Contrary to earlier findings, preperoxisomal membrane structures form in yeast cells lacking the peroxin Pex3 and are competent to mature into functional peroxisomes upon Pex3 reintroduction.
Abstract
We demonstrate that the peroxin Pex3 is not required for the formation of peroxisomal membrane structures in yeast pex3 mutant cells. Notably, pex3 mutant cells already contain reticular and vesicular structures that harbor key proteins of the peroxisomal receptor docking complex—Pex13 and Pex14—as well as the matrix proteins Pex8 and alcohol oxidase. Other peroxisomal membrane proteins in these cells are unstable and transiently localized to the cytosol (Pex10, Pmp47) or endoplasmic reticulum (Pex11). These reticular and vesicular structures are more abundant in cells of a pex3 atg1 double deletion strain, as the absence of Pex3 may render them susceptible to autophagic degradation, which is blocked in this double mutant. Contrary to earlier suggestions, peroxisomes are not formed de novo from the endoplasmic reticulum when the PEX3 gene is reintroduced in pex3 cells. Instead, we find that reintroduced Pex3 sorts to the preperoxisomal structures in pex3 cells, after which these structures mature into normal peroxisomes.
Introduction
Peroxisomes are ubiquitous cell organelles that are involved in a large variety of metabolic functions (Wanders and Waterham, 2006; Hu et al., 2012; Kohlwein et al., 2013). It is generally accepted that peroxisomes proliferate by fission or form de novo from the ER. Although the question of which mechanism of organelle multiplication prevails in wild-type (WT) cells is a matter of debate, data obtained in yeast indicate that peroxisome fission is the most likely mechanism of peroxisome proliferation in normal WT cells (Motley and Hettema, 2007; Nagotu et al., 2008; Saraya et al., 2011).
In pex3 mutant cells, which are reported to lack peroxisomal membrane structures, new organelles appear upon reintroduction of the PEX3 gene. A generally accepted view is that in these cells reintroduced Pex3 sorts to the ER, followed by the formation of preperoxisomal structures, which pinch off and develop into mature peroxisomes. It has been suggested that all peroxisomal membrane proteins (PMPs) accumulate at the ER in pex3 cells (van der Zand et al., 2010), and that upon reintroduction of Pex3 these PMPs are incorporated in two types of vesicles that fuse to form peroxisomes (van der Zand et al., 2012). According to this model, Pex3 is important for the exit of PMPs from the ER into preperoxisomal vesicles.
To date, relatively little is known about the molecular mechanisms involved in the reintroduction of peroxisomes in pex3 cells. Here, we reinvestigated this process, focusing on the ultrastructure of these cells and the subcellular localization of different PMPs before and after reintroduction of Pex3 using a Hansenula polymorpha pex3 atg1 double deletion strain. The rationale for this approach is that we have previously shown that removal of Pex3 from the peroxisomal membrane is an essential early step in selective autophagic degradation of peroxisomes (Bellu et al., 2002; Williams and van der Klei, 2013). This implies that the presence of Pex3 at the peroxisomal membrane protects the organelles against autophagy. Hence, if peroxisomal membrane structures develop in pex3 cells, they are likely to be rapidly degraded after their formation. To prevent autophagy, we deleted ATG1, a gene essential for this process, in an H. polymorpha pex3 strain. Our results show that pex3 atg1 cells contain preperoxisomal vesicles, which are the target for reintroduced Pex3, after which they mature into normal peroxisomes.
Results and discussion
H. polymorpha pex3 atg1 cells contain vesicular structures that harbor PMPs
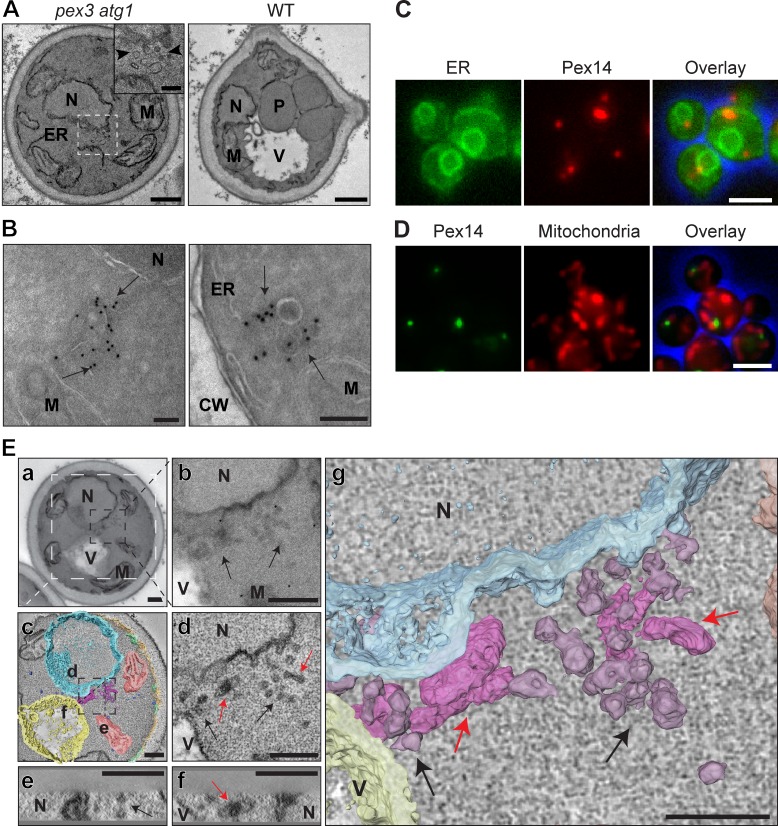
Careful EM analysis of H. polymorpha pex3 atg1 cells, grown at peroxisome-inducing conditions (mineral medium containing methanol and glycerol; MM-M/G) revealed that these cells contain clusters of vesicular structures, which measure up to 70 nm in diameter and have electron-dense contents. These structures were not detected in WT control cells (Fig. 1 A). Immuno-EM (iEM) indicated that these structures contain Pex14, a PMP involved in peroxisomal matrix protein import. The structures were generally observed in the vicinity of the nuclear envelope, lateral ER, and mitochondria (Fig. 1 B). In support of our EM results, mGFP- or mCherry-tagged Pex14 were observed as fluorescent spots adjacent to the nuclear envelope, ER (Fig. 1 C), or mitochondria (Fig. 1 D). Electron tomography analysis indicated that the clusters consist of reticular and vesicular structures (Fig. 1 E and Video 1). Distinct connections with other cell organelles were not detected.
