Abstract
Lipid droplets (LDs) are ubiquitous dynamic organelles that store and supply lipids in all eukaryotic and some prokaryotic cells for energy metabolism, membrane synthesis, and production of essential lipid-derived molecules. Interest in the organelle’s cell biology has exponentially increased over the last decade due to the link between LDs and prevalent human diseases and the discovery of new and unexpected functions of LDs. As a result, there has been significant recent progress toward understanding where and how LDs are formed, and the specific lipid pathways that coordinate LD biogenesis.
Introduction
Lipids are essential for life, and the existence of cells specialized in fat storage has been conserved from flies and worms to humans. However, almost all cells maintain the capability to retain and accumulate lipids. Cellular mechanisms of lipid storage are relatively conserved from unicellular organisms, such as yeast, to complex organisms such as plants and mammals (Chapman et al., 2012; Kühnlein, 2012; Mak, 2012; Walther and Farese, 2012). Indeed, in all eukaryotic and some prokaryotic cells, lipid droplets (LDs; Fig. 1, A and C) are the intracellular organelle specialized in assembling, storing, and then supplying lipids. For decades considered passive cytosolic inclusions, LDs are now understood as being complex and dynamic organelles, with a central role in the regulation of lipid homeostasis. LDs provide substrates for energy metabolism, membrane synthesis, and production of essential lipid-derived molecules such as lipoproteins, bile salts, or hormones.
Figure 1.
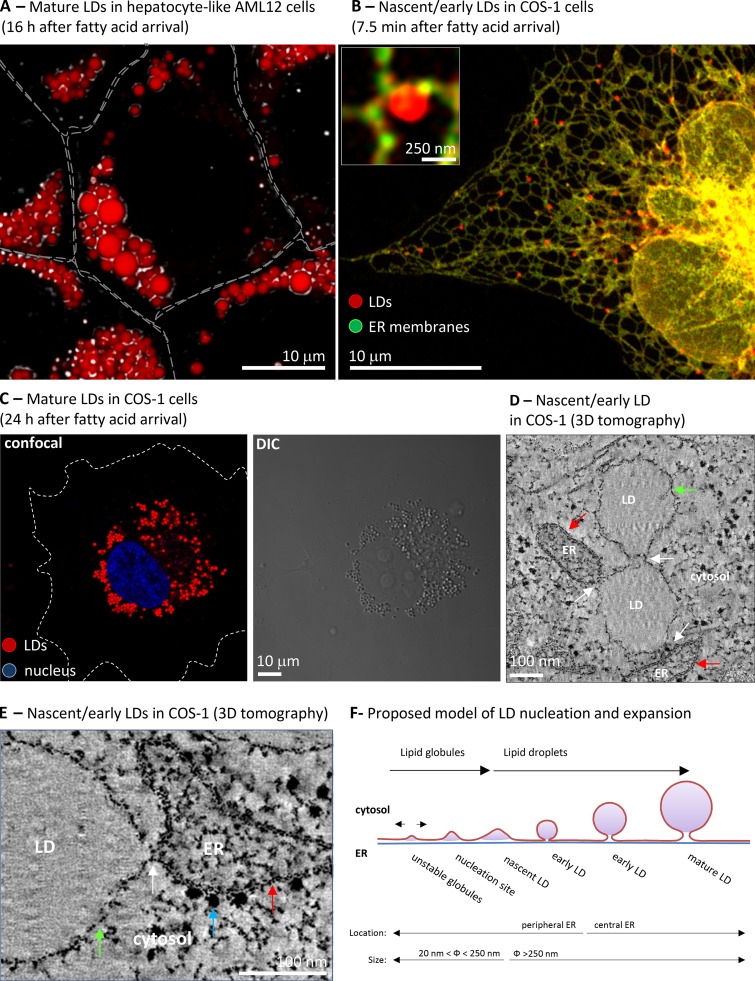
Morphology of LDs. (A) AML-12 cells were treated for 16 h with BSA-bound oleic acid. Cells were fixed and neutral lipids stained with Nile red. The image combines images of confocal microscopy (red) and differential interference contrast images (DIC, black and white). Gray lines indicate the contour of the cells. Image courtesy of Albert Herms. (B) COS-1 cells were cotransfected with OFP-HPos (an LD marker, red) and GFP-HNeu (ER marker, green) and cells were lipid starved during 24 h to reduce the presence of LDs. Then cells were loaded with BSA-bound oleic acid for 7.5 min and the segregation of the markers analyzed by confocal microscopy. The red globular structures correspond to the fatty acid–induced LDs and the inset shows a high magnification image of one of these LDs. The images are from Kassan et al. (2013). (C) Cells treated as in B, but for 24 h, were fixed and neutral lipids stained with Nile red and the nuclei with DAPI (blue). Lipid droplets are visible both by confocal microscopy (left) and DIC (right). White dotted lines indicate the contour of the cell. Image courtesy of Albert Herms. (D and E) LDs induced to form in COS-1 cells were analyzed by electron tomography. The images shows selected low magnification (D) and high magnification (E) sections in which the LD monolayer (green arrow) contacts (white arrow) with the other LDs or the ER bilayer (red arrow), which is recognized by the presence of electron-dense ribosomes (blue arrow). These LDs have an average diameter of 250 nm and thus are nascent or very early LDs. Image from Kassan et al. (2013). (F) Schematic representation of a model of LD biogenesis. Lipid globules produced during triacylglycerol synthesis and deposited between the two ER leaflets of the bilayer that laterally move unless they are nucleated. This nucleation marks the onset of LD biogenesis. From here, LD expansion (sizes are indicated) can be accomplished by recruitment of additional proteins, accumulation of new lipids locally and distally produced, and the synthesis of new phospholipids and lysophospholipids. Finally, the concave nascent LD will be converted into the globular early LD, move into central regions of the cell, and form a mature LD that shows a relatively constant size (Kassan et al., 2013).
Cellular lipid storage varies, and reflects a balance between lipid arrival and lipid consumption; excessive accumulation of LDs occurs in fat-related diseases, such as obesity and arteriosclerosis (Krahmer et al., 2013). However, although the balance between supply and consumption determines LD levels, LD accumulation is remarkably heterogeneous even between otherwise identical cells (Herms et al., 2013). Interestingly, factors such as intracellular and extracellular stresses trigger LD formation (Hapala et al., 2011), reflecting a role for LDs in processes not directly related to lipid metabolism, such as protein degradation or immunity. Indeed, accumulation of LDs also occurs during progression of pathologies not obviously related to lipids, such as cardiomyopathies, neuropathies, or during viral hepatitis caused by, among others, the human immunodeficiency virus (Vallet-Pichard et al., 2012).
In eukaryotes, LDs likely form de novo by progressive accumulation of neutral lipids in the ER (Fig. 1, B and D–F), although fission of preexisting LDs has been also suggested as a mechanism to generate new LDs in yeast (Long et al., 2012). Multiple enzymatic reactions contribute to LD assembly and numerous bioactive lipid intermediates are synthesized and transformed (Fig. 2). LD formation is regulated by complex and robust mechanisms, often conducted by apparently redundant activities of enzymes and structural proteins. For example, at least eleven mammalian acyl-CoA synthetases (ACSs) can activate major long-chain fatty acids, and thus potentially initiate the synthesis of neutral lipids (Watkins et al., 2007). At the other end of the pathway, in humans, at least twelve genes of three different acyltransferase families produce enzymes capable of catalyzing the last step of neutral lipid synthesis (Sturley and Hussain, 2012). Such complexity and functional compensation likely reflects the physiological significance of the LD, and enormously complicates determining how particular proteins and lipids contribute to LD assembly and metabolism. This review will summarize our current understanding of where and how LDs are formed, and in what way LD formation requires coordination of: (1) fatty acid activation, (2) synthesis of neutral lipids, (3) remodeling of phospholipids, (4) synthesis of new phospholipids, and (5) function of accessory proteins. Finally, these biochemical reactions will be integrated in a single model to describe mechanisms and sites of LD biogenesis.
Figure 2.
