Arp5 suppresses myocardin activity through both direct binding to myocardin and binding to SRF to prevent transcriptional activation of myogenic genes by the myocardin–SRF complex.
Abstract
Myocardin (Myocd) and Myocd-related transcription factors (MRTFs) are robust coactivators of serum response factor (SRF). RPEL motifs are monomeric globular actin (G-actin) binding elements that regulate MRTF localization and activity. However, the function of the RPEL motif in Myocd is largely unknown because of its low affinity for G-actin. Here, we demonstrated that the Myocd RPEL motif bound to actin-related protein 5 (Arp5) instead of conventional actin, resulting in a significant suppression of Myocd activity. In addition, Arp5 bound to a DNA binding domain of SRF via its C-terminal sequence and prevented the association of the Myocd–SRF complex with the promoter regions of smooth muscle genes. Well-differentiated smooth muscle cells mainly expressed a specific splicing variant of arp5; therefore, the protein level of Arp5 was markedly reduced by partial messenger RNA decay and translational suppression. In dedifferentiated smooth muscle cells, Arp5 knockdown restored the differentiated phenotype via Myocd activation. Thus, Arp5 is a key regulator of Myocd activity.
Introduction
Myocardin (Myocd) is a cardiac and smooth muscle–specific coactivator of serum response factor (SRF) that regulates the expression of cardiac and smooth muscle marker genes in an SRF-dependent manner (Wang et al., 2001; Chen et al., 2002). Myocd knockout mice die by embryonic day 10.5 and exhibit defective vascular smooth muscle cell (VSMC) differentiation (Li et al., 2003), illustrating that Myocd is a principal regulator of smooth muscle cell differentiation. MAL/MKL1/Myocd-related transcription factor-A (MRTF-A) and MAL16/MKL2/MRTF-B, the other members of the Myocd family, are distributed in a wide variety of cell types and tissues and function in various biological processes (Miralles et al., 2003; Oh et al., 2005; Li et al., 2006; Morita et al., 2007a,b; Medjkane et al., 2009). All the Myocd family proteins contain three tandem RPEL repeats, which are monomeric globular actin (G-actin) binding motifs, at their N-terminal RPEL domain. In MRTFs, G-actin tightly binds to the RPEL motifs to prevent the nuclear localization of MRTF proteins (Miralles et al., 2003). When actin polymerization is accelerated by the activation of Rho signaling, cytoplasmic G-actin is depleted, resulting in the translocation of free MRTFs from the cytoplasm to the nucleus and subsequent transcriptional activation. In contrast, Myocd continuously localizes to the nucleus because of a low affinity of Myocd RPEL motifs for G-actin (Guettler et al., 2008). Thus, in contrast to MRTFs, the functional importance of Myocd RPEL motifs remains largely unknown.
Several types of actin family proteins and actin-associated proteins exist in the nucleus and they participate in chromatin remodeling and nuclear organization (Dion et al., 2010; de Lanerolle and Serebryannyy, 2011). Recently, Vartiainen et al. (2007) reported that nuclear G-actin also interacts with MRTF RPEL motifs and suppresses MRTF activity via a mechanism that is different from that regulating the subcellular localization of MRTF. In vertebrates, there are at least eight homologous proteins of conventional actin, known as actin-related proteins (ARPs), and among these, four ARPs (Arp4, Arp5, Arp6, and Arp8) predominantly localize to the nucleus (Dion et al., 2010). Unlike conventional actin, however, nuclear ARPs cannot polymerize to form filamentous structures and they function as components of the chromatin-remodeling complex (Meagher et al., 2009; Dion et al., 2010). Although Arabidopsis thaliana nuclear Arps have been reported to be involved in multicellular development (Meagher et al., 2009), developmental roles of animal nuclear Arps remain unclear. In the present study, we identified Arp5 as a key regulator of Myocd. Arp5 bound more tightly to Myocd RPEL motifs than β-actin and significantly suppressed Myocd activity in the nucleus. Furthermore, Arp5 modulated the VSMC phenotype via Myocd–SRF signaling. This is the first report of the importance of RPEL motifs in the activity regulation of Myocd and a novel function of Arp5 in smooth muscle cell differentiation.
Results
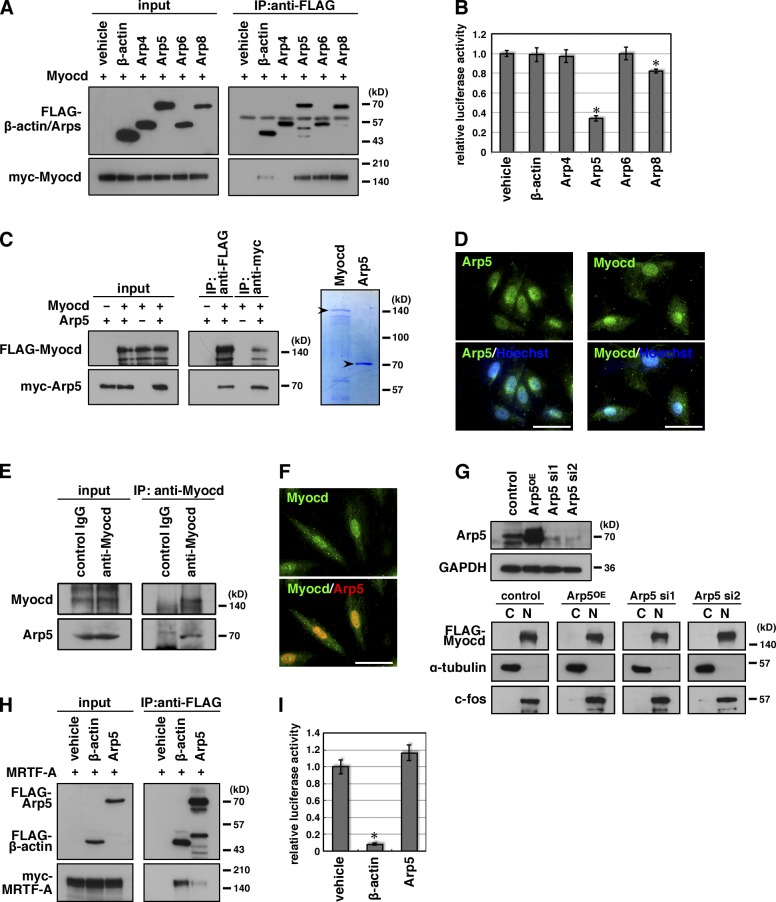
Arp5 is a negative regulator of Myocd
Compared with MRTFs, which shuttle between the cytoplasm and the nucleus, Myocd continuously localizes to the nucleus (Wang et al., 2001; Miralles et al., 2003). To determine whether nuclear-localized actin family proteins contribute to the regulation of Myocd activity, coimmunoprecipitation between Myocd and the nuclear ARPs was performed (Fig. 1 A). As reported previously (Guettler et al., 2008), β-actin weakly bound to Myocd, whereas Arp5, Arp6, and Arp8 more tightly bound to Myocd. Arp4 showed no affinity for Myocd, despite the fact that Arp4 has the highest similarity to conventional actin in the nuclear ARPs (Dion et al., 2010). We performed a luciferase reporter assay using an SRF binding cis-element CArG box in HeLa cells. Only Arp5 markedly suppressed Myocd-induced activation of SRF–CArG signaling (34.0 ± 2.7% of vehicle control; P < 0.001, Student’s t test; Fig. 1 B). Although the suppression by Arp8 was also statistically significant, the efficacy was considerably low (82.1 ± 1.7%; P = 0.008; Fig. 1 B). Thus, Arp5 is a promising candidate for the nuclear regulatory factor of Myocd. In addition, we confirmed that exogenous Myocd and Arp5 formed a complex in HeLa cells (Fig. S1) and that purified recombinant Myocd and Arp5 proteins directly interacted with each other in vitro (Fig. 1 C). In the A7r5 rat aortic smooth muscle cell line, endogenous Myocd and Arp5 were localized in nucleus (Fig. 1 D) and formed a complex (Fig. 1 E). Furthermore, neither overexpression nor knockdown of Arp5 affected the nuclear localization of Myocd (Fig. 1, F and G). This indicated that Arp5 directly bind to Myocd and suppress its activity in the nucleus. MRTF-A interacted with Arp5, although this interaction was very weak as compared with that with β-actin (Fig. 1 H). In addition, MRTF-A activity was remarkably suppressed by excess β-actin but not by Arp5 (Fig. 1 I), which suggested that Arp5 was not involved in regulating MRTF-A activity.
Figure 1.