Figure 1.
pex3 atg1 cells harbor Pex14-containing structures. (A) EM analysis of KMnO4-fixed pex3 atg1 and WT cells grown for 16 h on MM-M/G. The inset shows a cluster of vesicles (enlarged from the boxed region). (B) iEM analysis of pex3 atg1 cells using α-Pex14 antibodies. (C and D) FM images of pex3 atg1 cells producing Pex14-mCherry and the ER marker BiPN30-eGFP-HDEL (C), or Pex14-mGFP complemented with Mitotracker orange staining (D). (E) Electron tomography analysis of a serial-sectioned pex3 atg1 cell (a) containing a perinuclear membrane cluster (b, arrows). (d–f) 10-nm-thin digital slices through the tomogram reconstruction (viewing direction indicated in c) revealed vesicles (black arrows) and reticular structures (red arrows). The surface-rendered reconstruction in g shows the reticulovesicular structures in 3D viewed at right angles from d. CW, cell wall; M, mitochondrion; N, nucleus; P, peroxisome; V, vacuole. Bars: (A) 500 nm; (A, inset; and B) 100 nm; (C and D) 2.5 µm; (E) 250 nm.
The PMP-containing structures in pex3 cells are susceptible to autophagic degradation
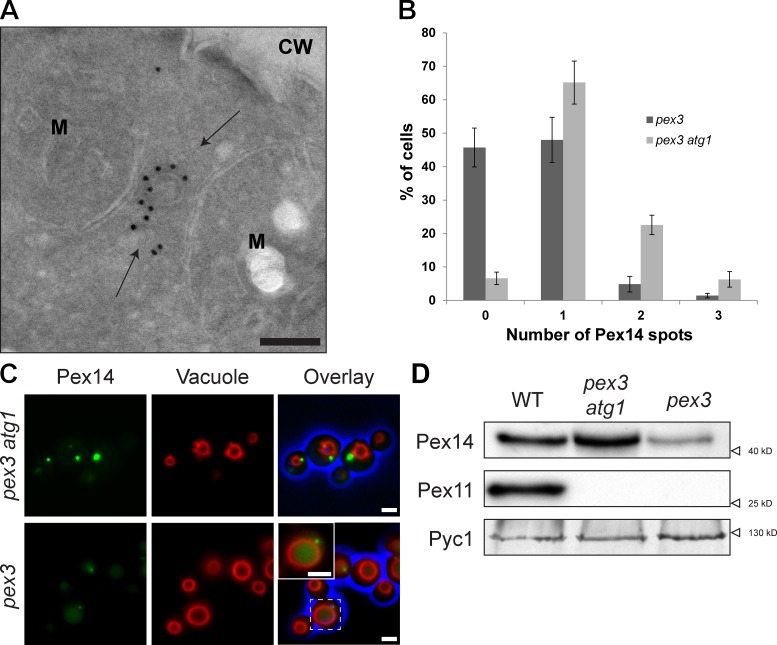
Although previous fluorescence microscopy (FM) studies suggested that, in H. polymorpha pex3 cells, Pex14-GFP is present in spots associated with mitochondria (Haan et al., 2006), iEM revealed that these spots also represent clusters of vesicles located adjacent to the nuclear envelope, ER (not depicted), or mitochondria at distances that cannot be resolved by FM (Fig. 2 A). The number of Pex14-mGFP spots is strongly reduced in pex3 cells, as was evident from quantitative analysis of FM images (1.3 ± 0.04 spots per cell in atg1 pex3 cells, relative to 0.6 ± 0.04 in pex3 cells; Fig. 2 B). In pex3 cells, but not in pex3 atg1 cells, mGFP fluorescence was also observed in vacuoles (Fig. 2 C), which indicates autophagic degradation of the structures. This was supported by Western blot (WB) analysis, which revealed that the level of Pex14 was strongly reduced in pex3 cells compared with WT and pex3 atg1 cells (Fig. 2 D).
Figure 2.
pex3 atg1 cells contain enhanced numbers of Pex14-containing structures. (A) iEM analysis of pex3 cells using α-Pex14 antibodies, identifying structures (arrows) in the vicinity of mitochondria. CW, cell wall; M, mitochondrion. (B) Quantification of Pex14-mGFP spots in pex3 and pex3 atg1 cells. (C) FM images of pex3 atg1 or pex3 cells, producing Pex14-mGFP complemented with FM4-64 vacuolar staining. The inset (enlarged from the boxed region) shows optimized intensities for pex3 cells, highlighting the Pex14-mGFP spot and vacuolar mGFP. (D) WB analysis of cells grown for 16 h on MM-M/G using α-Pex11 or α-Pex14 antibodies. Pyruvate carboxylase 1 (Pyc1) was used as a loading control. Error bars indicate SEM. Bars: (A) 100 nm; (C) 1 µm.
Several peroxisomal proteins colocalize with Pex14 in pex3 atg1 cells
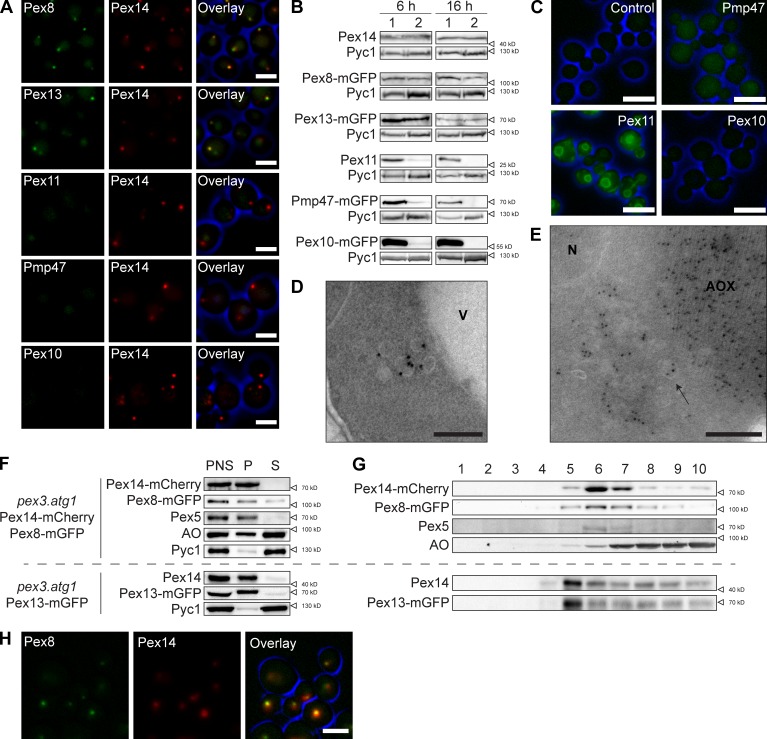
To examine whether other PMPs are also associated with the structures, we performed colocalization studies using pex3 atg1 strains producing Pex14-mCherry together with different PMP-mGFP fusion proteins, all under control of their endogenous promoter. We analyzed Pex8, Pex10, and Pex13, proteins of the importomer (Rucktäschel et al., 2011), as well as Pex11, a PMP involved in peroxisome fission (Thoms and Erdmann, 2005), and Pmp47, a peroxisomal carrier protein (Sakai et al., 1996). In WT cells grown for 16 h on MM-M/G, all mGFP fusion proteins were readily detected at peroxisomes (unpublished data). In pex3 atg1 cells, Pex8-mGFP and Pex13-mGFP colocalized with Pex14-mCherry, whereas the levels of Pex10-mGFP, Pex11-mGFP, and Pmp47-mGFP were below the limit of detection (Fig. 3 A).