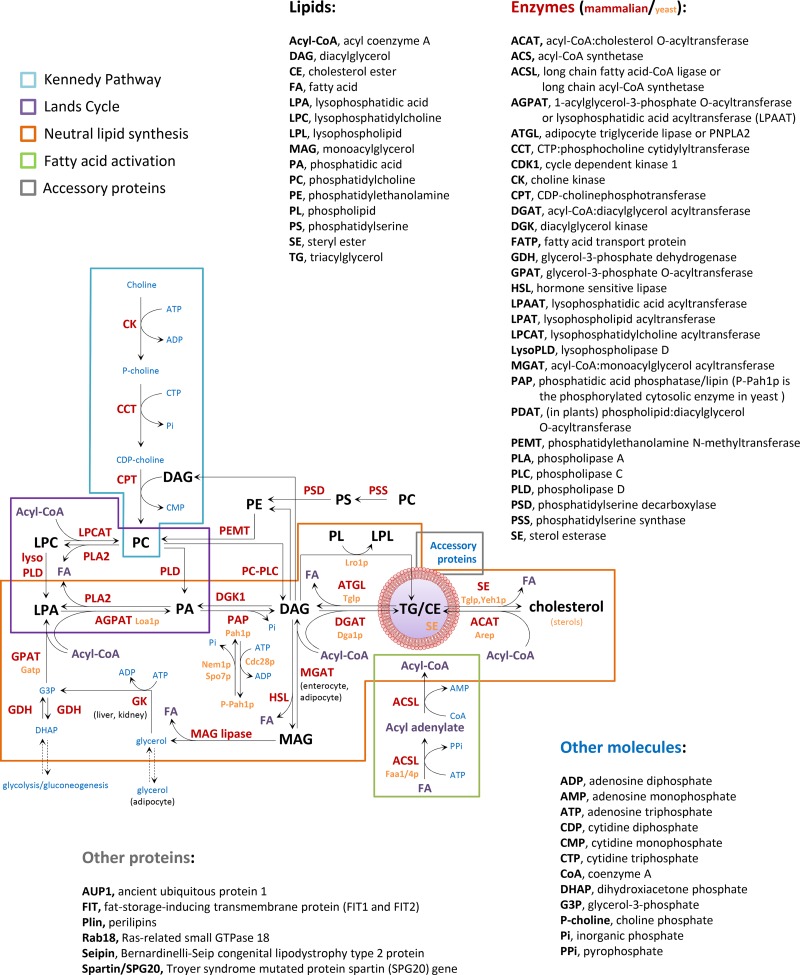
Lipid metabolism and LD formation. LD biogenesis requires coordination of fatty acid activation by acyl-CoA synthetases (green box), de novo synthesis of neutral lipids (orange box), the Lands cycle of phospholipid remodeling (purple box), the Kennedy pathway of phospholipid synthesis (blue box), and the function of accessory proteins (gray box). The enzymes involved in these pathways are indicated with red (mammal) and yellow letters (yeast), the acyl-CoA with purple letters, the lipid intermediates with black letters, and other required molecules with blue letters. See details of lipolysis mechanisms in Zechner et al. (2012).
Fatty acid activation and multi-enzymatic complexes on LDs
Many reactions occurring during LD biogenesis require an acyl-CoA. Fatty acids are chemically inert, and thus must be activated by esterification with coenzyme A (CoA). This occurs in an ATP-dependent two-step reaction, catalyzed by ACS (Fig. 2, green box; Ellis et al., 2010). The proposed LD-bound ACS members are ACSL1, ACSL3, and ACSL4. Inhibition of these ACSLs with Triacsin C (Tomoda et al., 1987) highly reduces triacylglycerol synthesis (Igal and Coleman, 1996), and thus completely inhibits the formation of LDs promoted by fatty acids (Kassan et al., 2013).
Interestingly, the ACSL family members are differentially regulated during 3T3-L1 adipocyte differentiation: ACSL1 is up-regulated, but ACSL3 is down-regulated (Sandoval et al., 2008), suggesting that each could be involved in different aspects of LD metabolism. Some studies differ regarding LD association of ACSL1 and ACSL4 (Brasaemle et al., 2004; Liu et al., 2004; Poppelreuther et al., 2012; Wilfling et al., 2013), but ACSL3 is definitely a bona fide LD protein (Brasaemle et al., 2004; Fujimoto et al., 2004, 2007). ACSL3 was linked to cellular lipid uptake, and a peptide comprising the N-terminal domain of ACSL3 was shown in early LDs (Poppelreuther et al., 2012). Indeed, in COS-1–cultured cells, endogenous ACSL3 is recruited early to LD assembly sites, and is required for efficient LD nucleation (Kassan et al., 2013). The absence of ACSL3 significantly reduces fatty acid–induced formation of LDs and the short- and long-term accumulation of neutral lipids. However, because ACSL3 is highly down-regulated in mature 3T3 adipocytes, it is likely that factors other than ACSL3 assist LD nucleation and expansion in specialized cells.
The roles of ACS proteins during LD biogenesis might go beyond their enzymatic activity (Kassan et al., 2013). Some ACS proteins are fatty acid transporters (FATPs; Doege and Stahl, 2006), some regulate fatty acid uptake (Füllekrug et al., 2012), and because ACS activity modifies the cellular AMP/ATP ratio, they are also likely involved in metabolic signaling via AMP kinases (Ellis et al., 2010). Further, ACS proteins form complexes with other proteins, and thus potentially organize specific microdomains in the ER. Complexes of neutral lipid synthesis enzymes and synthetase activities were demonstrated in early studies (Rao and Johnston, 1966); such complexes likely increase efficiency of neutral lipid production by rapidly transferring substrates from one enzyme to the next (Wilfling et al., 2013). For example, in Caenorhabditis elegans, a fatty acid transport protein 1 (FATP1)/diacylglycerol acyltransferase 2 (DGAT2) complex was located at the ER–LD interface, and proposed to couple neutral lipid synthesis and deposition (Xu et al., 2012). Finally, ACSL3 physically interacts with other LD proteins such as Spartin/SPG20 and the ancient ubiquitous protein 1 (AUP1), which are in turn crucial adaptor proteins that determine LD size and numbers (Eastman et al., 2009; Milewska et al., 2009; Klemm et al., 2011; Spandl et al., 2011).
Neutral lipid synthesis “within” and “outside” LDs
Although their relative amount varies between cell types, the most abundant neutral lipids in LDs are triacylglycerol and cholesterol esters (sterol esters in yeast and Drosophila). While in adipocytes, caveolae may contribute to triacylglycerol synthesis (Ost et al., 2005); in most cells neutral lipid synthesis is conducted by enzymes permanently or transiently located in the ER. De novo triacylglycerol synthesis occurs in four reactions, catalyzed by members of the glycerol-3-phosphate O-acyltransferase (GPAT), 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), phosphatidic acid phosphatase (PAP)/lipin, and DGAT enzyme families (Fig. 2, orange box). The final step in this pathway is esterification of diacylglycerol into triacylglycerol conducted by DGAT1 and DGAT2 (Dga1p in yeast; Sturley and Hussain, 2012), which accounts for nearly all triacylglycerol synthesis in mouse cells (Harris et al., 2011). Nevertheless, alternative mechanisms have been proposed (Harris et al., 2011; Sturley and Hussain, 2012), and in plants and yeast, phospholipid:diacylglycerol O-acyltransferase (PDTA) and Lro1p can also produce triacylglycerol through trans-esterification of a fatty acid from phospholipids (Chapman et al., 2012; Czabany et al., 2008). In addition, LDs accumulate free cholesterol in adipocytes (Krause and Hartman, 1984) and cholesterol esters in most cells, especially in macrophages and steroidogenic cells. The synthesis of cholesterol esters is mediated by acyl-CoA:cholesterol O-acyltransferase (ACAT1 and ACAT2, Are1p and Are2p in yeast). Yeast strains with deletions of all four acyltransferases (ARE1Δ, ARE2Δ, DGA1Δ, and LRO1Δ) completely lack LDs and although viable, have a marked sensitivity to the lipotoxicity caused by fatty acids (Garbarino et al., 2009).
In cells lacking LDs, these enzymes are distributed homogenously along the ER, with the exception of PAP (Pah1p in yeast), which is largely cytosolic and in animal cells localizes to the nucleus. When LD formation is promoted with fatty acids, enzyme segregation occurs, with some remaining in the ER and others dynamically accumulating on the LDs. Specifically, a permanent ER location has been shown for AGPAT1, AGPAT2, AGPAT4, and DGAT1 in Drosophila cells (Wilfling et al., 2013); for Lro1p, Are1p, and Are2p in yeast (Zweytick et al., 2000; Jacquier et al., 2011); and for DGAT1, ACAT1, and ACAT2 in mammalian cells (Khelef et al., 1998; Stone et al., 2009). However, these LD-excluded enzymes efficiently promote LD formation (Harris et al., 2011; Jacquier et al., 2011), suggesting that neutral lipids can be synthesized along the ER tubules, diffuse laterally, and somehow nucleate to assemble LDs.
Interestingly, triacylglycerol synthesis can also occur locally on LDs and at least one member of each family required for de novo synthesis is present on LDs. The accumulation on LDs has been described for GPAT4, AGPAT3, PAP, and DGAT2 in flies or mammals (Fig. 3, orange box; Kuerschner et al., 2008; Stone et al., 2009; Wang et al., 2011; Wilfling et al., 2013) and for Gat1p, Gat2p, Pah1p, and Dga1p in yeast (Athenstaedt and Daum, 1997; Adeyo et al., 2011; Jacquier et al., 2011). Thus, such accumulation reflects enzymes that laterally segregate from the ER into LDs. For Dga1p in yeast, such LD accumulation is independent of temperature and energy, and thus does not require vesicle formation (Jacquier et al., 2011). Although not completely understood, it is mediated by a hairpin topology of the hydrophobic regions of these proteins, or an equivalent favorable folding (Walther and Farese, 2012), in combination with additional information, such as sequences of positive amino acids (Ingelmo-Torres et al., 2009). In contrast, enzymes with membrane-spanning helices (Stevanovic and Thiele, 2013) or without positive sequences (Kassan et al., 2013) remain localized in the ER during LD formation. The length and hydrophobicity of the hydrophobic regions, and the proximity of flanking positive residues, also determine whether the tail- and signal-anchored proteins are targeted to peroxisomes, chloroplasts, mitochondria, or the ER (Borgese et al., 2007). Interestingly, specific deletions within some LD-resident proteins localize the mutants to mitochondria (Nakamura and Fujimoto, 2003; Suzuki et al., 2005). Intriguingly, DGAT2 simultaneously contains signals to be sorted into the ER–LDs or mitochondria (Stone et al., 2009). Whether the positive residues of LD proteins are recognized by specific receptors, chaperones, or lipid species (as demonstrated for tail- and signal-anchored proteins) remains to be elucidated.
Figure 3.