Arp5 interacts with Myocd and suppresses Myocd activity. (A) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (B) The indicated expression vectors were transfected into HeLa cells with the Myocd expression vector and 3× CArG-Luc, and the luciferase reporter assay was performed. Data represent the mean ± SEM of seven independent experiments. *, P < 0.05, Student’s t test. (C) Direct interaction between Myocd and Arp5 was determined by coimmunoprecipitation assay using bacterially synthesized recombinant Myocd and Arp5 proteins (left). Purified recombinant proteins were visualized on a Coomassie-stained gel (right). Arrowheads indicate the positions of full-length recombinant Myocd and Arp5 proteins. Because the Myocd protein was too large for bacterial expression, it was partially degraded. (D) Immunostaining of endogenous Arp5 (left, green) and Myocd (right, green) in A7r5 cells. The nuclei were visualized by Hoechst 33342 (blue). Bars, 50 µm. (E) Interaction between endogenous Myocd and Arp5 was determined by coimmunoprecipitation using anti-Myocd antibody in A7r5 cells. (F) A7r5 cells were transfected with the myc-Arp5 expression vector and then immunostained with anti-Myocd (green) and anti-myc (red) antibodies. Bar, 50 µm. (G) FLAG-Myocad expression vector was transfected into A7r5 cells with a myc-Arp5 expression vector (Arp5OE) or Arp5 siRNAs (Arp5 si1 and si2). Arp5 protein expression levels were determined by Western blotting (top panels). The subcellular distribution of FLAG-Myocd was determined by cell fractionation and Western blotting (bottom panels). C and N indicate the cytoplasmic fraction and the nuclear fraction, respectively. (H) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (I) The indicated expression vectors were transfected into HeLa cells with the MRTF-A expression vector and 3×CArG-Luc, and the luciferase reporter assay was performed. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test.
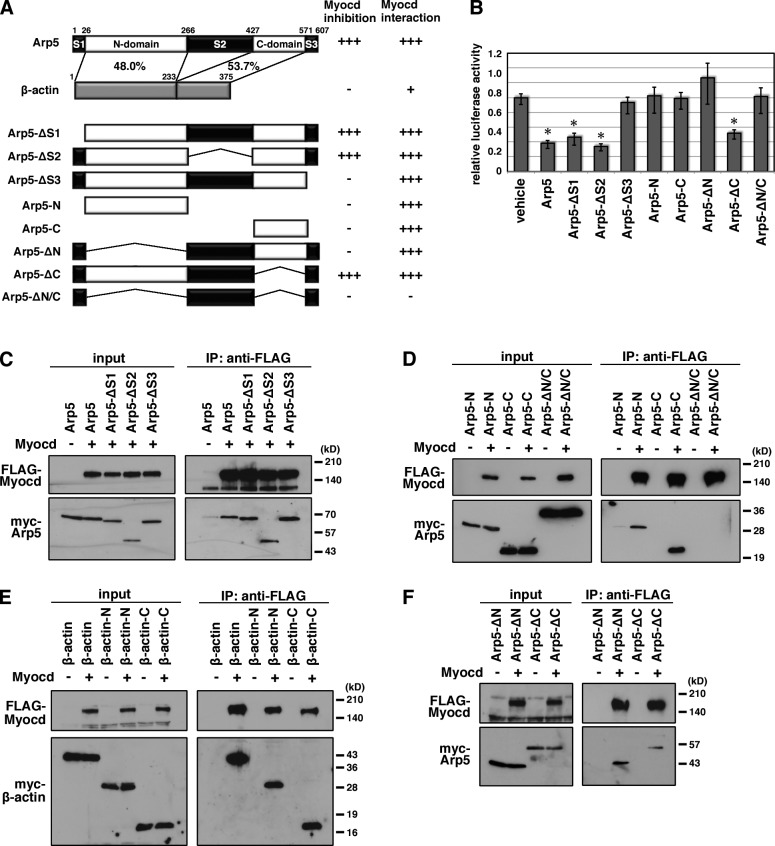
Identification of Myocd binding and inhibitory domains of Arp5
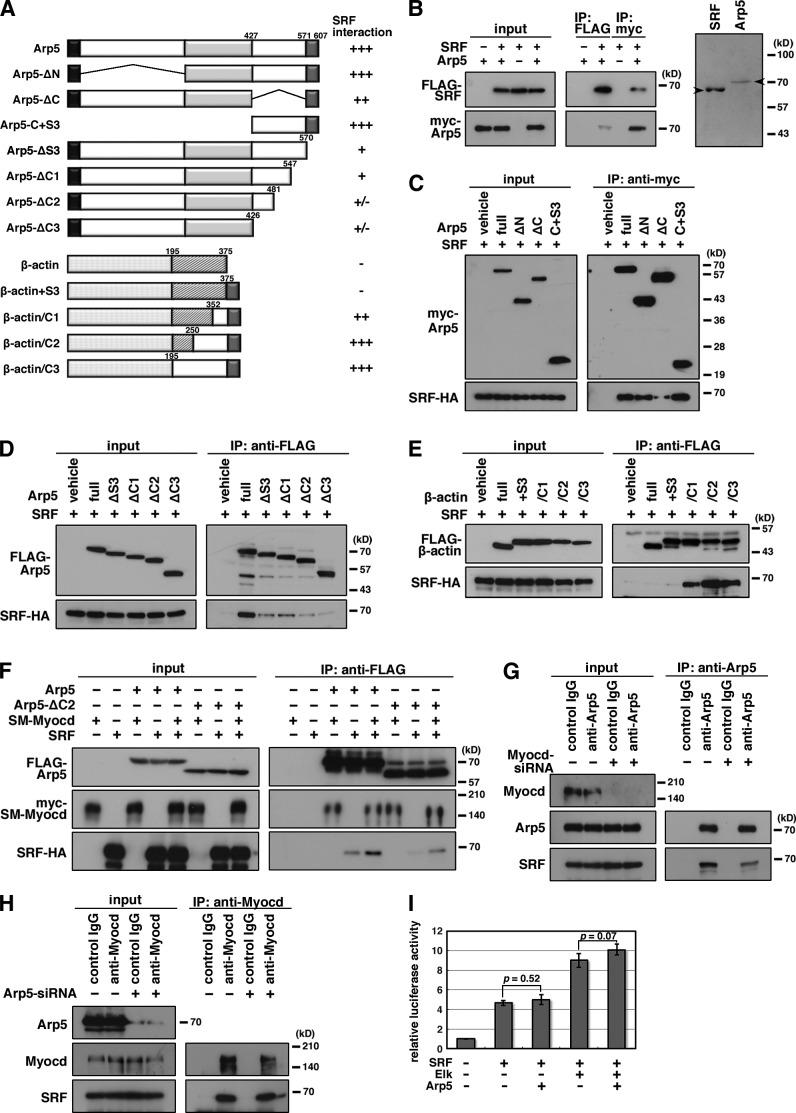
Arp5 has short unique sequences at its N terminus (S1) and C terminus (S3) and a long insertion sequence at its central region (S2; Figs. 2 A and S2). In the luciferase reporter assay, the S3 sequence was necessary for Arp5 to suppress Myocd activity, whereas deletion of S1 and S2 sequences did not affect the Arp5 function (Fig. 2 B). In contrast, full-length Arp5 and S1, S2, or S3 deletion mutants equivalently bound to Myocd (Fig. 2 C). N-terminal half (N-domain) and C-terminal half (C-domain) sequences, which are conserved regions in actin family proteins (Figs. 2 A and S2), bound to Myocd (Fig. 2 D), although they did not suppress Myocd activity (Fig. 2 B). We confirmed that N- and C-terminal half sequences of β-actin also bound to Myocd (Fig. 2 E). The N/C-domain deletion mutant (Arp5-ΔN/C) did not bind to Myocd and suppress its activity (Fig. 2, B and D). Although the N-domain deletion mutant (Arp5-ΔN) and the C-domain deletion mutant (Arp5-ΔC) contained the S3 sequence, bound to Myocd (Fig. 2 F), Arp5-ΔN did not suppress Myocd activity (Fig. 2 B). These data indicate that Arp5 interacts with Myocd through the N/C-domains, whereas the N-domain and S3 sequence are necessary for its inhibitory function against Myocd.
Figure 2.
Mapping the interaction domain of Arp5 with Myocd. (A) Schematic representation of the domain deletion constructs of Arp5. Numbers indicate amino acid positions. S1, S2, and S3 are N-terminal, central, and C-terminal sequences, respectively, unique to Arp5. N- and C-domain are conserved N- and C-terminal regions, respectively, between Arp5 and β-actin. The similarities between Arp5 N/C-domains and β-actin N/C-domains are indicated as percentages. (B) The indicated expression vectors were transfected into HeLa cells with the Myocd expression vector and 3× CArG-Luc, and the luciferase reporter assay was then performed. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test. (C–F) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed.