Figure 3.
Pex10, Pex11, and Pmp47 do not colocalize with Pex14. (A) FM images of pex3 atg1 cells grown for 16 h on MM-M/G. Cells produced Pex14-mCherry and C-terminal mGFP fusions of the indicated proteins. (B) WB analysis of WT (1) and pex3 atg1 cells (2), grown for 6 or 16 h on MM-M/G. (C) FM images showing mGFP-fluorescence in pex3 atg1 cells producing Pex14-mCherry (control) or Pex14-mCherry together with the indicated mGFP fusion protein. Cells were grown for 6 h on MM-M/G. (D and E) iEM of pex3 atg1 cells using α-Pex5 (D) or α-alcohol oxidase antibodies (E). (F) Cell fractionation of the two indicated strains (seperated by the horizontal broken line). Postnuclear supernatants (PNS) were subjected to differential centrifugation resulting in a 30,000 g organelle pellet (P) and supernatant fraction (S). (G) Flotation analysis of the organelle pellet showing the distribution of the indicated proteins in the top (1) to bottom (10) fractions. (H) Colocalization of Pex8-GFP and Pex14-mCherry in pex10 cells. Bars: (A) 2.5 µm; (C) 5 µm; (D and E) 100 nm. AOX, cytosolic alcohol oxidase crystalloid; N, nucleus; V, vacuole.
To precisely compare the levels of the above proteins, we monitored their induction after shifting cells from peroxisome-repressing (glucose; MM-Glu) to peroxisome-inducing conditions (MM-M/G). In pex3 atg1 cells, Pex8, Pex13, and Pex14 showed similar induction patterns (Fig. S1, A–C) and protein levels (Fig. 3 B) as WT controls. Conversely, Pex10, Pex11, and Pmp47 were only detected in pex3 atg1 cells at the initial stages after the shift, with the highest levels after 6 h of induction (Fig. S1, D–G) followed by a very strong reduction after prolonged cultivation. However, at 6 h their levels were still strongly reduced compared with the WT controls (Fig. 3 B). FM revealed that in these cells Pex11-mGFP is predominantly localized to the nuclear envelope and lateral ER, whereas Pmp47-mGFP was dispersed over the cytosol and Pex10-mGFP was below the limit of detection (Fig. 3 C).
The strong reduction in Pex10, Pex11, and Pmp47 levels cannot be (fully) explained by a sudden arrest in the synthesis of these PMPs, as growth was minimal between 6 and 8 h (Fig. S1 G), and hence must be caused by proteolytic degradation. Therefore, we conclude that in H. polymorpha pex3 atg1 cells two classes of PMPs can be discriminated: i.e., those that sort independently of Pex3 to vesicular structures, where they are relatively stable; and PMPs that require Pex3 for sorting and stability.
Pex13 and Pex14 are associated with membranes in pex3 atg1 cells
To study whether PMPs are membrane-bound in atg1 pex3 cells, a flotation analysis of an organelle pellet was performed. Pex8, Pex13, and Pex14 were detected in the organelle pellet and migrated to fractions of low density upon flotation centrifugation (Fig. 3, F and G). Pex10 and Pmp47 could not be analyzed because of strong degradation during the fractionation procedure (unpublished data). Interestingly, the PTS1 receptor Pex5, as well as a minor portion of the peroxisomal matrix protein alcohol oxidase, cofractionated with Pex14 (Fig. 3, F and G). The bulk of the pelleted AO represent cytosolic crystalloids, which do not float. Localization of Pex5 and AO at the vesicles was confirmed by iEM (Fig. 3, D and E). The accumulation of Pex5 at these structures can be explained by the presence of a functional receptor docking complex, and the absence of Pex10, which is essential for receptor recycling. Our observation that the structures contain matrix protein is supported by the electron density of their lumen (Fig. 1, A and B). Association of Pex8 with the Pex14-containing structures is in line with observations obtained in Pichia pastoris, which revealed that Pex8 import into peroxisomes only depends on PTS receptors and Pex14 (Zhang et al., 2006; Ma et al., 2009). Also, in H. polymorpha pex10 cells, Pex8 colocalizes with Pex14 (Fig. 3 H).
Pex14-containing structures in pex3 atg1 cells develop into peroxisomes upon reintroduction of Pex3
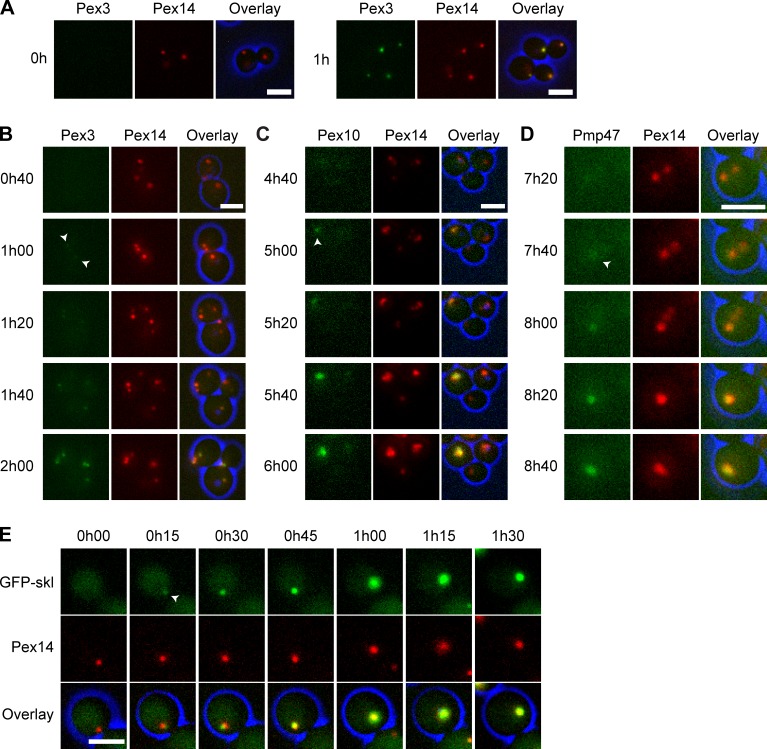
To analyze whether the membrane structures can develop into peroxisomes upon reintroduction of Pex3, we constructed a pex3 atg1 strain that contained PEX3-eGFP under control of the inducible amine oxidase promoter (PAMO). Cells were extensively precultivated on MM-Glu in the presence of ammonium sulfate to fully repress PAMO. Subsequently, cells were shifted to MM-M/G/methylamine to induce PAMO and peroxisome proliferation. Live cell imaging revealed that the first eGFP fluorescence invariably colocalized with the Pex14-mCherry spots (Fig. 4 B). The Pex14-mCherry spots present in pex3 single deletion cells also appeared to be the sole targets for reintroduced Pex3-eGFP (Fig. 4 A).
Figure 4.