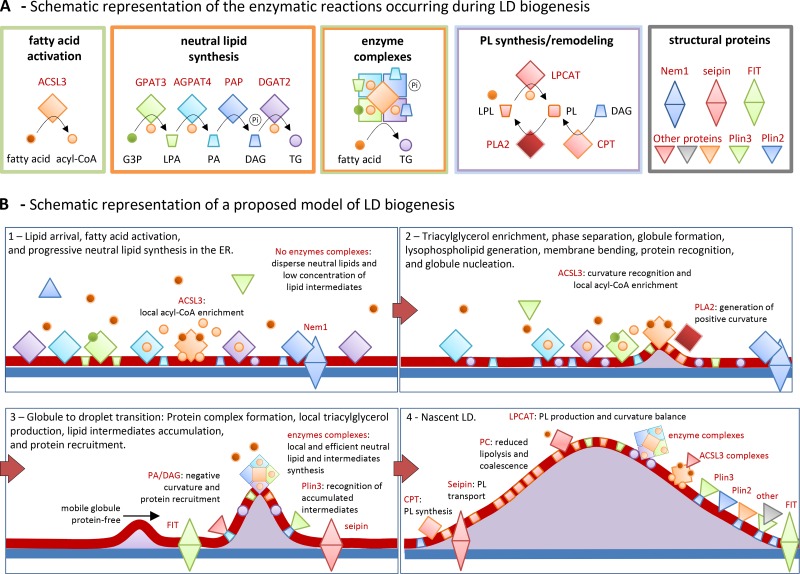
Proposed model of LD biogenesis. (A) Summary of the metabolic pathways detailed in Fig. 2 and occurring on LDs. Small circles and cones indicate lipids, squares enzymes, and triangles accessory protein. The color of each group follows the legend of Fig. 2. The green box illustrates the ACSL3-mediated conversion of the fatty acid into an acyl-CoA. The orange box includes the LD enzymes involved in the local synthesis of triacylglycerol. G3-P, glycerol 3-phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; DAG, diacylglycerol; TG, triacylglycerol; Pi, inorganic phosphate. The green/orange box illustrates the protein complexes of neutral lipid synthesis enzymes (orange box) and synthetase activities (green box) proposed to be a cellular mechanism to increase the efficiency of the process by rapidly transferring substrates from one enzyme to the next. The blue and purple box encloses reactions of phospholipid synthesis by the Kennedy pathway (using diacylglycerol as substrate, blue circle) and the Lands cycle of phospholipid remodeling by deacylation and reacylation of phospholipids (PL) and lysophospholipids (LPL). The gray box includes the accessory proteins (single triangles for cytosolic proteins and double triangles for ER-associated proteins). Additional proteins have been added to illustrate in B different mechanisms of protein association with LDs. (B) Proposed model of the stepwise formation of LDs. Shapes and colors follow the legend detailed in A. (B1) Before fatty acid arrival—likely bound to fatty acid transport proteins (not depicted)—neutral lipid synthesis enzymes and ACSL3 are homogenously distributed in the ER tubules. After lipid arrival PAP is recruited to the ER by interaction with Nem1p, perhaps directly to the assembly sites. Lipid intermediates and triacylglycerol are generated along the tubules. However, the enzymes in the proximity of ACSL3 will have availability of at least one required substrate. Finally, a triacylglycerol globule is generated and progressively expands. (B2) At sufficient concentration, the globule of triacylglycerol is not accommodated in the ER membrane, induces phase separation, and it is deposited between the two leaflets of the ER bilayer. The resulting membrane curvature—additionally produced/stabilized by local lysophospholipids or lipid demixing—is recognized by ER proteins with amphipathic helices, such as ACSL3, which stabilize the globule and mark the onset of LD biogenesis. (B3) Transition from a globule into a droplet rapidly occurs by accumulation of ACSL3 that locally increases the acyl-CoA concentration and thus efficiently provides to the neighboring enzymes with a substrate required for many reactions. Neutral synthesis enzymes form complexes at the sites of assembly, illustrated in A. Now triacylglycerol is rapidly produced at the sites of biogenesis, but in addition this pathway locally generates bioactive lipid intermediates. These lipids also determine membrane remodeling and protein recruitment. For example, Plin3 recognizes the accumulation of diacylglycerol and binds to the LD. (B4) The nascent LD rapidly expands into the cytosol by packing mobile globules of triacylglycerol arriving from the rest of the ER (B3, black arrow) and globules locally produced by the enzyme complexes. New phospholipids, to support LD expansion, are generated in neighboring tubules via CPT, and proteins such as seipin and FIT cooperate during the exchange. Conversion of conical lysophospholipids into cylindrical phospholipids, via LPCAT, progressively adjusts the growing structure to the new required curvature and, in addition, the cylindrical phospholipids protect the nascent globule from lipolytic activities and coalescence. Other proteins are anchored to LDs by interaction with lipid species such as phosphatidic acid and diacylglycerol, or by interaction with other LD proteins such as ACSL3. Finally, phosphatidic acid at the base of the structure recruits structural proteins and shapes the concave nascent LD into the globular early LD that will progressively expand and move to central regions of the cell to become a mature LD (Fig. 1 F).
In contrast to these enzymes arriving from the ER, PAP associates with LDs from the cytosol, regulated by phosphorylation. The yeast homologue, Pah1p, is dephosphorylated by the ER-resident phosphatase complex Nem1p–Spo7p, and is phosphorylated by Cdc28p kinase. When Pah1p is dephosphorylated it binds to the ER membrane through an amphipathic helix (Karanasios et al., 2010). Nem1p also localizes next to LDs, and it was suggested that it marks the ER sites of LD biogenesis (Adeyo et al., 2011). Overexpression of Nem1p–Spo7p recruits Pah1p in the vicinity of LDs and induces LD formation (Karanasios et al., 2013). Whether Dullard (Nem1p homologue) or the cyclin-dependent kinase 1 (CDK1, Cdc28p homologue) are involved in mammalian LD nucleation also remains to be analyzed. However, this scenario is much more complex in mammalian cells: mammals express at least three PAP paralogues (lipin 1, 2, and 3), with several isoforms, intracellular distributions, and expression patterns (Siniossoglou, 2013). Nevertheless, it has been shown that lipin 1γ has affinity for LDs (Wang et al., 2011) and a role of lipins in LD biogenesis has been recently proposed (Sembongi et al., 2013).
In addition to triacylglycerol, diacylglycerol is also present in LDs (Kuerschner et al., 2008). In early LD biogenesis, diacylglycerol likely generates both membrane curvature and provides a platform for protein binding. In yeast, diacylglycerol produced by Pah1p is critical for LD formation even when the LDs primarily contain sterol esters (Adeyo et al., 2011). In the absence of Pah1p, neutral lipids accumulate in the ER in a disorganized arrangement; this might also occur in mammalian cells (Sembongi et al., 2013). Interestingly, yeast strains with deletions in diacylglycerol kinase (ΔDGK1) have more LDs even when triacylglycerol synthesis is blocked. Similarly, the balance between diacylglycerol and phosphatidic acid regulates membrane tubulation and vesicle formation in the Golgi complex, by mechanisms that involve membrane remodeling and protein recruitment (Fei et al., 2011a; Malhotra and Campelo, 2011). On LDs, diacylglycerol could play an important role in recruitment of key proteins involved in LD formation and function including perilipin 3 (Plin3; Skinner et al., 2009), CTP:phosphocholine cytidylyltransferase (CCT; Krahmer et al., 2011), and protein kinase C (PKC; Chen et al., 2002; Suzuki et al., 2013).
The Lands cycle and membrane remodeling
The turnover of glycerophospholipid acyl chains in cycles of deacylation and reacylation catalyzed by phospholipase A (PLA) and lysophospholipid acyltransferase enzymes is known as the Lands cycle of phospholipid remodeling (Fig. 2, purple box; Lands, 2000). It is both an alternative source of phosphatidylcholine (see next section) and triacylglycerol (see previous section), and also contributes to dynamic remodeling of membranes. That the Lands cycle contributes to LD biogenesis is suggested by the fact that PLA2 inhibitors strongly reduce LD formation (Gubern et al., 2008; Du et al., 2009). Similar to the genetic deletion of yeast PAH1, loss of PLA2 activity inhibits LD assembly without impairing triacylglycerol formation. An LD-associated PLA2 has been described in hepatoma cell lines (Sato et al., 2006) and the yeast LD enzyme Tgl4p also exhibit PLA2 activity (Rajakumari and Daum, 2010b).
For the lysophospholipid acyltransferase portion of the Lands cycle, in addition to AGAPT3 (a lysophosphatidic acid acyltransferase, LPAAT), mammalian LDs also contain lysophosphatidylcholine acyltransferase (LPCAT) with activity on lysophosphatidylcholine (Moessinger et al., 2011). In yeast, Loa1p (a LPAAT) is also present on LDs; its overexpression results in fewer LDs, suggesting a direct role of lysophosphatidic acid during LD formation (Ayciriex et al., 2012). Further, likely indicating the importance of this cycle, the yeast LD enzymes Tgl3p and Tgl5p also catalyze acylation of lysophosphatidylethanolamine and lysophosphatidic acid (Rajakumari and Daum, 2010a). Dynamic turnover between lysophospholipids and phospholipids, conically and cylindrically shaped, respectively, could be required to curve initially the ER membrane and next to progressively adapt the membrane curvature in order to shield the underlying globule in each step of the biogenesis process.