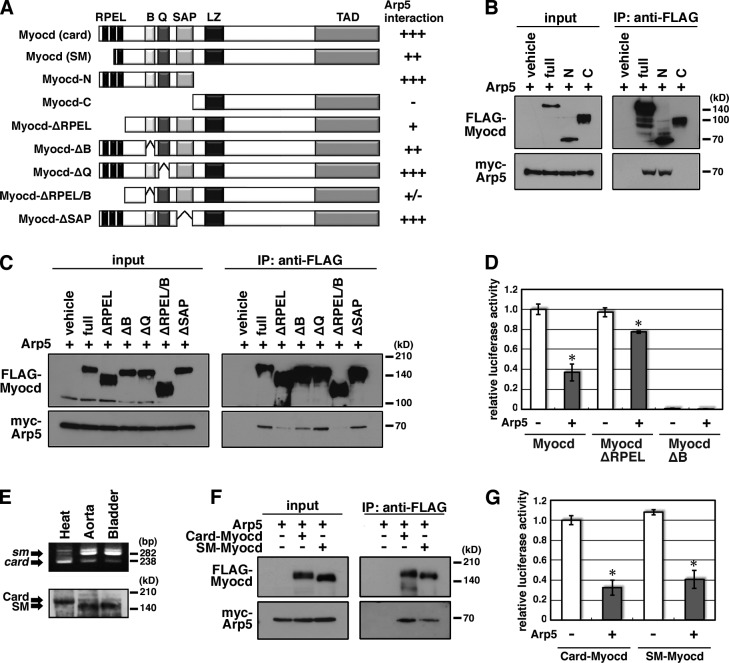
Identification of Arp5 binding domains of Myocd
To identify the interaction domain of Myocd with Arp5, coimmunoprecipitation was performed with various domain deletion mutants of Myocd (Fig. 3 A). Arp5 tightly bound to the Myocd N-terminal fragment (Myocd-N), containing RPEL domain, a basic region, a Q-rich region, and an SAP domain, but not to the C-terminal fragment (Myocd-C; Fig. 3 B). The RPEL domain deletion mutant (Myocd-ΔRPEL) exhibited considerably weak interaction with Arp5 (Fig. 3 C). The Q-rich region and SAP domain were not involved in the Arp5–Myocd interaction, whereas deletion of the basic region attenuated their interaction (Fig. 3 C). Double deletion of the RPEL motifs and basic region almost eliminated the interaction (Fig. 3 C). The luciferase reporter assay also showed the importance of RPEL domain in Arp5-mediated Myocd inhibition, although Myocd-ΔRPEL activity was still weakly suppressed by Arp5 (79.6 ± 1.3% of Arp5–Myocd-ΔN; P = 0.015), which may be caused by the remaining basic region (Fig. 3 D). The contribution of the basic region was not investigated because it is necessary for Myocd–SRF interaction (Fig. 3 D; Wang et al., 2001).
Figure 3.
Mapping the interaction domain of Myocd with Arp5. (A) Schematic representation of the domain deletion constructs of Myocd. RPEL, RPEL domain; B, basic region; Q, Q-rich region; SAP, SAP domain; LZ, leucine zipper; TAD, transcription activation domain. (B and C) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (D) The indicated expression vectors were transfected into HeLa cells with 3× CArG-Luc, and the luciferase reporter assay was performed. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test. (E) Expression pattern of SM/Card-Myocd in the rat heart, aorta, and bladder was determined by RT-PCR (top) and Western blotting (bottom). RT-PCR was performed with the primer pair for amplifying myocd exon 2–5, including (SM-type, top arrow) or excluding (Card-type, bottom arrow) exon 2a. In Western blotting, anti-Myocd antibody recognized high molecular weight Card-Myocd (top arrow) and low molecular weight SM-Myocd (bottom arrow). (F) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (G) The indicated expression vectors were transfected into HeLa cells with 3× CArG-Luc, and the luciferase reporter assay was performed. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test.
Creemers et al. (2006) reported two alternative splicing isoforms of Myocd, a cardiac isoform (Card-Myocd) and a smooth muscle isoform (SM-Myocd). sm-myocd mRNA contains an alternative exon 2a between exon 2 and 3, resulting in the synthesis of 79–amino acid–shorter proteins compared with Card-Myocd. We confirmed that SM-Myocd was mainly expressed in smooth muscle–rich tissues such as the aorta and bladder (Fig. 3 E). Although SM-Myocd lacks RPEL1 and RPEL2 motifs (Fig. 3 A), Arp5 bound to SM-Myocd and suppressed its activity (37.8 ± 1.5% of SM-Myocd/(−)Arp5; P = 0.003; Fig. 3, F and G).
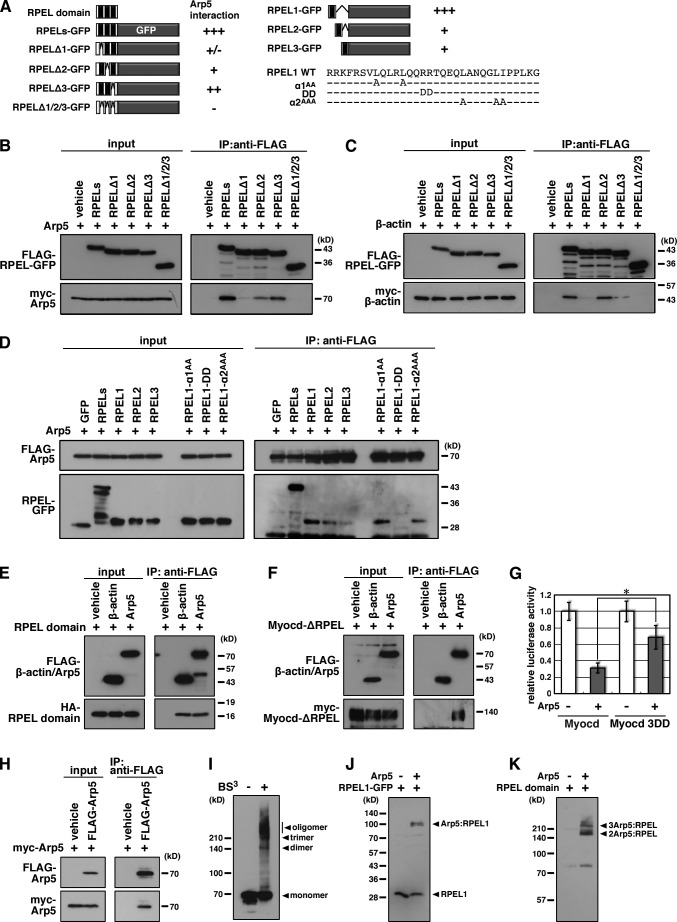
Arp5 binds to Myocd RPEL motifs
To characterize the interaction between Arp5 and Myocd RPEL domain in detail, we constructed the expression vectors of RPEL domain or RPEL motifs fused with the N terminus of GFP (Fig. 4 A). GFP-fused RPEL domain (RPELs-GFP) bound to Arp5, and the deletion of each of the RPEL1, 2, and 3 motifs diminished the interaction with Arp5 (Fig. 4 B). For β-actin, the deletion of RPEL2 motif did not affect its interaction with RPELs-GFP (Fig. 4 C), which was consistent with a previous study showing that the RPEL2 motif dose not contribute to the interaction of β-actin with Myocd RPEL domain (Guettler et al., 2008). Arp5 also bound to GFP-fused 32–amino acid RPEL motif peptides (RPEL1-, RPEL2-, and RPEL3-GFP), but not to GFP only (Fig. 4 D), which indicated that all three RPEL motifs were binding sites for Arp5. As shown in Fig. 1 A, full-length Myocd bound more tightly to Arp5 than to β-actin, but RPEL domain binding to Arp5 and β-actin was comparable (Fig. 4 E). These differences were probably because Arp5 but not β-actin could bind to other regions of Myocd than the RPEL domain (Figs. 3 C and 4 F).
Figure 4.
