasp Nasonia. Development 133:3973–3982. 10.1242/dev.02576 [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. 1997. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91:639–648. 10.1016/S0092-8674(00)80451-1 [DOI] [PubMed] [Google Scholar]
- Patel NH, Ball EE, Goodman CS. 1992. Changing role of even-skipped during the evolution of insect pattern formation. Nature 357:339–342. 10.1038/357339a0 [DOI] [PubMed] [Google Scholar]
- Patel NH, Condron BG, Zinn K. 1994. Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature 367:429–434. 10.1038/367429a0 [DOI] [PubMed] [Google Scholar]
- Peel AD, Chipman AD, Akam M. 2005. Arthropod segmentation: beyond the Drosophila paradigm. Nature Reviews Genetics 6:905–916. 10.1038/nrg1724 [DOI] [PubMed] [Google Scholar]
- Pourquie O. 2003. Vertebrate somitogenesis: a novel paradigm for animal segmentation? International Journal of Developmental Biology 47:597–603 [PubMed] [Google Scholar]
- Pourquie O. 2011. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145:650–663. 10.1016/j.cell.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pultz MA, Pitt JN, Alto NM. 1999. Extensive zygotic control of the anteroposterior axis in the wasp Nasonia vitripennis. Development 126:701–710 [DOI] [PubMed] [Google Scholar]
- Pultz MA, Zimmerman KK, Alto NM, Kaeberlein M, Lange SK, Pitt JN, Reeves NL, Zehrung DL. 2000. A genetic screen for zygotic embryonic lethal mutations affecting cuticular morphology in the wasp Nasonia vitripennis. Genetics 154:1213–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pultz MA, Westendorf L, Gale SD, Hawkins K, Lynch J, Pitt JN, Reeves NL, Yao JC, Small S, Desplan C, Leaf DS. 2005. A major role for zygotic hunchback in patterning the Nasonia embryo. Development 132:3705–3715. 10.1242/dev.01939 [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Lynch JA, Desplan C. 2009. Heads and tails: evolution of antero-posterior patterning in insects. Biochimica et Biophysica Acta 1789:333–342. 10.1016/j.bbagrm.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin AF, Peel AD, Averof M. 2012. A segmentation clock with two-segment periodicity in insects. Science 336:338–341. 10.1126/science.1218256 [DOI] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. 2004. Transcriptional control in the segmentation gene network of Drosophila. PLOS Biology 2:E271. 10.1371/journal.pbio.0020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Greer C, Gaul U. 2011. How to make stripes: deciphering the transition from non-periodic to periodic patterns in Drosophila segmentation. Development 138:3067–3078. 10.1242/dev.062141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy TD, Ronshaugen M, Cande J, He J, Beeman RW, Levine M, Brown SJ, Denell RE. 2008. Analysis of the Tribolium homeotic complex: insights into mechanisms constraining insect Hox clusters. Development Genes and Evolution 218:127–139. 10.1007/s00427-008-0213-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Levine M. 1991. The initiation of pair-rule stripes in the Drosophila blastoderm. Current Opinion In Genetics & Development 1:255–260. 10.1016/S0959-437X(05)80079-6 [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. The EMBO Journal 11:4047–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. 1996. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Developmental Biology 175:314–324. 10.1006/dbio.1996.0117 [DOI] [PubMed] [Google Scholar]
- Sommer RJ, Tautz D. 1993. Involvement of an orthologue of the Drosophila pair-rule gene hairy in segment formation of the short germ-band embryo of Tribolium (Coleoptera). Nature 361:448–450. 10.1038/361448a0 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57:758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Schoppmeier M, Damen WG. 2003. Involvement of Notch and Delta genes in spider segmentation. Nature 423:863–865. 10.1038/nature01682 [DOI] [PubMed] [Google Scholar]