Pex14-containing vesicular structures mature into peroxisomes upon reintroduction of PEX3. (A) FM images of pex3 cells with Pex14-mCherry upon Pex3-eGFP reintroduction after shifting cells from MM-Glu with ammonium sulfate to MM-M/G with methylamine. (B–E) Live cell FM images of pex3 atg1 cells upon Pex3 reintroduction. Shown are pex3 atg1 cells producing Pex14-mCherry and PAMOPEX3-eGFP (B), PAMOPEX3.PEX10-GFP (C), PAMOPEX3.PMP47-GFP (D), or PAMOPEX3.PTEFGFP-SKL (E). Cells were grown similar to the method in A. The arrows in B–D indicate the first detectable GFP signal. Bars, 2.5 µm.
We then examined Pex10-mGFP and Pmp47-mGFP upon reintroduction of Pex3 using strains that also produced Pex14-mCherry and PAMO-driven PEX3. In cells precultivated on MM-Glu with ammonium sulfate, these PMPs (with the exception of Pex14-mCherry) were below the limit of detection (unpublished data). Upon induction of PEX3 expression, the first Pex10-mGFP fluorescence signal appeared after 5 h, and invariably colocalized with Pex14-mCherry (Fig. 4 C). A similar result was observed for Pmp47-mGFP, except that the first fluorescence was detected after 8 h (Fig. 4 D).
Finally, we tested whether the Pex14-containing vesicles are capable of importing the matrix marker GFP-SKL upon PEX3 induction. As shown in Fig. 4 E, GFP-SKL was cytosolic before Pex3 reintroduction, but was found to be concentrated at the Pex14-mCherry spots when Pex3 synthesis was induced.
These results indicate that in pex3 atg1 cells the Pex14-containing structures, rather than the ER, are the target for reintroduced Pex3. Subsequently, Pex10 and Pmp47 also sort to these structures, which mature into normal peroxisomes that import GFP-SKL.
Pex10, Pex11, and Pmp47 are stabilized upon Pex19 overproduction
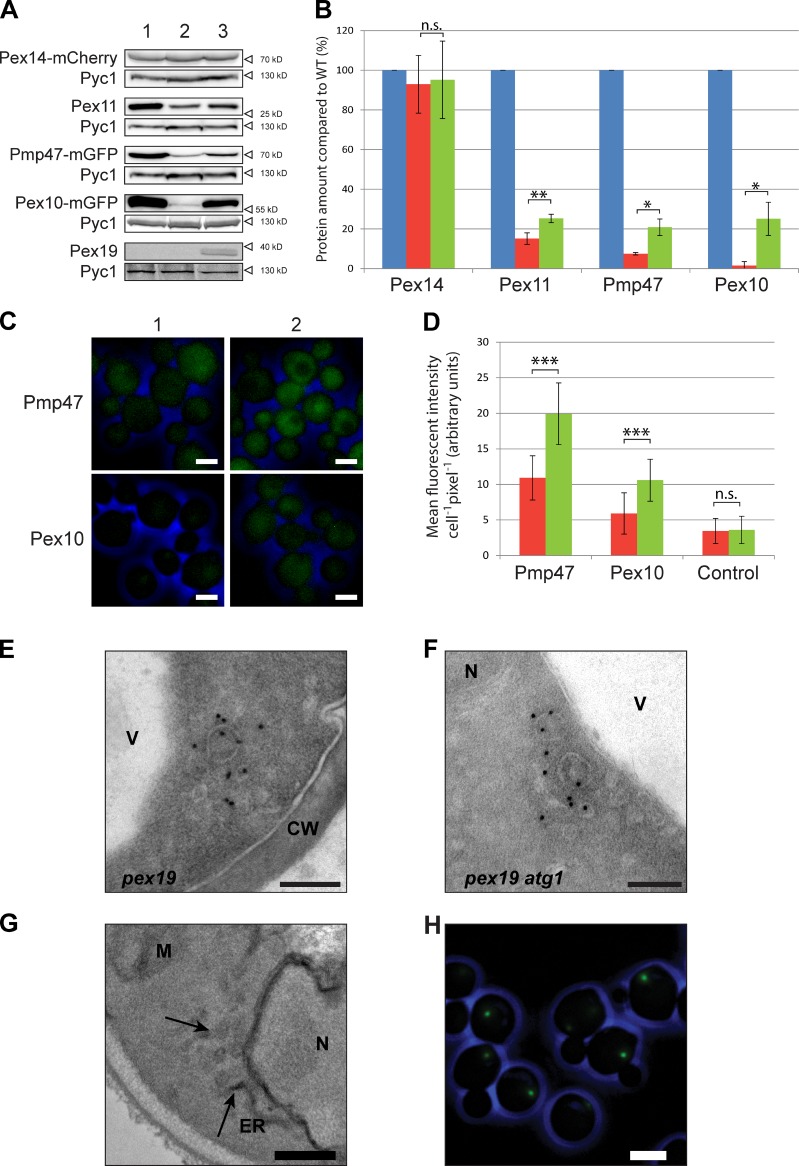
One of the models of Pex19 function proposes that cytosolic Pex19 binds newly synthesized PMPs, followed by recruitment of the complex by Pex3, and subsequent insertion of the PMPs into the peroxisomal membrane (Schliebs and Kunau, 2004). This led us to speculate that in the absence of Pex3, Pex19 may become saturated with PMPs that are dependent on the Pex3–Pex19 machinery, as cargo release is abolished. As a consequence, additionally synthesized PMPs cannot bind to Pex19 and may become susceptible to degradation. To test this, we analyzed the effect of Pex19 overproduction, indeed finding an increase of the levels of Pex10, Pex11, and Pmp47, but not of Pex14 (Fig. 5, A and B). The enhanced protein levels allowed visualization of Pex10 and Pmp47 by FM, which revealed that they both are cytosolic (Fig. 5, C and D).
Figure 5.
Pex19 overproduction and PEX19 or PEX25 deletion. (A and B) WB analysis (A) and protein quantification (B) in WT (lane 1; blue), pex3 atg1 (lane 2; red), and pex3 atg1-PAOXPEX19 (lane 3; green) cells grown for 6 h on MM-M/G. The protein levels in WT were set to 100%. (C and D) FM images (C) and quantification (D) of Pmp47-mGFP and Pex10-mGFP in pex3 atg1 (1; red) and pex3 atg1-PAOXPex19 (2; green) cells grown for 6 h on MM-M/G. Control cells in D did not produce mGFP. Significance indications: n.s., P < 0.10; *, 0.10 > P > 0.05; **, 0.05 > P > 0.01; ***, P < 0.01. Error bars indicate SD. (E and F) iEM analysis of pex19 (E) and pex19 atg1 (F) cells using α-Pex14 antibodies. (G) EM analysis of KMnO4-fixed pex3 atg1 pex25 cells grown for 16 h on MM-M/G–showing membrane vesicles (arrows). (H) FM image of pex3 atg1 pex25 cells producing Pex14-mGFP. CW, cell wall; M, mitochondrion; N, nucleus; V, vacuole. Bars: (C) 2 µm; (E and F) 100 nm; (G) 250 nm; (H) 2.5 µm.