Membrane remodeling by the Lands cycle is best characterized in the Golgi complex (Ha et al., 2012), where dynamic equilibrium between the activities of PLA2 and AGPAT3 determines the relative abundance of vesicles and tubules (Yang et al., 2011). This might be analogous to a role in LDs, promoting LD formation from ER tubules. Consistent with this general picture, in the Golgi complex, the final step of COPI vesicle fission requires activity of phospholipase D (PLD) to hydrolyze phosphatidylcholine into phosphatidic acid; for LDs, the phosphatidic acid produced by PLD is needed for LD generation in a microsome-based cell-free system (Marchesan et al., 2003). How mechanisms for shaping phospholipid bilayers also function on monolayers clearly deserves further investigation.
Phospholipid synthesis for LD expansion and to avoid coalescence
When LDs expand, the phospholipid content of the LD must also increase. This is likely critical during the initial stages of the LD growth process, in which the nascent structures grow rapidly. In yeast and mammalian LDs, phosphatidylcholine is the major phospholipid (50–60%), followed by phosphatidylethanolamine (20–30%), phosphatidylinositol, and ether-linked species with choline and ethanolamine headgroups (Tauchi-Sato et al., 2002; Bartz et al., 2007). In contrast, Drosophila LDs contain more phosphatidylethanolamine (50–60%) than phosphatidylcholine (20–25%; Krahmer et al., 2011). Such an important difference, in addition to the proposal that expansion of mature LDs requires very few phospholipids (Penno et al., 2013), suggests that rather than the initial phospholipid composition, a dynamic turnover between different species of phospholipids and lysophospholipids is important during LD formation.
Two major pathways contribute to phosphatidylcholine synthesis: the Kennedy pathway for de novo synthesis of phospholipids (Fig. 2, blue box) and the Lands cycle (Fig. 2, purple box). In the Kennedy pathway, the rate-limiting enzyme is CCT. Before induction of LD formation, CCT1 localizes to the nucleus. However, after loading cells with fatty acids, CCT1 and CCT2 (but not choline kinase or CDP-cholinephosphotransferase [CPT]) localize to LDs (Krahmer et al., 2011). CCT binds to membranes depleted of phosphatidylcholine (Jamil et al., 1990), and in vitro CCT is activated by diacylglycerol and phosphatidic acid (Cornell and Northwood, 2000). Thus, as an LD grows, a relative depletion of phosphatidylcholine and the enrichment in diacylglycerol and phosphatidic acid will promote CCT binding and activation. In Drosophila cells, CCT1 accumulates on LDs containing complexes of neutral lipid synthesis enzymes (Wilfling et al., 2013). However, the final step in the pathway—using diacylglycerol as a substrate—is catalyzed by CPT, an integral membrane protein of the ER (Krahmer et al., 2011). Thus, the newly synthesized phosphatidylcholine is actually produced in the ER and needs to be transferred from the ER to the LD.
Like triacylglycerol, phosphatidylcholine can also be produced locally on LDs, now by the Lands cycle. The Lands cycle and the Kennedy pathway are connected by LPCAT1; overexpression of LPCAT1 increases degradation of CPT (Butler and Mallampalli, 2010). In various mammalian cell lines, LPCAT1 and LPCAT2 have been localized on LDs (Moessinger et al., 2011). The LPCAT-mediated synthesis of phosphatidylcholine requires an acyl-CoA and lysophosphatidylcholine. The acyl-CoA can be produced on LDs by ACSL3 and the lysophosphatidylcholine by PLA2. The conversion of lysophospholipids into phospholipids could be required to protect the nascent globule from lipolysis, to avoid coalescence with other LD, or to adapt the membrane curvature during LD expansion.
Finally, in adipocytes and hepatocytes, phosphatidylethanolamine methyltransferase (PEMT) also contributes to synthesis of phosphatidylcholine (Vance and Vance, 2004). In cultured adipocytes, PEMT localizes to LDs and assembles methyl groups to the phosphatidylethanolamine synthesized by decarboxylation of the phosphatidylserine produced in mitochondria. In adipocytes, this pathway can produce a significant portion of the LD’s phosphatidylcholine (Hörl et al., 2011). In addition, the biosynthesis of ether-linked phospholipids, present in internal neutral lipids and in the phospholipid monolayer of LDs, occurs only in peroxisomes (van den Bosch and de Vet, 1997). Thus, although most components required for LD biogenesis seem to be ER derived, activity of mitochondria and peroxisomes is also required for LD formation.
Accessory proteins with unclear roles
The number of proteins described on LDs has grown exponentially (Yang et al., 2012). Nevertheless, a protein essential for LD formation has not been identified. However, the existence of functional compensation between structural proteins (Sztalryd et al., 2006) makes identification of a single indispensable scaffold unlikely.
Two ER proteins have been proposed to play structural roles during LD formation; seipin (Fld1p in yeast) and the fat storage–inducing transmembrane protein (FITM/FIT; Fig. 3, gray box). Both have membrane-spanning helices (two for seipin and six for FIT) and thus in principle might function from outside the emerging LD (Gross et al., 2010; Cartwright and Goodman, 2012). Indeed, reduced levels of FIT2 in cultured adipocytes or zebrafish decreases triacylglycerol storage in LDs, and overexpression of FIT1/FIT2 in cells, liver, and muscle increases accumulation of triacylglycerol in LDs (Kadereit et al., 2008; Miranda et al., 2011). On the other hand, ablation of seipin in yeast and in fibroblasts results in small, irregular LDs, but also in supersized LDs (Szymanski et al., 2007; Fei et al., 2008). Due to conflicting results from different systems, the role of seipin in LD formation is still unclear. For example seipin overexpression causes the opposite effect, reducing accumulation of triacylglycerol in HeLa cells and adipocytes (Fei et al., 2011b; Cui et al., 2012). Both of these proteins have been hypothesized to contribute to filling LDs with neutral lipids, phospholipids, or proteins (Gross et al., 2011; Cartwright and Goodman, 2012). Indeed, seipin is located at ER–LD junctions in yeast (Szymanski et al., 2007). However, Dga1p moves into LDs even in seipin mutant cells (Jacquier et al., 2011), suggesting that protein transfer is not affected, and that triacylglycerol can be still produced locally on LDs. In contrast, because the last step of the Kennedy pathway mediated by CPT occurs in the ER, but not locally on LDs, deficient ER–LD transport could modify the incorporation of phospholipids into the LD (Fei et al., 2011a). Alternatively, it was also suggested that seipin regulates lipolysis (Fei et al., 2011b).
The accumulation of LDs is the result of a dynamic balance between neutral lipid synthesis and neutral lipid lipolysis (Herms et al., 2013). Regulation of lipolysis seems to be the primary function of perilipins, a family of five related members collectively termed Plin proteins (Kimmel et al., 2010). The best-characterized Plin proteins control lipolysis by recruiting or preventing access of lipases on LDs (Bickel et al., 2009). In adipocytes, emerging LDs are coated first with Plin3 and Plin4, next with Plin2, and finally with Plin1 (Wolins et al., 2006). This interesting observation demonstrates, as previously with ACSL, that LD formation and expansion follows a stepwise and synchronized maturation process. However, Plin1, Plin4, and Plin5 expression is limited to a few tissues (Brasaemle and Wolins, 2012). In contrast, Plin2 and Plin3 are ubiquitously expressed and coat LDs in the majority of cells. In mammalian cells, Plin3 is recruited to the ER during LD biogenesis by recognition of the initial accumulation of diacylglycerol (Skinner et al., 2009) and regulates efficient LD formation (Bulankina et al., 2009). However, many organisms (e.g., yeast) do not have a Plin3 orthologue, and certain cell types (e.g., Drosophila S2 cells) do not express Plin3, suggesting alternative mechanisms of LD formation (Walther and Farese, 2012), or functional compensation between Plin proteins (Sztalryd et al., 2006). Again, redundancy complicates the analysis of how each protein regulates LD metabolism (Sztalryd et al., 2006; Bi et al., 2012).
Interestingly, proteins not directly involved in lipid metabolism also regulate LD formation, e.g., proteins related to ER-associated degradation of proteins, such as AUP1 or the yeast Ubx2/Ubxd8 (Klemm et al., 2011; Spandl et al., 2011; Wang and Lee, 2012). Similarly, LD biogenesis is promoted by unknown mechanisms by many pathogens (Stehr et al., 2012); viruses and bacteria often target LDs for nutritional purposes or as an anti-immunity strategy (Herker and Ott, 2012; Saka and Valdivia, 2012). For example, hepatitis C virus assembles on hepatic LDs (Roingeard and Depla, 2011). Intriguingly, hepatic cells actively respond to this infection by accumulating anti-viral proteins on LDs (Hinson and Cresswell, 2009), and we recently demonstrated that histones on LDs protect Drosophila against bacterial infections (Anand et al., 2012).
Sites and mechanisms of assembly
So far, we have reviewed biochemical reactions mostly occurring in the ER, and connected to LD formation. In this last section, we integrate these data into a single working model describing LD biogenesis. We propose a synchronized, stepwise, and self-reinforcing process involving the ER and triggered by fatty acids.