Interaction between Arp5 and Myocd RPEL motifs. (A) Schematic representation of the constructs of the RPEL domain and 32–amino acid RPEL motif peptides that were fused with the N terminus of GFP. (B and C) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (D) The GFP-fused RPEL domain and RPEL motifs were synthesized in vitro. His-FLAG-Arp5 was synthesized in Hek293T cells and purified using TALON resin. These proteins were coincubated for 1 h at room temperature, after which coimmunoprecipitation was done. (E) The HA-tagged RPEL domain was synthesized in vitro. His-FLAG-β-actin and His-FLAG-Arp5 were synthesized in Hek293T cells and purified using TALON resin. These proteins were coincubated for 1 h at room temperature, after which coimmunoprecipitation was done. (F) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (G) The indicated expression vectors were transfected into HeLa cells with 3× CArG-Luc, and the luciferase reporter assay was performed. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test. (H) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (I) His-FLAG-Arp5 was synthesized in Hek293T cells and purified using TALON resin. The purified FLAG-Arp5 was chemically cross-linked using 0.5 mM BS3 in a PBS cross-linking buffer and then separated and detected by Western blotting using an anti-FLAG antibody. (J) The RPEL1-GFP proteins were synthesized in vitro. The RPEL1-GFP and purified FLAG-Arp5 proteins were coincubated in a Hepes cross-linking buffer for 1 h and then cross-linked using 0.5 mM BS3. The cross-linked complexes were separated and detected by Western blotting using anti-GFP antibody. (K) HA-RPEL domain was synthesized in vitro. HA-RPEL domain and purified FLAG-Arp5 proteins were coincubated in a Hepes cross-linking buffer for 1 h and then cross-linked using 0.5 mM BS3. The cross-linked complexes were separated and detected by Western blotting using anti-HA antibody.
A previous structural study determined the critical residues in RPEL motifs for binding to β-actin (Mouilleron et al., 2008). Two leucine residues in helix α1, two arginine or proline residues in the R-loop, and three leucine or isoleucine residues in helix α2 are critical for the interactions between RPEL motifs and β-actin. However, in the case of Arp5, mutations in helix α1 or α2 of the Myocd RPEL1 motif (α1AA or α2AAA) retained the interactions with Arp5, whereas mutations in the R-loop (DD) eliminated this interaction (Fig. 4, A and D). This experiment therefore shows that although Arp5 can bind Mycd RPEL1 sequence, its mode of interaction must be significantly different from that of actin itself. Based on these results, we constructed a Myocd-3DD mutant in which all of the critical residues in the R-loop of Myocd RPEL1, 2, and 3 motifs were substituted by aspartate. Similar to Myocd-ΔRPEL (Fig. 3 D), the activity of Myocd-3DD was only modestly suppressed by Arp5 (Fig. 4 G).
Unlike conventional actin, nuclear ARPs do not form filamentous structure by polymerization. However, Arp4 and Arp8 have been reported to form homodimers or heterodimers in vitro (Fenn et al., 2011; Saravanan et al., 2012). Coimmunoprecipitation assay revealed that Arp5 bound tightly to itself (Fig. 4 H). Furthermore, homooligomer formation of purified recombinant Arp5 protein was found in vitro by chemical cross-linking using bis(sulfosuccinimidyl) suberate (BS3; Fig. 4 I), indicating that Arp5 also formed homodimers/homooligomers, at least under the experimental conditions. The chemical cross-linking between purified Arp5 and RPEL1-GFP contained a 30-kD RPEL1-GFP and an ∼100-kD complex that was predicted to be between RPEL1-GFP and monomeric Arp5 with a 1:1 stoichiometry, which were detected by Western blotting using an anti-GFP antibody (Fig. 4 J). Thus, RPEL motifs appear to interact with only the monomeric form of Arp5 protein, similar to that of β-actin. Cross-linking between 17-kD RPEL domain peptides and Arp5 resulted in ∼160- and 240-kD complexes, which were predicted to comprise two or three molecules of Arp5 and RPEL domain peptide (Fig. 4 K). Consistent with the Arp5 binding studies using isolated RPEL motif peptides, this experiment suggests that the Mycd RPEL domain binds multiple Arp5 molecules.
Arp5 S3 sequence is necessary for Arp5–SRF interaction
In a previous study, the physical interaction between conventional actin and SRF was not detectable (Posern et al., 2002). In contrast, the interaction between Arp5 and SRF was detected by coimmunoprecipitation (Fig. 5, A–C; and Fig. S1). The C-terminal region of Arp5, particularly the S3 sequence, was important for this interaction (Fig. 5, C and D). When β-actin C-terminal sequences were replaced by equivalent C-terminal regions of Arp5 containing the S3 sequence, these chimeric proteins bound tightly to SRF (Fig. 5 E), indicating that the Arp5 C-terminal region is critical for SRF binding. To determine whether Arp5 forms a ternary complex with SRF and Myocd, coimmunoprecipitation was performed in the presence of Myocd. Arp5 simultaneously bound to SRF and Myocd, and the Arp5–SRF interaction was remarkably enhanced in the presence of Myocd (Fig. 5 F). Consistent with this, the depletion of endogenous Myocd by RNA interference reduced the Arp5–SRF interaction in A7r5 cells, whereas Arp5 knockdown did not affect the Myocd–SRF interaction (Fig. 5, G and H). Furthermore, Arp5-ΔC2 bound to SRF only in the presence of Myocd (Fig. 5 F), which suggested that Myocd formed a bridge between SRF and Arp5-ΔC2. Thus, Arp5 could form a ternary complex with SRF and Myocd.
Figure 5.
Mapping the interaction domain of Arp5 with SRF. (A) Schematic representation of the deletion and chimeric constructs of Arp5 and β-actin. Numbers indicate amino acid positions. (B) The direct interaction between SRF and Arp5 was determined by coimmunoprecipitation using bacterially synthesized recombinant SRF and Arp5 proteins (left). Purified proteins were visualized on a Coomassie-stained gel (right). Arrowheads indicate the positions of full-length recombinant SRF and Arp5 proteins. (C–F) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (G and H) A7r5 cells were transfected with Myocd siRNA (G) or Arp5 siRNA1 (H) for 3 d, after which a coimmunoprecipitation assay was done using the indicated antibodies. (I) The indicated expression vectors were transfected into HeLa cells with the c-fos promoter reporter construct, and the luciferase reporter assay was performed. Data represent the mean ± SEM of four independent experiments. P-values were calculated using Student’s t test.
The formation of the Arp5–SRF complex raises the possibility that Arp5 directly suppresses SRF activity independently of Myocd. The promoter region of c-fos contains a CArG box and a ternary complex factor binding site. The c-fos promoter is reportedly enhanced by SRF with its cofactor Elks but not by Myocd (Zhou and Herring, 2005). SRF and Elk-1 coordinately increased the c-fos promoter activity, and Arp5 did not suppress them (Fig. 5 I), suggesting that Arp5 cannot directly suppress SRF activity in the absence of Myocd.
Arp5 suppresses CArG-dependent expression of smooth muscle genes
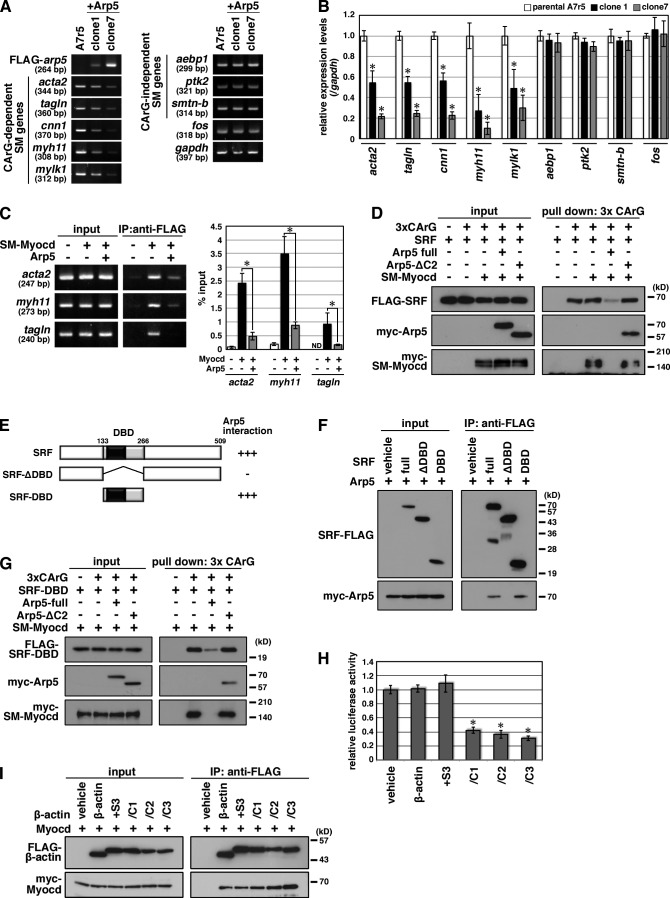
To determine the effect of excess Arp5 on the endogenous expression of smooth muscle genes, stable cell lines overexpressing Arp5 were established in A7r5 cells (A7r5 + Arp5). The expression levels of smooth muscle marker genes that were driven by the SRF–CArG box were decreased in these clones, particularly in clone 7, which highly expressed exogenous Arp5 (Fig. 6, A and B). In contrast, there was no difference in the expression levels of CArG-independent smooth muscle marker genes and c-fos between A7r5 + Arp5 cells and parental A7r5 cells (Fig. 6, A and B). A chromatin immunoprecipitation (ChIP) assay showed that Arp5 inhibited SM-Myocd recruitment to the CArG box–containing promoter regions of acta2, myh11, and tagln in A7r5 cells (Fig. 6 C). In addition, DNA–protein binding assay revealed that Arp5 prevented the SM-Myocd–SRF complex from binding to a 3× CArG DNA probe but Arp5-ΔC2 did not (Fig. 6 D). Arp5 bound to a DNA binding domain (DBD) of SRF and inhibited SRF-DBD binding to the CArG probe (Fig. 6, E–G), which suggested that the Arp5 C-terminal region masked the DNA binding site of SRF. The chimeric proteins β-actin/C1, C2, and C3, which included the C-terminal SRF binding domain of Arp5 (Fig. 5 A), suppressed Myocd activity (Fig. 6 H), whereas β-actin and β-actin + S3 did not, despite that they also bound to Myocd (Fig. 6 I). This supported our hypothesis that the Arp5 C-terminal region is critical for Myocd–SRF inhibition.