These findings are consistent with a model that newly synthesized Pex10, Pex11, and Pmp47 directly insert into the peroxisomal membrane by a process that requires Pex3 and Pex19.
Pex19 and Pex25 are not required for vesicle formation in pex3 atg1 cells
Because in vitro assays suggested that Pex19, rather than Pex3, is essential to form peroxisomal vesicles from the ER (Lam et al., 2010; Agrawal et al., 2011), we also analyzed H. polymorpha pex19 and pex19 atg1 cells. As shown in Fig. 5 (E and F), Pex14-containing structures, similar to those observed in pex3 cells, are also present in these cells.
Pex25 is required for the reintroduction of peroxisomes in H. polymorpha pex3 cells (Saraya et al., 2011). However, vesicular structures and Pex14-mGFP spots were also observed in cells of a pex3 atg1 pex25 triple deletion strain (Fig. 5, G and H), which suggests that Pex25 is not required for the formation of these vesicles.
Conclusions
Because peroxisomal membranes to which common marker PMPs colocalize were not detected in yeast (Baerends et al., 1996; Wiemer et al., 1996; Hettema et al., 2000) or mammalian (Shimozawa et al., 2000) cells lacking a functional PEX3 gene, it is generally accepted that cells lacking Pex3 are unable to form peroxisomal membranes. Instead, FM analysis suggested that PMPs were localized to the ER, mitochondria, or were below the limit of detection, depending on the marker PMP examined (Hettema et al., 2000; South et al., 2000; Haan et al., 2006; van der Zand et al., 2010). Here, we show that in the absence of Pex3, the PMPs Pex13 and Pex14 colocalize at membrane structures that are often located adjacent to other cell organelles at distances that cannot be resolved by FM. Apparently, these PMPs can insert in membranes independent of Pex3 (Fig. S2).
Based on FM, van der Zand et al. (2010) concluded that, in Saccharomyces cerevisiae pex3 cells, Pex13 and Pex14 are present in foci at the ER. We consider it likely that these foci represented similar structures. Indeed, our iEM analyses on S. cerevisiae pex3 atg1 cells revealed that these cells also harbor Pex14-containing vesicles (unpublished data). Our observations are furthermore supported by the presence of PMP-containing membrane structures in P. pastoris pex3 cells (Hazra et al., 2002).
In contrast to Pex13 and Pex14, Pex10, Pex11, and Pmp47 apparently do require the Pex3–Pex19 machinery for insertion into these membrane structures, given that in cells lacking Pex3 they do not colocalize with Pex14 and are very unstable. Instead, they are stabilized and sorted to the structures upon Pex3 reintroduction. In addition, their levels increase in pex3 atg1 cells upon PEX19 overexpression, which suggests that Pex19 serves as cytosolic receptor for these PMPs (Fig. S2). Pex10 and Pmp47 were invariably cytosolic in pex3 atg1 cells, which is consistent with the cytosolic localization of the mammalian Ant1 (a homologue of Pmp47) in PEX3 mutant cells (Fang et al., 2004).
Pex11 was the only PMP that we (transiently) observed at the ER, but only in minor amounts, with the protein being very instable. The latter finding is consistent with pulse-chase experiments using S. cerevisiae pex3 cells, which showed that Pex11 is normally synthesized, but, unlike in the WT control, rapidly degraded (Hettema et al., 2000). This instability suggests that localization at the ER may not be an intermediate stage of its normal sorting pathway. However, at this stage it cannot be excluded that Pex11 traffics via the ER to peroxisomes and is degraded in pex3 cells because of its inability to exit the ER. We note, however, that this pathway is not consistent with our observation that Pex11 levels increase upon Pex19 overproduction in pex3 cells.
The most pressing question is the nature of the vesicles in pex3 cells. Our data indicate that they have several properties in common with normal peroxisomal membranes as they appear to contain a functional receptor docking site to which Pex5 associates, and are capable of importing matrix proteins (Pex8, alcohol oxidase). This property is shared with peroxisomal membrane ghosts that are present in H. polymorpha PEX deletion strains, which are defective in receptor recycling, e.g., pex4 or pex10 (Koek et al., 2007). Thus, they may represent peroxisomal ghosts, an assumption that is reinforced by the finding that they mature into normal peroxisomes upon Pex3 reintroduction.
According to our model (Fig. S2), the vesicles may proliferate from a preexisting peroxisomal membrane structure. Alternatively, they may form from other membranes. If so, they are most likely formed from the ER (Fakieh et al., 2013; Tabak et al., 2013), possibly by a similar mechanism as the in vitro generated vesicles reported by Lam et al. (2010) and Agrawal et al. (2011). Importantly, our current data demonstrate that, if these structures indeed derive from the ER, their formation does not require Pex3.
Materials and methods
Strains and growth conditions
The H. polymorpha strains used in this study are listed in Table S1. Yeast cultures were grown at 37°C, on (1) YPD media containing 1% yeast extract, 1% peptone, and 1% glucose; (2) selective media containing 0.67% yeast nitrogen base without amino acids (YNB; Difco; BD); or (3) mineral media (MM; Van Dijken et al., 1976) supplemented with 0.5% glucose (MM-Glu), 0.5% methanol, or a mixture of 0.5% methanol and 0.05% glycerol (MM-M/G) as carbon sources and 0.25% ammonium sulfate or 0.25% methylamine as nitrogen sources. If required, amino acids, uracil, or leucine were added to a final concentration of 30 µg/ml. For growth on agar plates, the medium was supplemented with 2% agar. For the selection of resistant transformants, YPD plates containing 100 µg/ml zeocin (Invitrogen), 300 µg/ml hygromycin B (Invitrogen), or 100 µg/ml nourseothricin (WERNER BioAgents) were used.
For cloning purposes, Escherichia coli DH5α were used. Cells were grown at 37°C in Luria broth (LB) media supplemented with 100 µg/ml ampicillin or 50 µg/ml kanamycin, when required.
Molecular and biochemical techniques
Standard recombinant DNA techniques and transformation of H. polymorpha was performed by electroporation as described previously (Faber et al., 1994). Cell extracts of TCA-treated cells were prepared for SDS-PAGE as detailed previously (Baerends et al., 2000). SDS-PAGE and WB were performed by established methods. Equal amounts of protein were loaded per lane and blots were probed with rabbit polyclonal antisera against H. polymorpha alcohol oxidase, Pex5, Pex11, Pex14, Pex19, or pyruvate carboxylase 1 (Pyc1). mGFP fusion proteins of Pex8, Pex10, Pex13, and Pmp47 were detected using mouse monoclonal antiserum against GFP (sc-9996; Santa Cruz Biotechnology, Inc.). Secondary goat anti–rabbit or goat anti–mouse antibodies conjugated to horseradish peroxidase (Thermo Fisher Scientific) were used for detection. Pyc1 was used as a loading control. Blots were scanned by using a densitometer (GS-710; Bio-Rad Laboratories) and quantified using ImageJ. From two individual blots per sample, the total intensity of the band of interest was measured and corrected for background intensity and Pyc1 loading amount.