The ER is a dynamic system of connected membranes forming the nuclear envelope, central rough sheet-like structures, and a polygonal network of tubules extending into the cell periphery (Fig. 1 B; Park and Blackstone, 2010). Functionally, the ER organizes into microdomains specialized for functions such as protein and lipid synthesis, quality control, protein and lipid secretion, or detoxification. These microdomains interact physically, and highly selective yet poorly understood mechanisms allow their functional segregation. Using model peptides to simultaneously visualize the ER and LDs, we have observed that when cultured mammalian cells are treated with fatty acids, LDs preferentially originate in the network of tubules of the cell periphery (Fig. 1 B; Kassan et al., 2013). If lipid arrival continues, these LDs expand and, although apparently in contact with the ER, progressively move into central regions of the cell (Fig. 1 C). A particular lipid composition (phospholipids or free cholesterol) or the presence of specific proteins (ACSL3, Plin2, Nem1p, or seipin) have each been proposed as the mechanism resulting in the lateral segregation of these microdomains (Tauchi-Sato et al., 2002; Robenek et al., 2006; Szymanski et al., 2007; Ohsaki et al., 2009; Adeyo et al., 2011; Kassan et al., 2013).
Regardless of the precise ER sites, and although the following model requires much more experimental testing, the current belief is that neutral lipid deposition occurs between the two leaflets of the ER bilayer (Murphy and Vance, 1999). The 3D tomograms of early LDs confirmed that LDs are encircled by a single layer (Fig. 1, D and E; green arrow) and demonstrated multiple but discrete LD–LD and LD–ER interactions (Fig. 1, C and D; white arrow; Kassan et al., 2013). In principle, this morphology rules out vesicle formation models (Walther and Farese, 2009), being more consistent with the ‘‘blister’’-like organization of lipid globules in the bilayer, shown using simulation models (Khandelia et al., 2010) and described in early electron microscopy studies of the ER of plants (Wanner et al., 1981). Because the yeast Lro1p and Dga1p likely have opposite topological orientations, neutral lipids produced in both leaflets of the ER bilayer can promote LD nucleation (Choudhary et al., 2011).
Initially, newly synthesized neutral lipids seem to be integrated in the ER membrane. However, it is likely that a bilayer only accommodates few molar percent of triacylglycerol or cholesterol esters (Gorrissen et al., 1980, 1982). Thus, at higher concentrations the neutral lipids likely induce phase separation, and the resulting globule is deposited between the two leaflets of the bilayer (Fig. 3 B; Suzuki et al., 2011). These globules, with a predicted diameter of ∼20 nm (Khandelia et al., 2010), are highly mobile and likely move laterally within the membrane (Mountford and Wright, 1988). Such a lateral mobility explains why lipid globules produced by enzymes excluded from the LD, such as Lro1p or ACAT, can efficiently generate LDs. Because triacylglycerol can be enriched in the ER bilayer without inducing LD assembly (Chang et al., 2006; Gubern et al., 2008; Adeyo et al., 2011), additional mechanisms to nucleate LDs are required.
In vitro, a small lipid globule of a few nanometers is sufficient to generate ER membrane curvature stable for at least a few microseconds (Khandelia et al., 2010). Also in vitro, it was suggested that curvature produced by these globules is supported, and likely even driven, by gradual demixing of the phospholipids of the monolayer (Zanghellini et al., 2010). However, in vivo, local formation of other lipid species such as conical lysophospholipids likely generates stronger and more stable membrane bending. For example, although it has a very short half-life, lysophosphatidic acid has the most positive spontaneous curvature of any membrane lipid (Kooijman et al., 2005). During LD assembly, lysophosphatidic acid can be produced by the combined activity of GPAT, PLA2, and PLD (Fig. 2). In addition, the lysophosphatidylcholine produced by PLA2 also positively curves membranes (Fuller and Rand, 2001). Because GPATs have not yet been described in nascent LDs, we favor the two lipases as the most likely candidates involved at this early stage of biogenesis. Indeed, although the precise role of lysophospholipids needs to be experimentally addressed, both PLA and PLD are essential for LD formation in vivo and in vitro (Marchesan et al., 2003; Andersson et al., 2006; Gubern et al., 2008; Du et al., 2009).
Amphipathic helices—which can produce membrane bending by themselves—also function as membrane curvature sensors (McMahon and Gallop, 2005). Thus, once the lipid globule has generated bending of the membrane, proteins with amphipathic helices and anchored to the ER (and thus in proximity to the globule) will likely recognize this curvature. Indeed, a significant number of proteins are anchored on LDs by amphipathic helices (Walther and Farese, 2012), and thus it is in principle an efficient mechanism for recognizing nascent LDs. The amphipathic helices are predicted to sit flat on the membrane surface, with the hydrophobic residues dipping into the hydrophobic phase of the membrane. The hydrophobic interaction of the helix with the membrane is short-range but strong, and thus will provide the thermodynamic equilibrium required to stabilize a budding structure and mark the onset of LD biogenesis (Fig. 3 B). Consistent with this model, in vitro studies using ER microsomes from Helianthus annuus show formation of globules in the ER bilayer that contained mobile triacylglycerol and that were able to recruit proteins such as oleosin (Lacey et al., 1999). 1H-NMR spectroscopy analysis was consistent with deposition of these globules in the ER bilayer, and suggested the existence of two populations of globules: a highly mobile fraction free of significant amounts of associated proteins and a second population with lower mobility and containing associated proteins. Further, it was recently shown that oleosins (and Plin proteins) promote the sequestration of neutral lipids from the ER bilayer and induce LD formation (Jacquier et al., 2013).
Interestingly, ACSL3 can function similarly to oleosins, arriving very early to the assembly sites. ACSL3 contains amphipathic helices in the proximity of the globule because it is anchored to the ER membrane (Poppelreuther et al., 2012; Kassan et al., 2013). In the nucleation sites, ACSL3 can interact with other proteins and simultaneously produce a concomitant local enrichment of the acyl-CoA required in many reactions of the biogenesis process. The enzymes described in Fig. 3 A will be particularly efficient at the sites of assembly and organize in complexes (Wilfling et al., 2013). From here, neutral lipids will be rapidly and locally produced and deposited in the underlying globule to generate the nascent LD. Simultaneously, numerous bioactive lipid intermediates, such as phosphatidic acid and diacylglycerol, will be now locally generated, accumulated, and transformed. These lipids will shape the membranes and function as platforms for protein recruitment. New phospholipids to support LD expansion can be generated in the neighboring tubules via CPT, using the diacylglycerol and the CDP-choline produced by PAP and CCT on LDs (Fig. 3 A). Proteins such as seipin and FIT could be involved during the exchange. Next, conversion of conical lysophospholipids into cylindrical phospholipids, occurring on the LD by the activity of LPCAT, will progressively adjust the growing structure to the new required curvature (Fig. 3 A).
Finally, transition from a concave structure (nascent LD) into a globular organelle (early LD) will require generation of negative curvature at the base of the LD (Fig. 1 F). As in the Golgi complex, this can be achieved by generation of phosphatidic acid, which has a considerable negative spontaneous curvature (Kooijman et al., 2005). Phosphatidic acid can be produced by AGPAT, DGK1, or PLD at the ER–LD interface (Fig. 2), or even displaced to these sites during phospholipid demixing of the LD monolayer.
Because the accumulation of LDs is the result of the balance between neutral lipid synthesis and lipolysis (Herms et al., 2013), and some yeast lipases are recruited very early in LDs (Jacquier et al., 2011), preventing nascent LD lipolysis is likely required during biogenesis (Fig. 2, orange box; Zechner et al., 2012). This could be achieved by accumulation of phosphatidylcholine, which has a larger head group and is more cylindrical than other phospholipids (Vance and Vance, 2009), and thus will efficiently shield the LD and additionally avoid coalescence between neighboring LDs. Further, the presence of seipin and the recruitment of Plin2 and Plin3 could also control access of lipases to the nucleation sites.
Once formed, although some models proposed bicelle excision (Ploegh, 2007), the expanding LD likely remains connected with the ER. Real-time microscopy of nascent LD formation did not show perceptible budding or detachment processes involving ER membranes (Kassan et al., 2013). An intimate association of mature LDs with the ER was observed in early studies (Novikoff et al., 1980; Blanchette-Mackie et al., 1995). Physical bridges may permanently connect LDs and the ER (Ohsaki et al., 2008; Fujimoto and Parton, 2011; Jacquier et al., 2011; Wilfling et al., 2013) and allow active partitioning of proteins and lipids between both organelles. Further, the ER–LD contact can be regulated by proteins such as Rab18 and Plin2 (Martin et al., 2005; Ozeki et al., 2005). However, how nascent and early LDs move from the periphery and accumulate as mature LDs in the cell center will require additional analysis. Further, because some LDs have a bidirectional saltatory movement (Pol et al., 2004; Welte, 2009), it seems likely that these ER bridges are released under some circumstances.
In conclusion, the assembly of LDs is driven by complex but highly robust mechanisms, likely reflecting the physiological significance of this organelle even beyond its function in fat storage and lipid homeostasis. Indeed, LDs can dynamically exchange lipids and proteins with other organelles, and thus provide timely additional space for sequestration to avoid lipid and protein degradation, to keep them inactive, or even to control the protein folding state. Their rapid assembly and remodeling make LDs well suited to contribute to these processes, and it is thus not surprising that altered LD accumulation frequently correlates with disease. Further understanding of the molecules and mechanisms involved in LD formation will undoubtedly provide additional insights into these disease conditions.
Acknowledgments
We would like to thank M. Bosch and the rest of the members of the Pol laboratory for the critical reading of the Review, and to A. Herms for providing the confocal images shown in Fig. 1, A and C.