Figure 6.
Arp5 suppresses the expression of CArG-dependent smooth muscle–specific genes. (A) Total RNA was extracted from FLAG-Arp5–expressing stable cell lines (A7r5 + Arp5, clone 1 and clone 7) and parental A7r5 cells, and RT-PCR was performed with the indicated gene-specific primer pairs. (B) Smooth muscle genes’ expression levels in A7r5 cells and A7r5 + Arp5 cell lines were quantitated by real-time RT-PCR. Data represent the mean ± SEM of three independent experiments. *, P < 0.05, Student’s t test. (C) A7r5 cells were transfected with the indicated combinations of expression vectors, after which a ChIP assay was done. Immunoprecipitated DNA was visualized by PCR using primer pairs for amplifying a part of the promoter region of acta2, myh11, or tagln (left) or quantitated by real-time PCR (righ). ND, not detected. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test. (D) A DNA–protein binding assay was done using a biotinylated 3× CArG DNA probe. The DNA–protein complexes were pulled down using Dynabeads M-280 Streptavdin. (E) Schematic representation of the domain deletion constructs for SRF. Gray box, DBD; black box, MADS domain. Numbers indicate amino acid positions. (F) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed. (G) A DNA–protein binding assay was performed using a biotinylated 3× CArG DNA probe. The DNA–protein complex was pulled down using Dynabeads M-280 Streptavdin. (H) The indicated expression vectors were transfected into HeLa cells with 3× CArG-Luc, and the luciferase reporter assay was performed. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test. (I) Hek293T cells were transfected with the indicated combinations of expression vectors and coimmunoprecipitation was performed.
Quantitative regulation of Arp5 expression by smooth muscle–specific alternative splicing
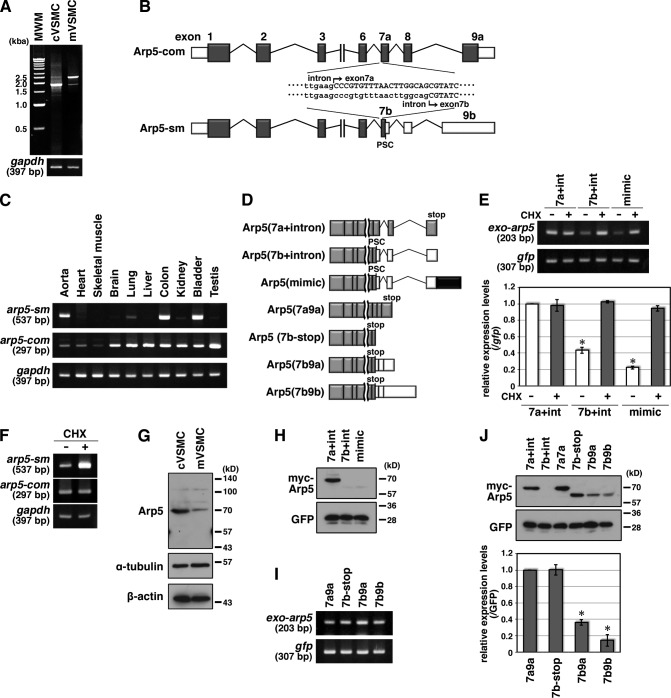
Cultured rat VSMCs expressed a common variant of arp5 mRNA with a 1,827-bp open reading frame (arp5-com; Fig. 7 A). In medial VSMC layers from rat abdominal aortae, however, ∼2,500-bp arp5 mRNA, referred to as arp5-sm, was majorly expressed (Fig. 7 A). cDNA sequence analysis revealed that arp5-sm mRNA contained a 19-bp-short exon 7 (exon 7b) and a 661-bp-long exon 9 (exon 9b) compared with arp5-com mRNA (Figs. 7 B and S3). The short exon 7b leads to a frameshift and premature stop codon (PSC) in exon 7b; therefore, arp5-sm mRNA is predicted to encode a 456–amino acid protein (Figs. 7 B and S3). When the expression profile of these splicing variants was investigated in various tissues, arp5-sm expression was highly restricted to smooth muscle–rich tissues such as the aorta, colon, and bladder (Fig. 7 C). arp5-com mRNA was broadly expressed; however, its expression level was comparably low in the aorta, heart, and skeletal muscle (Fig. 7 C).
Figure 7.
Quantitative regulation of Arp5 in smooth muscle cells by alternative splicing. (A) Total RNA was extracted from cultured VSMCs (cVSMC) and medial VSMC layers of abdominal aortae (mVSMC), and RT-PCR was performed with a primer pair for amplifying full-length arp5 mRNA. (B) Schematic representation of the alternative splicing of rat arp5. White box, 5′ and 3′UTR; gray box, exon; bar, intron. (C) Tissue-expression pattern of arp5-sm and arp5-com mRNA. Total RNA was extracted from the indicated rat tissues, and RT-PCR was performed with arp5 variant-specific primer pairs. (D) Schematic representation of expression constructs for arp5 variants. White box, 3′UTR; gray box, exon; black box, exon 9b characteristic sequence; bar, intron. (E) NMD of exogenous arp5 variants. HeLa cells were cotransfected with the indicated Arp5 variants and GFP expression vectors and treated with 100 µg/ml CHX or DMSO as a vehicle control for 4 h. Total RNA was extracted and RT-PCR was performed with a primer pair for amplifying exogenous arp5 (exo-arp5) or gfp (top panels). The amounts of PCR products were quantified by densitometry with normalization to gfp mRNA and statistically analyzed (bottom). Data represent the mean ± SEM of four independent experiments.*, P < 0.05, Student’s t test. (F) NMD of endogenous arp5 mRNA in cultured VSMCs. Total RNA was extracted from cultured VSMCs treated with CHX or DMSO for 4 h. RT-PCR was performed with arp5 variant-specific primer pairs. (G) Total proteins were extracted from cultured VSMCs (cVSMC) and medial VSMC layers of abdominal aortae (mVSMC), and Western blotting was performed with anti-Arp5 antibody and anti–α-tubulin and anti–β-actin antibodies as loading controls. (H) HeLa cells were cotransfected with the indicated Arp5 variant and GFP expression vectors, and Western blotting was performed with anti-myc and anti-GFP antibodies. (I) HeLa cells were cotransfected with the indicated Arp5 variant and GFP expression vectors. Total RNA was extracted and RT-PCR was performed with the indicated gene-specific primer pairs. (J) HeLa cells were cotransfected with the indicated Arp5 variant and GFP expression vectors, and Western blotting was performed with anti-myc and anti-GFP antibodies (top panels). The amounts of Arp5 variant proteins were quantified by densitometry with normalization to the GFP protein and statistically analyzed (bottom). Data represent the mean ± SEM of three independent experiments. *, P < 0.05, Student’s t test.