Construction of H. polymorpha strains
The plasmids and primers used in this study are listed in Tables S2 and S3. All integrations were confirmed by PCR. All deletions were confirmed by PCR and Southern blotting.
Construction of the pex3 atg1 and pex19 atg1 double deletion strain and the pex3 atg1 pex25 triple deletion strain
The pex3 atg1 double deletion strain was obtained by crossing an H. polymorpha pex3 strain (Baerends et al., 1996) with an atg1 strain (Komduur et al., 2003). Diploids were subjected to random spore analysis and prototrophic segregants were subjected to complementation analysis to determine their genotypes (Sudbery et al., 1988). The pex3 atg1 pex25 triple deletion strain was made as follows. A PCR fragment of 2,912 bp was obtained by PCR using primers Pex25-F and Pex25-R and plasmid pRSA018 as a template (Saraya et al., 2011). This PCR fragment was transformed to the pex3 atg1 double deletion strain. For the pex19 atg1 double deletion strain, a PEX19-deletion cassette plasmid (pHOR30b) was digested with BglII and EcoRI to replace the URA3 gene with the LEU2 gene, which was obtained after digestion of pBS-CaLeu2 with BamHI and EcoRI. The final deletion PEX19-deletion plasmid (pSEM188) was digested with BamHI and the resulting 4,434-bp fragment was integrated in the genome of atg1 cells (Komduur et al., 2003).
Construction of other strains
All pHIP plasmids used in this study are derivatives of the pOK12 cloning vector. Plasmids pHIPZ-PEX8-mGFP (pMCE4), pHIPZ-PEX10-mGFP (pMCE5), pHIPZ-PMP47-mGFP (pMCE7; Cepińska et al., 2011), pHIPZ5-PEX3-eGFP, and pHIPZ4-BiPN30-eGFP-HDEL (pRSA017; Saraya et al., 2010) were linearized and integrated in the endogenous promoter regions in the pex3 atg1 strain producing Pex14-mCherry essentially as described previously (Saraya et al., 2010; Cepińska et al., 2011).
For the construction of plasmid pSEM01, a PCR fragment of 563 bp was obtained by using primers Pex14-F and Pex14-R on genomic DNA. After digestion with HindIII and BglII, the resulting fragment was inserted between the HindIII and BglII sites of pMCE02, resulting in pSEM01 (5,488 bp) containing pHIPN-PEX14-mCherry. For stable integration in the PEX14 promoter region, XhoI linearized plasmid was transformed to the pex3 atg1 double mutant, resulting in a strain producing Pex14-mCherry under control of the endogenous promoter.
Plasmid pSEM02 (pHIPZ-PEX11-mGFP) was obtained as follows: Digestion of the pHIPZ-mGFP fusinator plasmid with HindIII and BglII yields a fragment of 5,077 bp. Similarly, the pHIPN-PEX11-mCherry plasmid (pMCE3) was digested with HindIII and BglII to obtain a fragment of 772 bp. Ligation of the 772 bp and 5,077 bp fragments resulted in pSEM02 of 5,849 bp. The plasmid was linearized using Pst1 and integrated in the genome of pex3 atg1 producing Pex14-mCherry.
To construct plasmid pSEM03 (pHIPZ-PEX13-mGFP), PCR was performed on genomic DNA using the primers Pex13-F and Pex13-R. The PCR product of 1,146 bp was digested with HindIII and BglII, and the resulting fragment was inserted between the HindIII and BglII sites of pHIPZ-mGFP fusinator plasmid. The resulting plasmid of 6,223 bp, designated pSEM03, was linearized with ApaI and transformed to H. polymorpha pex3 atg1, producing Pex14-mCherry.
For the construction of plasmid pSEM04, a PCR fragment of 2,547 bp was obtained using plasmid pHIPZ5-PEX3-eGFP (Table S2) as a template and primers H5-F and H5-R. The PCR fragment was digested with NotI and PspXI and the resulting fragment was ligated in NotI- and SalI-digested pHIPH4, resulting in plasmid pSEM04, which contains pHIPH5-PAMO-PEX3. The plasmid was linearized with BsiWI and integrated in strain pex3 atg1.Pex14-mCherry, producing Pex10-mGFP or Pmp47-mGFP.
Similarly, plasmid pSEM05 was made by PCR amplification of an 885-bp fragment using genomic DNA and primers Pex19-F and Pex19-R (Table S3). After digestion with HindIII and XbaI, the resulting fragment was ligated in HindIII- and XbaI-digested pHIPH4, resulting in plasmid pSEM05 containing pHIPH4-PAOX-PEX19. For stable integration, StuI linearized plasmid was transformed to H. polymorpha pex3 atg1.Pex14-mCherry, producing Pex10-mGFP or Pmp47-mGFP.
For the construction of plasmid pAKW27, a vector of 5,831 bp was obtained by BamHI and SalI digestion of pHIPZ7, whereas the 736-bp eGFP-SKL insert was obtained by BamHI and SalI digestion of pFEM35 followed by gel extraction. Ligation resulted in the plasmid pAKW27 containing pHIPZ7-PTEF1-GFPSKL. For stable integration, stuI linearized plasmid was transformed to H. polymorpha pex3 atg1.Pex14-mCherry producing Pex3 under control of the inducible PAMO.
Cell fractionation and membrane flotation
Crude extracts were prepared as described previously (Baerends et al., 1997). In brief, protoplasts were prepared with Zymolyase (Brunschwig Chemie) and homogenized using a Potter homogenizer. To remove cell debris, the homogenate was centrifuged twice at 3,000 g (10 min, 4°C). The supernatant (PNS) was then subjected to centrifugation at 30,000 g (30 min, 4°C) to separate the soluble fraction (supernatant [S]) from the membrane pellet (P).
The 30,000 g organelle pellet was used for flotation centrifugation as described previously (Baerends et al., 1997). In brief, the pellet was dissolved in 50% sucrose and layered over with 40%, 30%, and 20% sucrose. Centrifugation was performed at 140,000 g for 16 h at 4°C. 10 fractions of 200 µl were collected from the top and analyzed by SDS-PAGE and WB.
FM
All images were made using a 100× 1.30 NA Plan-Neofluar objective (Carl Zeiss). For wide-field microscopy, the GFP signal was visualized with a 470/40-nm band-pass excitation filter, a 495-nm dichromatic mirror, and a 525/50-nm band-pass emission filter. mCherry fluorescence was visualized with a 587/25-nm band pass excitation filter, a 605-nm dichromatic mirror, and a 647/70-nm band-pass emission filter. DsRed, FM4-64, and MitoTracker orange fluorescence were visualized with a 546/12-nm band-pass excitation filter, a 560-nm dichromatic mirror, and a 575–640-nm band-pass emission filter. Images were captured using a fluorescence microscope (Axioskop 50; Carl Zeiss) using MetaVue software and a digital camera (1300Y; Princeton Instruments). The images were captured in the media in which the cells were grown.