A. Pol is supported by grants from MICINN and Marató de TV3 (BFU2011-23745 and CSD2009-00016); R.G. Parton by a fellowship from the National Health and Medical Research Council of Australia (grant no. 569542); and S.P. Gross by a grant from the National Institutes of Health (GM64624).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ACS
- acyl-CoA synthetase
- AGPAT
- 1-acylglycerol-3-phosphate O-acyltransferase
- CCT
- CTP:phosphocholine cytidylyltransferase
- CoA
- coenzyme A
- CPT
- cholinephosphotransferase
- DGAT
- diacylglycerol acyltransferase
- FIT
- fat storage–inducing transmembrane
- GPAT
- glycerol-3-phosphate O-acyltransferase
- LD
- lipid droplet
- LPCAT
- lysophosphatidylcholine acyltransferase
- PAP
- phosphatidic acid phosphatase
References
- Adeyo O., Horn P.J., Lee S., Binns D.D., Chandrahas A., Chapman K.D., Goodman J.M. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192:1043–1055 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P., Cermelli S., Li Z., Kassan A., Bosch M., Sigua R., Huang L., Ouellette A.J., Pol A., Welte M.A., Gross S.P. 2012. A novel role for lipid droplets in the organismal antibacterial response. Elife. 1:e00003 10.7554/eLife.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L., Boström P., Ericson J., Rutberg M., Magnusson B., Marchesan D., Ruiz M., Asp L., Huang P., Frohman M.A., et al. 2006. PLD1 and ERK2 regulate cytosolic lipid droplet formation. J. Cell Sci. 119:2246–2257 10.1242/jcs.02941 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G. 1997. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J. Bacteriol. 179:7611–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayciriex S., Le Guédard M., Camougrand N., Velours G., Schoene M., Leone S., Wattelet-Boyer V., Dupuy J.W., Shevchenko A., Schmitter J.M., et al. 2012. YPR139c/LOA1 encodes a novel lysophosphatidic acid acyltransferase associated with lipid droplets and involved in TAG homeostasis. Mol. Biol. Cell. 23:233–246 10.1091/mbc.E11-07-0650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R., Li W.H., Venables B., Zehmer J.K., Roth M.R., Welti R., Anderson R.G., Liu P., Chapman K.D. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48:837–847 10.1194/jlr.M600413-JLR200 [DOI] [PubMed] [Google Scholar]
- Bi J., Xiang Y., Chen H., Liu Z., Grönke S., Kühnlein R.P., Huang X. 2012. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J. Cell Sci. 125:3568–3577 10.1242/jcs.101329 [DOI] [PubMed] [Google Scholar]
- Bickel P.E., Tansey J.T., Welte M.A. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791:419–440 10.1016/j.bbalip.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette-Mackie E.J., Dwyer N.K., Barber T., Coxey R.A., Takeda T., Rondinone C.M., Theodorakis J.L., Greenberg A.S., Londos C. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 36:1211–1226 [PubMed] [Google Scholar]
- Borgese N., Brambillasca S., Colombo S. 2007. How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19:368–375 10.1016/j.ceb.2007.04.019 [DOI] [PubMed] [Google Scholar]
- Brasaemle D.L., Wolins N.E. 2012. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J. Biol. Chem. 287:2273–2279 10.1074/jbc.R111.309088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle D.L., Dolios G., Shapiro L., Wang R. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279:46835–46842 10.1074/jbc.M409340200 [DOI] [PubMed] [Google Scholar]
- Bulankina A.V., Deggerich A., Wenzel D., Mutenda K., Wittmann J.G., Rudolph M.G., Burger K.N., Höning S. 2009. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185:641–655 10.1083/jcb.200812042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P.L., Mallampalli R.K. 2010. Cross-talk between remodeling and de novo pathways maintains phospholipid balance through ubiquitination. J. Biol. Chem. 285:6246–6258 10.1074/jbc.M109.017350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright B.R., Goodman J.M. 2012. Seipin: from human disease to molecular mechanism. J. Lipid Res. 53:1042–1055 10.1194/jlr.R023754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B.H., Li L., Paul A., Taniguchi S., Nannegari V., Heird W.C., Chan L. 2006. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 26:1063–1076 10.1128/MCB.26.3.1063-1076.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K.D., Dyer J.M., Mullen R.T. 2012. Biogenesis and functions of lipid droplets in plants: Thematic review series: lipid droplet synthesis and metabolism: from yeast to man. J. Lipid Res. 53:215–226 10.1194/jlr.R021436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Greenberg A.S., Wang S.M. 2002. Oleic acid-induced PKC isozyme translocation in RAW 264.7 macrophages. J. Cell. Biochem. 86:784–791 10.1002/jcb.10266 [DOI] [PubMed] [Google Scholar]
- Choudhary V., Jacquier N., Schneiter R. 2011. The topology of the triacylglycerol synthesizing enzyme Lro1 indicates that neutral lipids can be produced within the luminal compartment of the endoplasmatic reticulum: Implications for the biogenesis of lipid droplets. Commun. Integr. Biol. 4:781–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell R.B., Northwood I.C. 2000. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci. 25:441–447 10.1016/S0968-0004(00)01625-X [DOI] [PubMed] [Google Scholar]
- Cui X., Wang Y., Meng L., Fei W., Deng J., Xu G., Peng X., Ju S., Zhang L., Liu G., et al. 2012. Overexpression of a short human seipin/BSCL2 isoform in mouse adipose tissue results in mild lipodystrophy. Am. J. Physiol. Endocrinol. Metab. 302:E705–E713 10.1152/ajpendo.00237.2011 [DOI] [PubMed] [Google Scholar]
- Czabany T., Wagner A., Zweytick D., Lohner K., Leitner E., Ingolic E., Daum G. 2008. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283:17065–17074 10.1074/jbc.M800401200 [DOI] [PubMed] [Google Scholar]
- Doege H., Stahl A. 2006. Protein-mediated fatty acid uptake: novel insights from in vivo models. Physiology (Bethesda). 21:259–268 10.1152/physiol.00014.2006 [DOI] [PubMed] [Google Scholar]
- Du L., Hickey R.W., Bayir H., Watkins S.C., Tyurin V.A., Guo F., Kochanek P.M., Jenkins L.W., Ren J., Gibson G., et al. 2009. Starving neurons show sex difference in autophagy. J. Biol. Chem. 284:2383–2396 10.1074/jbc.M804396200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman S.W., Yassaee M., Bieniasz P.D. 2009. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J. Cell Biol. 184:881–894 10.1083/jcb.200808041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.M., Frahm J.L., Li L.O., Coleman R.A. 2010. Acyl-coenzyme A synthetases in metabolic control. Curr. Opin. Lipidol. 21:212–217 10.1097/MOL.0b013e32833884bb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., Yang H. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Du X., Yang H. 2011a. Seipin, adipogenesis and lipid droplets. Trends Endocrinol. Metab. 22:204–210 10.1016/j.tem.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Fei W., Li H., Shui G., Kapterian T.S., Bielby C., Du X., Brown A.J., Li P., Wenk M.R., Liu P., Yang H. 2011b. Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J. Lipid Res. 52:2136–2147 10.1194/jlr.M017566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T., Parton R.G. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3:a004838 10.1101/cshperspect.a004838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 1644:47–59 10.1016/j.bbamcr.2003.10.018 [DOI] [PubMed] [Google Scholar]
- Fujimoto Y., Itabe H., Kinoshita T., Homma K.J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., Takano T. 2007. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 48:1280–1292 10.1194/jlr.M700050-JLR200 [DOI] [PubMed] [Google Scholar]
- Füllekrug J., Ehehalt R., Poppelreuther M. 2012. Outlook: membrane junctions enable the metabolic trapping of fatty acids by intracellular acyl-CoA synthetases. Front Physiol. 3:401 10.3389/fphys.2012.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller N., Rand R.P. 2001. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophys. J. 81:243–254 10.1016/S0006-3495(01)75695-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J., Padamsee M., Wilcox L., Oelkers P.M., D’Ambrosio D., Ruggles K.V., Ramsey N., Jabado O., Turkish A., Sturley S.L. 2009. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 284:30994–31005 10.1074/jbc.M109.050443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrissen H., Tulloch A.P., Cushley R.J. 1980. Deuterium magnetic resonance of selectively deuterated cholesteryl esters in phosphatidylcholine vesicles. Biochemistry. 19:3422–3429 10.1021/bi00556a003 [DOI] [PubMed] [Google Scholar]
- Gorrissen H., Tulloch A.P., Cushley R.J. 1982. Deuterium magnetic resonance of triacylglycerols in phospholipid bilayers. Chem. Phys. Lipids. 31:245–255 10.1016/0009-3084(82)90060-3 [DOI] [PubMed] [Google Scholar]
- Gross D.A., Snapp E.L., Silver D.L. 2010. Structural insights into triglyceride storage mediated by fat storage-inducing transmembrane (FIT) protein 2. PLoS ONE. 5:e10796 10.1371/journal.pone.0010796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D.A., Zhan C., Silver D.L. 2011. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. USA. 108:19581–19586 10.1073/pnas.1110817108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubern A., Casas J., Barceló-Torns M., Barneda D., de la Rosa X., Masgrau R., Picatoste F., Balsinde J., Balboa M.A., Claro E. 2008. Group IVA phospholipase A2 is necessary for the biogenesis of lipid droplets. J. Biol. Chem. 283:27369–27382 10.1074/jbc.M800696200 [DOI] [PubMed] [Google Scholar]