In general, mRNA containing PSC is degraded by an mRNA decay pathway, known as nonsense-mediated mRNA decay (NMD; Kervestin and Jacobson, 2012). We constructed Arp5 expression vectors containing introns 7 and 8 (Fig. 7 D) because introns downstream of PSC are required for mRNAs to undergo NMD (Kervestin and Jacobson, 2012). The Arp5(7a + intron) construct, which contained exons 1–7a, 8, and 9a and introns 7 and 8, was transcribed into arp5-com mRNA. The Arp5(7b + intron) construct contained exon 7b instead of exon 7a, and it was transcribed into chimeric mRNA containing exons 7b and 9a because intron 8 was spliced in the same manner as arp5-com even in cultured VSMCs and A7r5 cells. To mimic the splicing product of arp5-sm, the 661-bp sequence specific for exon 9b (Fig. S3) was inserted upstream of exon 9a or downstream of exon 8; however, the inserted sequence interfered with the normal splicing of intron 8. Therefore, the specific sequence for exon 9b was inserted downstream of exon 9a (Arp5[mimic]), resulting in the splicing into arp5-sm–like mRNA containing exon 7b and 9b–like exon 9 (Fig. 7 D). When equal amounts of these vectors were transfected into HeLa cells, the amounts of mRNAs transcribed from Arp5(7b + intron) and Arp5(mimic) were lesser than that transcribed from Arp5(7a + intron) (40.7 ± 2.1% of Arp5[7a + intron]; P = 0.001, in Arp5(7b + intron); 20.5 ± 1.0%; P < 0.001, in Arp5[mimic]; Fig. 7 E). This reduced expression was restored by treatment with cycloheximide (CHX), which inhibits the NMD pathway (Carter et al., 1995), although the expression levels of cotransfected GFP were not changed (Fig. 7 E). In cultured VSMCs, arp5-sm mRNA was slightly expressed (Fig. 7 A) and its expression levels were markedly increased by CHX treatment, whereas the amount of arp5-com mRNA remained unchanged (Fig. 7 F). These data strongly suggest that arp5-sm mRNA is a target of NMD and is partially degraded in smooth muscle cells.
As shown in Fig. 7 A, the expression of arp5-sm mRNA was sufficiently detected in medial VSMC layers, although its expression level was lower than that of arp5-com mRNA in cultured VSMCs. This may have been caused by incomplete degradation by NMD as in the case of Arp5(7b + intron) and Arp5(mimic). However, the expression of Arp5-sm protein, which is predicted to have a lower molecular mass than 70-kD Arp5-com proteins, was not detected in medial VSMC layers (Fig. 7 G). Furthermore, Arp5(7b + intron) and Arp5(mimic) constructs were also scarcely translated, although there were transcribed in HeLa cells (Fig. 7, E and H), raising the possibility that besides NMD, translational control is involved in Arp5 expression. Without introns and 3′UTR sequences, Arp5-com and Arp5-sm expression constructs (Arp5[7a9a] and Arp5[7b − stop]) were equally transcribed and translated as 70- and 63-kD proteins, respectively (Fig. 7, I and J). To investigate the involvement of 3′UTR in translational suppression, we constructed Arp5-sm expression vectors with 3′UTR, including exon 9a (Arp5[7b9a]) or exon 9b (Arp5[7b9b]). These constructs were transcribed at the same level as Arp5(7b-stop) (Fig. 7 I); however, their translational levels were significantly reduced (36.2 ± 1.9% of Arp5[7a9a], P < 0.001, in Arp5[7b9a]; 14.5 ± 4.1%, P = 0.002, in Arp5[7b9b]; Fig. 7 J). These data indicate that Arp5-sm expression is strongly suppressed by a dual mechanism: PSC-mediated NMD and 3′UTR-mediated translational suppression. The degree of these suppressive effects was more severe in exon 9b–containing mRNAs compared with that in exon 9a–containing mRNAs (Fig. 7, E and J), indicating that an alternative exon 7b was critical for the quantitative regulation of Arp5-sm and that an alternative exon 9b also modestly contributed to this regulation.
Arp5 knockdown restores smooth muscle phenotype in VSMCs
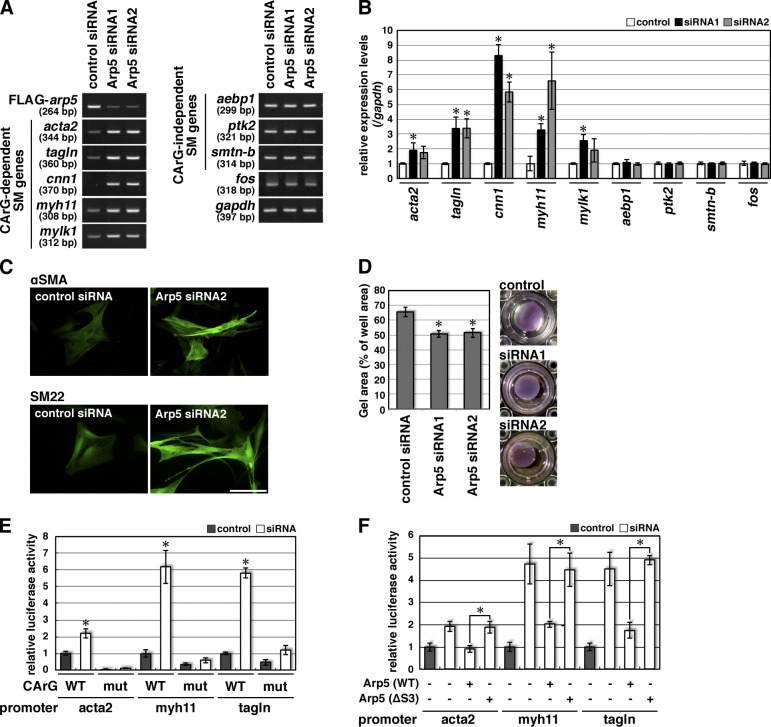
As shown in Fig. 7 G, the expression levels of Arp5 proteins were extremely low in well-differentiated VSMCs, caused by the splicing switch of arp5. To investigate the contribution of Arp5–Myocd signaling in the modulation of the smooth muscle phenotype, Arp5 expression was knocked down by RNA interference in cultured VSMCs. In Arp5-knockdown VSMCs, the expression levels of CArG-dependent smooth muscle marker genes were significantly increased, whereas those of CArG-independent marker genes and c-fos were not altered (Fig. 8, A and B). Arp5-knockdown VSMCs showed a more spindle-shaped morphology with enhanced expression of contractile proteins αSMA and SM22 (Fig. 8 C), and therefore increased contractility as shown in a collagen gel contraction assay (77.7% of control siRNA, P < 0.001, in Arp5 siRNA1; 78.5%, P < 0.001, in Arp5 siRNA2; Fig. 8 D). Thus, decreased expression of Arp5 is involved in the maintenance of the differentiation phenotype of VSMCs. The luciferase promoter assay using CArG containing the promoter regions of acta2, myh11, and tagln demonstrated that Arp5 knockdown strongly enhanced activities of these promoters but not those of CArG-mutated ones in A7r5 cells (Fig. 8 E). These enhanced promoter activities were reversed by reconstitution with wild-type Arp5 expression but not with Arp5-ΔS3 expression (Fig. 8 F). These data support the hypothesis that Arp5 modulates the smooth muscle phenotype by regulating the expression levels of smooth muscle genes via Myocd–SRF–CArG signaling.
Figure 8.
Knockdown of Arp5 restores smooth muscle phenotypes in dedifferentiated VSMCs. (A) Total RNA was extracted from VSMCs transfected with control or Arp5 siRNA for 4 d, and RT-PCR was performed with the indicated gene-specific primer pairs. (B) Smooth muscle genes’ expression levels in the control or Arp5-knockdown VSMCs were quantified by real-time RT-PCR. Data represent the mean ± SEM of three independent experiments. *, P < 0.05, Student’s t test. (C) Cultured VSMCs were transfected with control or Arp5 siRNA2 for 4 d, and immunostaining was performed with anti-αSMA (top) or anti-SM22 (bottom) antibodies. Bar, 100 µm. (D) VSMC contractility was determined by a collagen gel lattice contraction assay. VSMCs were transfected with control or Arp5 siRNA for 3 d and then seeded into collagen gels. The contractile response was initiated by stimulation with 100 µM carbachol. The well area was set as 100%. Data represent the mean ± SEM of three independent experiments. *, P < 0.05, Student’s t test. (E) A7r5 cells were transfected with control or Arp5 siRNA2 for 5 d, and the luciferase reporter assay was performed using the indicated promoter–reporter constructs, which contained wild-type (WT) or mutated (mut) CArG boxes. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test. (F) Human Arp5 or Arp5-ΔS3 expression vector was transfected into A7r5 cells with the indicated promoter–reporter constructs. Subsequently, these cells were transfected for 5 d with control or Arp5 siRNA1, which was designed against endogenous rat Arp5 but not against human Arp5, and then the luciferase reporter assay was performed. Data represent the mean ± SEM of four independent experiments. *, P < 0.05, Student’s t test.