Mitochondria were stained by incubation of intact cells for 30 min at 37°C with 0.5 µg/ml MitoTracker orange (Invitrogen) followed by extensive washing with medium. For vacuolar staining, 1 ml of cell culture was supplemented with 1 µl FM4-64 (Invitrogen), incubated for 60 min at 37°C, and analyzed.
Live cell imaging was performed on an inverted microscope (Observer Z1; Carl Zeiss) using AxioVision software (Carl Zeiss) and a digital camera (CoolSNAP HQ2; Photometrics). Cells were grown on 1% agar containing growth medium, and the temperature of the heating chamber XL was set at 37°C. Three z axis planes were acquired for each time interval using 0.5-s exposure times for both GFP and mCherry.
Confocal images were captured with a confocal microscope (LSM510; Carl Zeiss) equipped with photomultiplier tubes (Hamamatsu Photonics) and Zen 2009 software (Carl Zeiss). For live cell imaging, the temperature of the objective and object slide was kept at 37°C and the cells were grown on 1% agar in medium. GFP fluorescence was analyzed by excitation of the cell with a 488-nm argon ion laser (Lasos), and emission was detected using a 500–550-nm band-pass emission filter. During simultaneous GFP and FM4-64 detection, both probes were excited with a 488-nm argon ion laser, GFP was detected using a 500–530-nm band-pass emission filter, FM4-64 was detected using a 560-nm long-pass emission filter. Six z-axis planes were acquired for each time interval.
Image analysis was performed using ImageJ, and figures were prepared using Photoshop CS4 (Adobe). Unless otherwise indicated, the intensity minimum and maximum of the image were set to be equal for all images represented within a single figure panel, thus facilitating direct fluorescence intensity comparison between different strains.
For quantitative analysis of Pex14-mGFP fluorescent spots, z stacks were made of randomly chosen fields. Quantification was done on four images per culture, containing at least 65 cells per image. Cells were stained with FM4-64 to allow discrimination between vacuolar mGFP and cytosolic mGFP spots. The mean number of spots was calculated from 350 cells per culture. The error bars indicate the standard error of the mean.
For the quantification of cytosolic mGFP intensity, single plane images were acquired on a fluorescence microscope (Axioskop 50), after which the total intensity of ∼100 individual cells was measured and corrected for the background intensity. The error bars indicate the standard deviation between individual cells.
Electron microscopy
H. polymorpha pex3 atg1 were fixed in 1.5% potassium permanganate, stained en block with 0.5% uranyl acetate, and embedded in epon 812 (21045; Serva). For morphological studies, ultrathin sections were viewed with a transmission EM microscope (CM12; Philips). For electron tomography, serial sections were cut to be 150 nm thick. The serial images of whole cells were stacked and aligned using MIDAS (Kremer et al., 1996), after which individual cells could be scrutinized for peroxisomal remnants. 10-nm gold beads were layered on top of the serial sections and acted as fiducial markers for electron tomography. Two single-axis tilt series, each containing 141 images with 1° tilt increments, were acquired at a magnification of 42,000 with a pixel size of 0.7 nm on a transmission EM microscope (Tecnai 12; FEI) at 120 kV using the SerialEM acquisition software (Mastronarde, 2005) and a cooled slow-scan charge-coupled device camera (4k Eagle; FEI) in 2 × 2 binned mode. The tilt series were aligned and reconstructed using the IMOD software package and analyzed using the Amira visualization package (TGS Europe). To generate 3D surface-rendered models in Amira, masks of organelles were first drawn manually and then improved by nonlinear anisotropic diffusion filtering followed by thresholding.
Cryosectioning and immuno-gold labeling
For iEM, cells were fixed in 3% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2, for 1 h on ice and treated afterward with 0.4% sodium periodate (15 min) and 1% NH4Cl (15 min). Upon embedding in 12% gelatin in phosphate buffer, pH 7.4, ∼0.5 mm3 cubes were infiltrated overnight in 2.3 M sucrose in the same buffer. Cryosections of 60 nm were cut using a cryo diamond knife (Diatome) at −120°C in an ultramicrotome (Ultracut; Reichert). Sections were mounted on carbon-coated Formvar nickel grids. Gelatin was removed by incubating the grids for 30 min on 2% gelatin in phosphate buffer, pH 7.4, at 30°C. Pex14, Pex5, and alcohol oxidase were localized using polyclonal antibodies raised against Pex14, Pex5, and alcohol oxidase, respectively, and goat anti–rabbit antibodies conjugated to 10 nm gold (Aurion). Sections were stained with 2% uranyl oxalate, pH 7.0, for 10 min, briefly washed on three drops of distilled water, and embedded in 0.5% methylcellulose and 0.5% uranyl acetate on ice for 10 min before viewing them with a transmission EM microscope (CM12; Slot and Geuze, 2007).
Online supplemental material
Fig. S1 shows induction of PMPs in WT and pex3 atg1 cells after a shift from MM-Glu to MM-M/G and the corresponding growth curves. Fig. S2 shows a schematic of peroxisome maturation after Pex3 reintroduction. Video 1 shows a tilt series, reconstructed tomogram, and surface rendering of a pex3 atg1 cell. Tables S1, S2, and S3 contain the H. polymorpha strains, plasmids, and primers used in this study, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201310148/DC1.
Supplementary Material
Acknowledgments
We thank Ruchi Saraya, Geerke Maathuis, and Chris Williams for their valuable contributions and Abraham Koster (Leiden University Medical Center) for making the electron tomography facilities available.