- Suen G, Teiling C, Li L, Holt C, Abouheif E, Bornberg-Bauer E, Bouffard P, Caldera EJ, Cash E, Cavanaugh A, Denas O, Elhaik E, Favé MJ, Gadau J, Gibson JD, Graur D, Grubbs KJ, Hagen DE, Harkins TT, Helmkampf M, Hu H, Johnson BR, Kim J, Marsh SE, Moeller JA, Muñoz-Torres MC, Murphy MC, Naughton MC, Nigam S, Overson R, Rajakumar R, Reese JT, Scott JJ, Smith CR, Tao S, Tsutsui ND, Viljakainen L, Wissler L, Yandell MD, Zimmer F, Taylor J, Slater SC, Clifton SW, Warren WC, Elsik CG, Smith CD, Weinstock GM, Gerardo NM, Currie CR. 2011. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLOS Genetics 7:e1002007. 10.1371/journal.pgen.1002007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston IA, Anderson KV. 1996. Embryonic patterning mutants of Tribolium castaneum. Development 122:805–814 [DOI] [PubMed] [Google Scholar]
- Warrior R, Levine M. 1990. Dose-dependent regulation of pair-rule stripes by gap proteins and the initiation of segment polarity. Development 110:759–767 [DOI] [PubMed] [Google Scholar]
- Werren JH Richards S Desjardins CA Niehuis O Gadau J Colbourne JK, Nasonia Genome Working Group, Werren JH Richards S Desjardins CA Niehuis O Gadau J Colbourne JK Beukeboom LW Desplan C Elsik CG Grimmelikhuijzen CJ Kitts P Lynch JA Murphy T Oliveira DC Smith CD van de Zande L Worley KC Zdobnov EM Aerts M Albert S Anaya VH Anzola JM Barchuk AR Behura SK Bera AN Berenbaum MR Bertossa RC Bitondi MM Bordenstein SR Bork P Bornberg-Bauer E Brunain M Cazzamali G Chaboub L Chacko J Chavez D Childers CP Choi JH Clark ME Claudianos C Clinton RA Cree AG Cristino AS Dang PM Darby AC de Graaf DC Devreese B Dinh HH Edwards R Elango N Elhaik E Ermolaeva O Evans JD Foret S Fowler GR Gerlach D Gibson JD Gilbert DG Graur D Gründer S Hagen DE Han Y Hauser F Hultmark D Hunter HC IV Hurst GD Jhangian SN Jiang H Johnson RM Jones AK Junier T Kadowaki T Kamping A Kapustin Y Kechavarzi B Kim J Kim J Kiryutin B Koevoets T Kovar CL Kriventseva EV Kucharski R Lee H Lee SL Lees K Lewis LR Loehlin DW Logsdon JM Jnr Lopez JA Lozado RJ Maglott D Maleszka R Mayampurath A Mazur DJ McClure MA Moore AD Morgan MB Muller J Munoz-Torres MC Muzny DM Nazareth LV Neupert S Nguyen NB Nunes FM Oakeshott JG Okwuonu GO Pannebakker BA Pejaver VR Peng Z Pratt SC Predel R Pu LL Ranson H Raychoudhury R Rechtsteiner A Reese JT Reid JG Riddle M Robertson HM Romero-Severson J Rosenberg M Sackton TB Sattelle DB Schlüns H Schmitt T Schneider M Schüler A Schurko AM Shuker DM Simões ZL Sinha S Smith Z Solovyev V Souvorov A Springauf A Stafflinger E Stage DE Stanke M Tanaka Y Telschow A Trent C Vattathil S Verhulst EC Viljakainen L Wanner KW Waterhouse RM Whitfield JB Wilkes TE Williamson M Willis JH Wolschin F Wyder S Yamada T Yi SV Zecher CN Zhang L Gibbs RA. 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327:343–348. 10.1126/science.1178028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MJ, Dearden PK. 2011. Diversity in insect axis formation: two orthodenticle genes and hunchback act in anterior patterning and influence dorsoventral organization in the honeybee (Apis mellifera). Development 138:3497–3507. 10.1242/dev.067926 [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Dearden PK. 2012. Pair-rule gene orthologues have unexpected maternal roles in the honeybee (Apis mellifera). PLOS ONE 7:e46490. 10.1371/journal.pone.0046490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MJ, Havler M, Dearden PK. 2010. Giant, Kruppel, and caudal act as gap genes with extensive roles in patterning the honeybee embryo. Developmental Biology 339:200–211. 10.1016/j.ydbio.2009.12.015 [DOI] [PubMed] [Google Scholar]
- Xu X, Xu PX, Amanai K, Suzuki Y. 1997. Double-segment defining role of even-skipped homologs along the evolution of insect pattern formation. Development Growth & Differentiation 39:515–522. 10.1046/j.1440-169X.1997.t01-3-00013.x [DOI] [PubMed] [Google Scholar]