Discussion
Previous studies have elucidated the regulatory mechanism of MRTFs via interaction with G-actin through their N-terminal RPEL motifs (Miralles et al., 2003; Guettler et al., 2008). In contrast, the functional significance of Myocd RPEL motifs was not understood because of their considerably low affinity for G-actin (Guettler et al., 2008). In the present study, we demonstrated that Myocd RPEL motifs bound to Arp5 by alternative mechanism, which was partly different from the conventional actin binding mechanism (Fig. 4). We have summarized the interaction domains and functional mechanisms of Arp5 in smooth muscle cell differentiation in Fig. S4. Arp5 interacts with Myocd through its N- and C-domain, whereas it interacts with SRF through the C-terminal region, which contains the C-domain and S3 sequence. The N-domain–mediated interaction with Myocd is necessary for inhibiting Myocd–SRF activity, and the C-domain does not substitute for it. Both the C-domain and the S3 sequence are required for the full extent of the SRF binding, but the S3 sequence is only essential for the Myocd–SRF inhibition. Thus, the N-domain and the S3 sequence are critical regions for the inhibitory role of Arp5 during Myocd–SRF signaling. In differentiated VSMCs, the smooth muscle–specific splicing isoform of arp5 (arp5-sm) is primarily expressed, although its mRNA is largely degraded by the NMD pathway. In addition, its expression is remarkably suppressed at the translational level, resulting in a low expression level of Arp5 protein in differentiated VSMCs. In contrast, dedifferentiated VSMCs highly express the common variant of Arp5 (Arp5-com), which forms a ternary complex with Myocd and SRF and inhibits their DNA binding; this results in low Myocd–SRF signaling activity.
Arp5 has a unique S3 sequence that is not conserved among other actin family proteins. This sequence was essential for the Arp5-mediated Myocd inhibition (Figs. 5 and 6), and the chimeric β-actin proteins that have the Arp5 C-terminal region acquired the ability to inhibit Myocd–SRF signaling (Fig. 6 H). Thus, the distinct function of Arp5 among actin family proteins is defined by this C-terminal sequence. Although Arp5 and SRF were directly associated, Arp5 did not suppress SRF activity in the absence of Myocd (Fig. 5 I). The binding of Arp5 to SRF was rather weak as compared with that to Myocd (Fig. S1). Therefore, Myocd reinforced the Arp5–SRF interaction by forming a bridge between Arp5 and SRF (Fig. 5, F and G). This suggested that the interaction between Arp5 and SRF in the absence of Myocd may be too weak to suppress SRF activity.
Although Arp5 functions have been poorly understood, some studies revealed that Arp5 is one of the components of the chromatin-remodeling complex INO80 (Shen et al., 2003; Kitayama et al., 2009). Arp5 is required for the recruitment of INO80 complex to chromatin, and the depletion of Arp5 impairs the DNA repair process after DNA double-strand breaks (Kitayama et al., 2009). Considering that chromatin remodeling by INO80 complex is also involved in the transcriptional regulation of several yeast genes (Ebbert et al., 1999; Shen et al., 2000), there remains the possibility that Arp5 regulates smooth muscle differentiation via the INO80 complex–mediated chromatin-remodeling pathway. INO80 complex generally enhances transcriptional activity by opening chromosomal structure (Ebbert et al., 1999; Shen et al., 2000; Neumann et al., 2012). In contrast, Arp5 knockdown, which impairs INO80 complex function, increased the expression levels of SRF-dependent smooth muscle genes accompanied by morphological changes and enhanced contractility of VSMCs (Fig. 8). These changes were also reported for cells that exogenously overexpressed Myocd (Yin et al., 2011). In addition, the luciferase reporter assay using episomal plasmids revealed increase in the CArG-dependent promoter activity by Arp5 knockdown, which was reversed by reconstitution with exogenous wild-type Arp5 but not with mutant Arp5-ΔS3 that could not bind to SRF (Fig. 8, E and F). Considering this in addition to our other data, it is probable that Arp5 contributes in regulating the Myocd–SRF signaling pathway by directly interfering with the formation of the Myocd–SRF–CArG complex rather than by INO80 complex–mediated chromatin remodeling.
In addition to actin family proteins, several transcription factors were involved in the regulation of Myocd activity. FOXO4 and Msx1/2 have been reported as transcription factors that directly interact with Myocd (Liu et al., 2005b; Hayashi et al., 2006). They bind to Myocd through its SAP domain and/or basic region but not RPEL motifs. Both proteins form a ternary complex with Myocd and SRF and suppress Myocd activity. GATA4 also binds to Myocd and SRF and stimulates or suppresses Myocd activity in a target gene-specific manner (Oh et al., 2004). Some transcription factors indirectly suppress Myocd activity by binding to SRF (Doi et al., 2005; Liu et al., 2005a; Tanaka et al., 2008). Thus, Arp5 and these transcription factors would cooperatively function in Myocd regulation.
In smooth muscle cells, many kinds of contractile genes such as caldesmon, tropomyosin, vinculin, and myosin heavy chain are expressed as smooth muscle–specific variants generated by tissue-specific alternative splicing (Koteliansky et al., 1992; Aikawa et al., 1993; Kashiwada et al., 1997). The differentiation state of smooth muscle cells is highly plastic, and the expression of these variants is immediately lost after dedifferentiation. In the present study, we isolated a novel smooth muscle–specific splicing variant of arp5, arp5-sm (Fig. 7, A and B). Well-differentiated VSMCs in the aortic medial layer majorly expressed arp5-sm but scarcely expressed arp5-com. Similar to other smooth muscle genes, dedifferentiated cultured VSMCs showed a variant switch from arp5-sm to arp5-com. arp5-sm mRNA contains PSC created by frameshift, and thereby it was partially degraded by NMD, which would result in the lower expression of arp5-sm mRNA in media VSMCs than that of arp5-com mRNA in cultured VSMCs (Fig. 7, D–F). In addition to NMD, the expression level of Arp5-sm was also suppressed at the translational level (Fig. 7, H–J). The long 3′UTR downstream of PSC but not introns was required for this translational suppression. Such regulations were also reported in mutated TGFBR2 containing a frameshift mutation and PSC (You et al., 2007). mRNA of mutated TGFBR2 escapes from NMD-mediated degradation; however, its translation was suppressed depending on long 3′UTR, suggesting that the expression of some PSC-containing mRNAs is also regulated at the translational level. Thus, Arp5-sm protein expression was remarkably suppressed by the dual mechanism in differentiated VSMCs. Even if the suppression is not complete, leaking Arp5-sm protein was probably nonfunctional because it lacked the critical C-terminal sequence (Figs. S3 and S4). In differentiated VSMCs, some Arp5-com protein expression still remained (Fig. 7 G), which probably contributed in the inhibition of Myocd–SRF–dependent gene expression.
This is the first paper to identify Arp5 as a regulatory protein of Myocd via interacting with its RPEL motifs instead of conventional actin in MRTF regulation. We also identified a novel alternative splicing variant of Arp5 restricted to smooth muscle cells and broadened our understanding of Arp5 function. Although further studies are required to clarify the roles of Arp5 in smooth muscle cell differentiation during embryonic development and smooth muscle phenotypic modulation during atherosclerosis, our present results provide new insights into the regulatory mechanism of Myocd and Arp5 function.
Materials and methods
Materials
CHX and carbachol were purchased from Nacalai Tesque and Santa Cruz Biotechnology, Inc. The following antibodies were purchased: anti-Arp5 (Proteintech Group), anti-αSMA (Sigma-Aldrich), anti-SM22 (Leica), anti–α-tubulin (Sigma-Aldrich), anti–β-actin (Sigma-Aldrich), anti–c-fos (Santa Cruz Biotechnology, Inc.), anti-GFP (Invitrogen), anti-SRF (Santa Cruz Biotechnology, Inc.), anti-FLAG (Sigma-Aldrich), anti-HA (Roche), and anti-myc (Santa Cruz Biotechnology, Inc.). Anti-Myocd antibody was produced in New Zealand rabbits against a synthesized Myocd peptide (CSKSLGDSKNRHKKPKD) and purified by affinity chromatography using this peptide.
Cell culture, transfection, and RNA interference
A7r5, HeLa, and HEK293T cells were cultured in DMEM supplemented with 10% FCS. VSMCs were isolated from rat aortae as described previously (Nakamura et al., 2010). In brief, the abdominal aortae were removed from anesthetized 6-wk-old male Sprague–Dawley rats, and the medial VSMC layers were separated from the adventitia and vascular endothelium by collagenase treatment. VSMCs were dispersed by collagenase and elastase treatment and then cultured in DMEM with 10% FCS. Cultured cells were transfected using Lipofectamine LTX and Plus Reagent (Invitrogen). To establish stable cell lines expressing FLAG-Arp5, A7r5 cells were transfected with pcDNA3.1(+)-FLAG-Arp5 and cultured with 100 µg/ml geneticin (Invitrogen) to isolate drug-resistant clones. In RNA interference experiments, cultured VSMCs or A7r5 cells were transfected with siRNA against rat arp5 or control siRNA using Lipofectamine RNAi MAX (Invitrogen) and cultured for 4–5 d. MISSION siRNA Universal Negative Control (Sigma-Aldrich) was used as the control siRNA. Arp5 siRNA sequences used in this study are presented in Table S1.