This work was supported by an EU Marie Curie IEF grant to K. Knoops. (FP7-330150).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- FM
- fluorescence microscopy
- iEM
- immuno–electron microscopy
- PAMO
- amine oxidase promoter
- PMP
- peroxisomal membrane protein
- WB
- Western blot
- WT
- wild type
References
- Agrawal G., Joshi S., Subramani S. 2011. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 108:9113–9118 10.1073/pnas.1018749108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends R.J., Rasmussen S.W., Hilbrands R.E., van der Heide M., Faber K.N., Reuvekamp P.T., Kiel J.A., Cregg J.M., van der Klei I.J., Veenhuis M. 1996. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J. Biol. Chem. 271:8887–8894 10.1074/jbc.271.15.8887 [DOI] [PubMed] [Google Scholar]
- Baerends R.J., Salomons F.A., Faber K.N., Kiel J.A., Van der Klei I.J., Veenhuis M. 1997. Deviant Pex3p levels affect normal peroxisome formation in Hansenula polymorpha: high steady-state levels of the protein fully abolish matrix protein import. Yeast. 13:1437–1448 [DOI] [PubMed] [Google Scholar]
- Baerends R.J., Faber K.N., Kram A.M., Kiel J.A., van der Klei I.J., Veenhuis M. 2000. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J. Biol. Chem. 275:9986–9995 10.1074/jbc.275.14.9986 [DOI] [PubMed] [Google Scholar]
- Bellu A.R., Salomons F.A., Kiel J.A., Veenhuis M., Van Der Klei I.J. 2002. Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J. Biol. Chem. 277:42875–42880 10.1074/jbc.M205437200 [DOI] [PubMed] [Google Scholar]
- Cepińska M.N., Veenhuis M., van der Klei I.J., Nagotu S. 2011. Peroxisome fission is associated with reorganization of specific membrane proteins. Traffic. 12:925–937 10.1111/j.1600-0854.2011.01198.x [DOI] [PubMed] [Google Scholar]
- Faber K.N., Haima P., Harder W., Veenhuis M., Ab G. 1994. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 25:305–310 10.1007/BF00351482 [DOI] [PubMed] [Google Scholar]
- Fakieh M.H., Drake P.J., Lacey J., Munck J.M., Motley A.M., Hettema E.H. 2013. Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol. Open. 2:829–837 10.1242/bio.20134788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Morrell J.C., Jones J.M., Gould S.J. 2004. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164:863–875 10.1083/jcb.200311131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan G.J., Baerends R.J., Krikken A.M., Otzen M., Veenhuis M., van der Klei I.J. 2006. Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 6:186–194 10.1111/j.1567-1364.2006.00037.x [DOI] [PubMed] [Google Scholar]
- Hazra P.P., Suriapranata I., Snyder W.B., Subramani S. 2002. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 3:560–574 10.1034/j.1600-0854.2002.30806.x [DOI] [PubMed] [Google Scholar]
- Hettema E.H., Girzalsky W., van Den Berg M., Erdmann R., Distel B. 2000. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223–233 10.1093/emboj/19.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R.T., Reumann S., Zolman B.K. 2012. Plant peroxisomes: biogenesis and function. Plant Cell. 24:2279–2303 10.1105/tpc.112.096586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koek A., Komori M., Veenhuis M., van der Klei I.J. 2007. A comparative study of peroxisomal structures in Hansenula polymorpha pex mutants. FEMS Yeast Res. 7:1126–1133 10.1111/j.1567-1364.2007.00261.x [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D., Veenhuis M., van der Klei I.J. 2013. Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat—store ‘em up or burn ‘em down. Genetics. 193:1–50 10.1534/genetics.112.143362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komduur J.A., Veenhuis M., Kiel J.A. 2003. The Hansenula polymorpha PDD7 gene is essential for macropexophagy and microautophagy. FEMS Yeast Res. 3:27–34 [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Yoda N., Schekman R. 2010. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 107:21523–21528 10.1073/pnas.1013397107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Schumann U., Rayapuram N., Subramani S. 2009. The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol. Biol. Cell. 20:3680–3689 10.1091/mbc.E09-01-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Motley A.M., Hettema E.H. 2007. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178:399–410 10.1083/jcb.200702167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagotu S., Saraya R., Otzen M., Veenhuis M., van der Klei I.J. 2008. Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim. Biophys. Acta. 1783:760–769 10.1016/j.bbamcr.2007.10.018 [DOI] [PubMed] [Google Scholar]
- Rucktäschel R., Girzalsky W., Erdmann R. 2011. Protein import machineries of peroxisomes. Biochim. Biophys. Acta. 1808:892–900 10.1016/j.bbamem.2010.07.020 [DOI] [PubMed] [Google Scholar]
- Sakai Y., Saiganji A., Yurimoto H., Takabe K., Saiki H., Kato N. 1996. The absence of Pmp47, a putative yeast peroxisomal transporter, causes a defect in transport and folding of a specific matrix enzyme. J. Cell Biol. 134:37–51 10.1083/jcb.134.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraya R., Cepińska M.N., Kiel J.A., Veenhuis M., van der Klei I.J. 2010. A conserved function for Inp2 in peroxisome inheritance. Biochim. Biophys. Acta. 1803:617–622 10.1016/j.bbamcr.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Saraya R., Krikken A.M., Veenhuis M., van der Klei I.J. 2011. Peroxisome reintroduction in Hansenula polymorpha requires Pex25 and Rho1. J. Cell Biol. 193:885–900 10.1083/jcb.201012083 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schliebs W., Kunau W.H. 2004. Peroxisome membrane biogenesis: the stage is set. Curr. Biol. 14:R397–R399 10.1016/j.cub.2004.05.017 [DOI] [PubMed] [Google Scholar]
- Shimozawa N., Suzuki Y., Zhang Z., Imamura A., Ghaedi K., Fujiki Y., Kondo N. 2000. Identification of PEX3 as the gene mutated in a Zellweger syndrome patient lacking peroxisomal remnant structures. Hum. Mol. Genet. 9:1995–1999 10.1093/hmg/9.13.1995 [DOI] [PubMed] [Google Scholar]
- Slot J.W., Geuze H.J. 2007. Cryosectioning and immunolabeling. Nat. Protoc. 2:2480–2491 10.1038/nprot.2007.365 [DOI] [PubMed] [Google Scholar]
- South S.T., Sacksteder K.A., Li X., Liu Y., Gould S.J. 2000. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149:1345–1360 10.1083/jcb.149.7.1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P.E., Gleeson M.A., Veale R.A., Ledeboer A.M., Zoetmulder M.C. 1988. Hansenula polymorpha as a novel yeast system for the expression of heterologous genes. Biochem. Soc. Trans. 16:1081–1083 [DOI] [PubMed] [Google Scholar]
- Tabak H.F., Braakman I., van der Zand A. 2013. Peroxisome formation and maintenance are dependent on the endoplasmic reticulum. Annu. Rev. Biochem. 82:723–744 10.1146/annurev-biochem-081111-125123 [DOI] [PubMed] [Google Scholar]
- Thoms S., Erdmann R. 2005. Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272:5169–5181 10.1111/j.1742-4658.2005.04939.x [DOI] [PubMed] [Google Scholar]
- van der Zand A., Braakman I., Tabak H.F. 2010. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell. 21:2057–2065 10.1091/mbc.E10-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zand A., Gent J., Braakman I., Tabak H.F. 2012. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 149:397–409 10.1016/j.cell.2012.01.054 [DOI] [PubMed] [Google Scholar]
- Van Dijken J.P., Otto R., Harder W. 1976. Growth of Hansenula polymorpha in a methanol-limited chemostat. Arch. Microbiol. 111:137–144 10.1007/BF00446560 [DOI] [PubMed] [Google Scholar]
- Wanders R.J., Waterham H.R. 2006. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75:295–332 10.1146/annurev.biochem.74.082803.133329 [DOI] [PubMed] [Google Scholar]
- Wiemer E.A., Lüers G.H., Faber K.N., Wenzel T., Veenhuis M., Subramani S. 1996. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J. Biol. Chem. 271:18973–18980 10.1074/jbc.271.31.18973 [DOI] [PubMed] [Google Scholar]
- Williams C., van der Klei I.J. 2013. Pexophagy-linked degradation of the peroxisomal membrane protein Pex3p involves the ubiquitin-proteasome system. Biochem. Biophys. Res. Commun. 438:395–401 10.1016/j.bbrc.2013.07.086 [DOI] [PubMed] [Google Scholar]
- Zhang L., Léon S., Subramani S. 2006. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol. Biol. Cell. 17:690–699 10.1091/mbc.E05-08-0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.