- Ha K.D., Clarke B.A., Brown W.J. 2012. Regulation of the Golgi complex by phospholipid remodeling enzymes. Biochim. Biophys. Acta. 1821:1078–1088 10.1016/j.bbalip.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapala I., Marza E., Ferreira T. 2011. Is fat so bad? Modulation of endoplasmic reticulum stress by lipid droplet formation. Biol. Cell. 103:271–285 10.1042/BC20100144 [DOI] [PubMed] [Google Scholar]
- Harris C.A., Haas J.T., Streeper R.S., Stone S.J., Kumari M., Yang K., Han X., Brownell N., Gross R.W., Zechner R., Farese R.V., Jr 2011. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 52:657–667 10.1194/jlr.M013003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E., Ott M. 2012. Emerging role of lipid droplets in host/pathogen interactions. J. Biol. Chem. 287:2280–2287 10.1074/jbc.R111.300202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms A., Bosch M., Ariotti N., Reddy B.J., Fajardo A., Fernández-Vidal A., Alvarez-Guaita A., Fernández-Rojo M.A., Rentero C., Tebar F., et al. 2013. Cell-to-cell heterogeneity in lipid droplets suggests a mechanism to reduce lipotoxicity. Curr. Biol. 23:1489–1496 10.1016/j.cub.2013.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson E.R., Cresswell P. 2009. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc. Natl. Acad. Sci. USA. 106:20452–20457 10.1073/pnas.0911679106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörl G., Wagner A., Cole L.K., Malli R., Reicher H., Kotzbeck P., Köfeler H., Höfler G., Frank S., Bogner-Strauss J.G., et al. 2011. Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo. J. Biol. Chem. 286:17338–17350 10.1074/jbc.M111.234534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igal R.A., Coleman R.A. 1996. Acylglycerol recycling from triacylglycerol to phospholipid, not lipase activity, is defective in neutral lipid storage disease fibroblasts. J. Biol. Chem. 271:16644–16651 10.1074/jbc.271.28.16644 [DOI] [PubMed] [Google Scholar]
- Ingelmo-Torres M., González-Moreno E., Kassan A., Hanzal-Bayer M., Tebar F., Herms A., Grewal T., Hancock J.F., Enrich C., Bosch M., et al. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic. 10:1785–1801 10.1111/j.1600-0854.2009.00994.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., Schneiter R. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Jacquier N., Mishra S., Choudhary V., Schneiter R. 2013. Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J. Cell Sci. 126:5198–5209 10.1242/jcs.131896 [DOI] [PubMed] [Google Scholar]
- Jamil H., Yao Z.M., Vance D.E. 1990. Feedback regulation of CTP:phosphocholine cytidylyltransferase translocation between cytosol and endoplasmic reticulum by phosphatidylcholine. J. Biol. Chem. 265:4332–4339 [PubMed] [Google Scholar]
- Kadereit B., Kumar P., Wang W.J., Miranda D., Snapp E.L., Severina N., Torregroza I., Evans T., Silver D.L. 2008. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA. 105:94–99 10.1073/pnas.0708579105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Han G.S., Xu Z., Carman G.M., Siniossoglou S. 2010. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA. 107:17539–17544 10.1073/pnas.1007974107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E., Barbosa A.D., Sembongi H., Mari M., Han G.S., Reggiori F., Carman G.M., Siniossoglou S. 2013. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell. 24:2124–2133 10.1091/mbc.E13-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelia H., Duelund L., Pakkanen K.I., Ipsen J.H. 2010. Triglyceride blisters in lipid bilayers: implications for lipid droplet biogenesis and the mobile lipid signal in cancer cell membranes. PLoS ONE. 5:e12811 10.1371/journal.pone.0012811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelef N., Buton X., Beatini N., Wang H., Meiner V., Chang T.Y., Farese R.V., Jr, Maxfield F.R., Tabas I. 1998. Immunolocalization of acyl-coenzyme A:cholesterol O-acyltransferase in macrophages. J. Biol. Chem. 273:11218–11224 10.1074/jbc.273.18.11218 [DOI] [PubMed] [Google Scholar]
- Kimmel A.R., Brasaemle D.L., McAndrews-Hill M., Sztalryd C., Londos C. 2010. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51:468–471 10.1194/jlr.R000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm E.J., Spooner E., Ploegh H.L. 2011. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J. Biol. Chem. 286:37602–37614 10.1074/jbc.M111.284794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman E.E., Chupin V., Fuller N.L., Kozlov M.M., de Kruijff B., Burger K.N., Rand P.R. 2005. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 44:2097–2102 10.1021/bi0478502 [DOI] [PubMed] [Google Scholar]
- Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H.W., Schmidt-Supprian M., Vance D.E., Mann M., et al. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14:504–515 10.1016/j.cmet.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N., Farese R.V., Jr, Walther T.C. 2013. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 5:905–915 10.1002/emmm.201100671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause B.R., Hartman A.D. 1984. Adipose tissue and cholesterol metabolism. J. Lipid Res. 25:97–110 [PubMed] [Google Scholar]
- Kuerschner L., Moessinger C., Thiele C. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 9:338–352 10.1111/j.1600-0854.2007.00689.x [DOI] [PubMed] [Google Scholar]
- Kühnlein R.P. 2012. Thematic review series: Lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J. Lipid Res. 53:1430–1436 10.1194/jlr.R024299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey D., Beaudoin F., Dempsey C.E., Shewry P.R., Napier J.A. 1999. The accumulation of triacylglycerols within the endoplasmic reticulum of developing seeds of Helianthus annuus. Plant J. 17:397–405 10.1046/j.1365-313X.1999.00387.x [DOI] [Google Scholar]
- Lands W.E. 2000. Stories about acyl chains. Biochim. Biophys. Acta. 1483:1–14 10.1016/S1388-1981(99)00177-8 [DOI] [PubMed] [Google Scholar]
- Liu P., Ying Y., Zhao Y., Mundy D.I., Zhu M., Anderson R.G. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279:3787–3792 10.1074/jbc.M311945200 [DOI] [PubMed] [Google Scholar]
- Long A.P., Manneschmidt A.K., VerBrugge B., Dortch M.R., Minkin S.C., Prater K.E., Biggerstaff J.P., Dunlap J.R., Dalhaimer P. 2012. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic. 13:705–714 10.1111/j.1600-0854.2012.01339.x [DOI] [PubMed] [Google Scholar]
- Mak H.Y. 2012. Lipid droplets as fat storage organelles in Caenorhabditis elegans: Thematic review series: Lipid droplet synthesis and metabolism: From yeast to man. J. Lipid Res. 53:28–33 10.1194/jlr.R021006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V., Campelo F. 2011. PKD regulates membrane fission to generate TGN to cell surface transport carriers. Cold Spring Harb. Perspect. Biol. 3:a005280 10.1101/cshperspect.a005280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesan D., Rutberg M., Andersson L., Asp L., Larsson T., Borén J., Johansson B.R., Olofsson S.O. 2003. A phospholipase D-dependent process forms lipid droplets containing caveolin, adipocyte differentiation-related protein, and vimentin in a cell-free system. J. Biol. Chem. 278:27293–27300 10.1074/jbc.M301430200 [DOI] [PubMed] [Google Scholar]
- Martin S., Driessen K., Nixon S.J., Zerial M., Parton R.G. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280:42325–42335 10.1074/jbc.M506651200 [DOI] [PubMed] [Google Scholar]
- McMahon H.T., Gallop J.L. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438:590–596 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- Milewska M., McRedmond J., Byrne P.C. 2009. Identification of novel spartin-interactors shows spartin is a multifunctional protein. J. Neurochem. 111:1022–1030 10.1111/j.1471-4159.2009.06382.x [DOI] [PubMed] [Google Scholar]
- Miranda D.A., Koves T.R., Gross D.A., Chadt A., Al-Hasani H., Cline G.W., Schwartz G.J., Muoio D.M., Silver D.L. 2011. Re-patterning of skeletal muscle energy metabolism by fat storage-inducing transmembrane protein 2. J. Biol. Chem. 286:42188–42199 10.1074/jbc.M111.297127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. 2011. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 286:21330–21339 10.1074/jbc.M110.202424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountford C.E., Wright L.C. 1988. Organization of lipids in the plasma membranes of malignant and stimulated cells: a new model. Trends Biochem. Sci. 13:172–177 10.1016/0968-0004(88)90145-4 [DOI] [PubMed] [Google Scholar]
- Murphy D.J., Vance J. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24:109–115 10.1016/S0968-0004(98)01349-8 [DOI] [PubMed] [Google Scholar]
- Nakamura N., Fujimoto T. 2003. Adipose differentiation-related protein has two independent domains for targeting to lipid droplets. Biochem. Biophys. Res. Commun. 306:333–338 10.1016/S0006-291X(03)00979-3 [DOI] [PubMed] [Google Scholar]
- Novikoff A.B., Novikoff P.M., Rosen O.M., Rubin C.S. 1980. Organelle relationships in cultured 3T3-L1 preadipocytes. J. Cell Biol. 87:180–196 10.1083/jcb.87.1.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y., Cheng J., Suzuki M., Fujita A., Fujimoto T. 2008. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J. Cell Sci. 121:2415–2422 10.1242/jcs.025452 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Cheng J., Suzuki M., Shinohara Y., Fujita A., Fujimoto T. 2009. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim. Biophys. Acta. 1791:399–407 10.1016/j.bbalip.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Ost A., Ortegren U., Gustavsson J., Nystrom F.H., Strålfors P. 2005. Triacylglycerol is synthesized in a specific subclass of caveolae in primary adipocytes. J. Biol. Chem. 280:5–8 [DOI] [PubMed] [Google Scholar]
- Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118:2601–2611 10.1242/jcs.02401 [DOI] [PubMed] [Google Scholar]
- Park S.H., Blackstone C. 2010. Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 11:515–521 10.1038/embor.2010.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penno A., Hackenbroich G., Thiele C. 2013. Phospholipids and lipid droplets. Biochim. Biophys. Acta. 1831:589–594 10.1016/j.bbalip.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Ploegh H.L. 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 448:435–438 10.1038/nature06004 [DOI] [PubMed] [Google Scholar]
- Pol A., Martin S., Fernandez M.A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R.G. 2004. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell. 15:99–110 10.1091/mbc.E03-06-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppelreuther M., Rudolph B., Du C., Großmann R., Becker M., Thiele C., Ehehalt R., Füllekrug J. 2012. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 53:888–900 10.1194/jlr.M024562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Daum G. 2010a. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell. 21:501–510 10.1091/mbc.E09-09-0775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Daum G. 2010b. Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 285:15769–15776 10.1074/jbc.M109.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao G.A., Johnston J.M. 1966. Purification and properties of triglyceride synthetase from the intestinal mucosa. Biochim. Biophys. Acta. 125:465–473 10.1016/0005-2760(66)90035-X [DOI] [PubMed] [Google Scholar]
- Robenek H., Hofnagel O., Buers I., Robenek M.J., Troyer D., Severs N.J. 2006. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J. Cell Sci. 119:4215–4224 10.1242/jcs.03191 [DOI] [PubMed] [Google Scholar]
- Roingeard P., Depla M. 2011. The birth and life of lipid droplets: learning from the hepatitis C virus. Biol. Cell. 103:223–231 10.1042/BC20100119 [DOI] [PubMed] [Google Scholar]
- Saka H.A., Valdivia R. 2012. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu. Rev. Cell Dev. Biol. 28:411–437 10.1146/annurev-cellbio-092910-153958 [DOI] [PubMed] [Google Scholar]
- Sandoval A., Fraisl P., Arias-Barrau E., Dirusso C.C., Singer D., Sealls W., Black P.N. 2008. Fatty acid transport and activation and the expression patterns of genes involved in fatty acid trafficking. Arch. Biochem. Biophys. 477:363–371 10.1016/j.abb.2008.06.010 [DOI] [PubMed] [Google Scholar]
- Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. 2006. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J. Biochem. 139:921–930 10.1093/jb/mvj104 [DOI] [PubMed] [Google Scholar]
- Sembongi H., Miranda M., Han G.S., Fakas S., Grimsey N., Vendrell J., Carman G.M., Siniossoglou S. 2013. Distinct roles of the phosphatidate phosphatases lipin 1 and 2 during adipogenesis and lipid droplet biogenesis in 3T3-L1 cells. J. Biol. Chem. 288:34502–34513 10.1074/jbc.M113.488445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S. 2013. Phospholipid metabolism and nuclear function: roles of the lipin family of phosphatidic acid phosphatases. Biochim. Biophys. Acta. 1831:575–581 10.1016/j.bbalip.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Skinner J.R., Shew T.M., Schwartz D.M., Tzekov A., Lepus C.M., Abumrad N.A., Wolins N.E. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284:30941–30948 10.1074/jbc.M109.013995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandl J., Lohmann D., Kuerschner L., Moessinger C., Thiele C. 2011. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J. Biol. Chem. 286:5599–5606 10.1074/jbc.M110.190785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehr M., Elamin A.A., Singh M. 2012. Cytosolic lipid inclusions formed during infection by viral and bacterial pathogens. Microbes Infect. 14:1227–1237 10.1016/j.micinf.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Stevanovic A., Thiele C. 2013. Monotopic topology is required for lipid droplet targeting of ancient ubiquitous protein 1. J. Lipid Res. 54:503–513 10.1194/jlr.M033852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.J., Levin M.C., Zhou P., Han J., Walther T.C., Farese R.V., Jr 2009. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J. Biol. Chem. 284:5352–5361 10.1074/jbc.M805768200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturley S.L., Hussain M.M. 2012. Lipid droplet formation on opposing sides of the endoplasmic reticulum. J. Lipid Res. 53:1800–1810 10.1194/jlr.R028290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Shinohara Y., Ohsaki Y., Fujimoto T. 2011. Lipid droplets: size matters. J. Electron Microsc. (Tokyo). 60(Suppl 1):S101–S116 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Iio Y., Saito N., Fujimoto T. 2013. Protein kinase Cη is targeted to lipid droplets. Histochem. Cell Biol. 139:505–511 10.1007/s00418-013-1083-z [DOI] [PubMed] [Google Scholar]
- Suzuki R., Sakamoto S., Tsutsumi T., Rikimaru A., Tanaka K., Shimoike T., Moriishi K., Iwasaki T., Mizumoto K., Matsuura Y., et al. 2005. Molecular determinants for subcellular localization of hepatitis C virus core protein. J. Virol. 79:1271–1281 10.1128/JVI.79.2.1271-1281.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztalryd C., Bell M., Lu X., Mertz P., Hickenbottom S., Chang B.H., Chan L., Kimmel A.R., Londos C. 2006. Functional compensation for adipose differentiation-related protein (ADFP) by Tip47 in an ADFP null embryonic cell line. J. Biol. Chem. 281:34341–34348 10.1074/jbc.M602497200 [DOI] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., Goodman J.M. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J. Biol. Chem. 277:44507–44512 10.1074/jbc.M207712200 [DOI] [PubMed] [Google Scholar]
- Tomoda H., Igarashi K., Omura S. 1987. Inhibition of acyl-CoA synthetase by triacsins. Biochim. Biophys. Acta. 921:595–598 10.1016/0005-2760(87)90088-9 [DOI] [PubMed] [Google Scholar]
- Vallet-Pichard A., Mallet V., Pol S. 2012. Nonalcoholic fatty liver disease and HIV infection. Semin. Liver Dis. 32:158–166 10.1055/s-0032-1316471 [DOI] [PubMed] [Google Scholar]
- van den Bosch H., de Vet E.C. 1997. Alkyl-dihydroxyacetonephosphate synthase. Biochim. Biophys. Acta. 1348:35–44 10.1016/S0005-2760(97)00107-0 [DOI] [PubMed] [Google Scholar]
- Vance D.E., Vance J.E. 2009. Physiological consequences of disruption of mammalian phospholipid biosynthetic genes. J. Lipid Res. 50(Suppl):S132–S137 10.1194/jlr.R800048-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J.E., Vance D.E. 2004. Phospholipid biosynthesis in mammalian cells. Biochem. Cell Biol. 82:113–128 10.1139/o03-073 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Farese R.V., Jr 2009. The life of lipid droplets. Biochim. Biophys. Acta. 1791:459–466 10.1016/j.bbalip.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Farese R.V., Jr 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81:687–714 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Lee S.C. 2012. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J. Cell Sci. 125:2930–2939 10.1242/jcs.100230 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang J., Qiu W., Han G.S., Carman G.M., Adeli K. 2011. Lipin-1γ isoform is a novel lipid droplet-associated protein highly expressed in the brain. FEBS Lett. 585:1979–1984 10.1016/j.febslet.2011.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner G., Formanek H., Theimer R.R. 1981. The ontogeny of lipid bodies (spherosomes) in plant cells: Ultrastructural evidence. Planta. 151:109–123 10.1007/BF00387812 [DOI] [PubMed] [Google Scholar]
- Watkins P.A., Maiguel D., Jia Z., Pevsner J. 2007. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 48:2736–2750 10.1194/jlr.M700378-JLR200 [DOI] [PubMed] [Google Scholar]
- Welte M.A. 2009. Fat on the move: intracellular motion of lipid droplets. Biochem. Soc. Trans. 37:991–996 10.1042/BST0370991 [DOI] [PubMed] [Google Scholar]
- Wilfling F., Wang H., Haas J.T., Krahmer N., Gould T.J., Uchida A., Cheng J.X., Graham M., Christiano R., Fröhlich F., et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24:384–399 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins N.E., Brasaemle D.L., Bickel P.E. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580:5484–5491 10.1016/j.febslet.2006.08.040 [DOI] [PubMed] [Google Scholar]
- Xu N., Zhang S.O., Cole R.A., McKinney S.A., Guo F., Haas J.T., Bobba S., Farese R.V., Jr, Mak H.Y. 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198:895–911 10.1083/jcb.201201139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.S., Valente C., Polishchuk R.S., Turacchio G., Layre E., Moody D.B., Leslie C.C., Gelb M.H., Brown W.J., Corda D., et al. 2011. COPI acts in both vesicular and tubular transport. Nat. Cell Biol. 13:996–1003 10.1038/ncb2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ding Y., Chen Y., Zhang S., Huo C., Wang Y., Yu J., Zhang P., Na H., Zhang H., et al. 2012. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 53:1245–1253 10.1194/jlr.R024117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghellini J., Wodlei F., von Grünberg H.H. 2010. Phospholipid demixing and the birth of a lipid droplet. J. Theor. Biol. 264:952–961 10.1016/j.jtbi.2010.02.025 [DOI] [PubMed] [Google Scholar]
- Zechner R., Zimmermann R., Eichmann T.O., Kohlwein S.D., Haemmerle G., Lass A., Madeo F. 2012. FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15:279–291 10.1016/j.cmet.2011.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweytick D., Leitner E., Kohlwein S.D., Yu C., Rothblatt J., Daum G. 2000. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267:1075–1082 10.1046/j.1432-1327.2000.01103.x [DOI] [PubMed] [Google Scholar]