Immunostaining and microscopy
VSMCs cultured on coverslips were fixed using 4% paraformaldehyde and then incubated with a blocking solution (0.1% Triton X-100, 0.2% BSA, and 10% normal goat serum, in PBS [Nacalai Tesque]). The cells were incubated with the primary antibodies diluted in the Can Get Signal immunostain reagent (TOYOBO) and then incubated with Alexa 488– or Alexa 568–conjugated secondary antibodies (Invitrogen) diluted in the blocking solution. To label the nuclei, Hoechst 33342 (Invitrogen) was added to the secondary antibody solution. The stained cells were mounted with Fluoromount (Diagnostic BioSystems) and observed using an all-in-one fluorescence microscope (BZ-9000; Keyence) with a CFI Plan Apochromat λ 20× lens (Nikon) at room temperature. Fluorescent images were acquired with an internal charge coupled device camera using BZ-II Analyzer software (Keyence) and processed with Photoshop Element 10 software (Adobe).
Expression plasmids
The coding regions for human β-actin, Arp4, Arp5, Arp6, Arp8, SRF, Elk-1, rat Arp5, mouse card-myocd, sm-myocd, MRTF-A, and the intronic sequences of rat arp5 were amplified by PCR and subcloned into the highly efficient mammalian expression plasmids pCAGGS, pCS2+, or pcDNA3.1(+) (Invitrogen) for the establishment of stable cell lines. FLAG, myc, or HA tag sequences were fused to the 5′ end of the coding sequences. Expression plasmids for SRF derivatives, SRF-DBD (amino acid 133–266) and SRF-ΔDBD, were constructed by PCR-mediated method and subcloned into the mammalian expression plasmid pcDNA3.1(+) (Invitrogen).
Luciferase reporter assay
The promoter region of human acta2 (−419/−1), myh11 (−1548/+63), tagle (−1343/+62), and c-fos (−588/+155) genes was amplified by PCR and subcloned into pGL3-basic (Promega). The CArG mutant constructs were designed with site-directed mutation in the CArG boxes (CC[A/T]6GG [wild type] to AA(A/T)6GG or CC(A/T)6AA [mutant]). The 3× CArG-Luc vector was constructed as reported previously (Morita et al., 2007b). These constructs were introduced into HeLa or A7r5 cells along with pSV-βGal (Promega), which was used to normalize the transfection efficiency. 2 d after transfection, luciferase and β-galactosidase activities were measured using the Luciferase Assay System (Promega) and Luminescent β-Galactosidase Detection Kit II (Takara Bio Inc.), respectively. For the reporter assay in Arp5-knockdown cells, A7r5 cells were transfected with reporter constructs for 24 h and then transfected with Arp5 siRNA2. 5 d after siRNA transfection, luciferase and β-galactosidase activities were measured.
Coimmunoprecipitation
HEK293T cells transfected with expression vectors were lysed with an NP-40 buffer (0.5% NP-40, 1 mM MgCl2, and protease inhibitor cocktail for use with mammalian cell and tissue extracts [Nacalai Tesque], in 1× PBS). Immunoprecipitation was performed using FLAG Affinity Gels (Sigma-Aldrich). For coimmunoprecipitation between endogenous proteins, cells were lysed with the NP-40 buffer, and immunoprecipitation was performed using the indicated antibody and Protein G Sepharose 4 Fast Flow (GE Healthcare). To demonstrate the direct interaction, His-tagged proteins were synthesized in Escherichia coli cells using cold-shock expression vector pCold I DNA (Takara Bio Inc.) or in Hek293T cells using pCAGGS vector and then purified using TALON Metal Affinity Resin (Takara Bio Inc.). For in vitro synthesis of recombinant proteins, we used a TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega) with pCS2+ vector. The purified recombinant proteins were diluted in a binding buffer (0.1% NP-40 and 50 mM imidazole, in 0.5× PBS, pH 7.4) and coimmunoprecipitation was performed. The resulting beads were boiled in SDS sample buffer to elute the immunocomplexes.
Chemical cross-linking
Chemical cross-linking was performed as previously described (Weitz et al., 2002) with some modifications. Purified recombinant proteins were incubated in a Hepes cross-linking buffer (5 mM Hepes, pH. 8.0, 0.5 mM MgCl2, 0.5 mM ATP, 0.2 mM EGTA, and 0.5 mM DTT) or a PBS cross-link buffer (1× PBS, 0.5 mM MgCl2, 0.5 mM ATP, 0.2 mM EGTA, and 0.5 mM DTT) for 30 min at room temperature. The chemical cross-linker BS3 (Thermo Fisher Scientific) was added to the mixture at a final concentration of 0.5 mM. After 30-min incubation, the cross-linking reaction was terminated by adding SDS sample buffer.
Cell fractionation assay
Cell fractionation assay was performed using a ProteoExtract Subcellular Proteome Extraction Kit (EMD Millipore). Fractions 1 and 3 were collected as the cytoplasmic fraction and nuclear fraction, respectively. Anti–α-tubulin (cytoplasm) and anti–c-fos (nucleus) antibodies were used as subcellular marker antibodies.
DNA–protein binding assay
myc-Arp5, myc-SM-Myocd, FLAG-SRF, and FLAG-SRF-DBD proteins were synthesized in Hek293T cells and diluted with the NP-40 buffer containing 20 µg/ml of salmon testes DNA (Sigma-Aldrich). A biotinylated DNA probe containing 3× CArG sequences was synthesized by PCR using the 3× CArG-Luc vector as a template, and it was coupled to Dynabeads M-280 Streptavdin (Invitrogen). The DNA-Dynabeads were incubated in the NP-40 buffer containing 5% BSA to block nonspecific binding. The protein solution was added to the beads, followed by incubation for 3 h at 4°C with gentle rotation. The beads were washed with the NP-40 buffer, and the bound proteins were eluted with SDS sample buffer for Western blotting.
RT-PCR and real-time PCR
Total RNA was extracted from cells using RNAiso Plus (Takara Bio Inc.) or illustra RNAspin Mini (GE Healthcare) and reverse transcribed using SuperScript III Reverse transcription (Invitrogen) with a random hexamer primer or gene-specific primer. cDNA was amplified using gene-specific PCR primer pairs. Real-time PCR was performed using THUNDERBIRD SYBR qPCR Mix (TOYOBO) and a 7900HY Fast Real-Time PCR System (Applied Biosystems). Primer sequences are presented in Table S1.
ChIP assay
A7r5 cells transfected with FLAG-Myocd and myc-Arp5 expression vectors were fixed using 1% paraformaldehyde and used in the ChIP assay. The ChIP assay was performed using the ChIP Assay Kit (EMD Millipore) and FLAG affinity gels. PCR primer sequences used in this assay are presented in Table S1.
Collagen gel lattice contraction assay
VSMCs transfected with control or Arp5 siRNA for 3 d were trypsinized and suspended in collagen type I-A/MEM solution at a density of 0.5 × 106 cells/ml using the Cellmatrix gel culturing kit (Nitta Gelatin, Inc.). A total of 500 µl of the mixture was pipetted into each well of a 24-well plate, followed by incubation for 30 min at 37°C to polymerize the collagen gels. After the polymerization, 500 µl of DMEM with 10% FCS was added into each well and incubated for 24 h. The contraction assay was initiated by detaching the gels from the wells with a pipette tip, followed by the addition of 100 µM carbachol into the medium. After 3 h of carbachol stimulation, the gel areas were measured using ImageJ software (National Institutes of Health).
Online supplemental material
Fig. S1 shows the Arp5–SRF and Arp5–Myocd interactions in HeLa cells. Fig. S2 shows amino acid alignment between human Arp5 and β-actin. Fig. S3 shows nucleotide alignment between rat arp5-com and arp5-sm. Fig. S4 shows a summary of the functional mechanisms for Arp5. Table S1 shows sequences of PCR primers and siRNAs used in this manuscript. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201307158/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Enago for the English language review.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (23770225 to T. Morita).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ARP
- actin-related protein
- BS3
- bis(sulfosuccinimidyl) suberate
- ChIP
- chromatin immunoprecipitation
- CHX
- cycloheximide
- DBD
- DNA binding domain
- G-actin
- globular actin
- MRTF
- Myocd-related transcription factor
- Myocd
- myocardin
- NMD
- nonsense-mediated mRNA decay
- PSC
- premature stop codon
- SRF
- serum response factor
- VSMC
- vascular smooth muscle cell
References
- Aikawa M., Sivam P.N., Kuro-o M., Kimura K., Nakahara K., Takewaki S., Ueda M., Yamaguchi H., Yazaki Y., Periasamy M., et al. 1993. Human smooth muscle myosin heavy chain isoforms as molecular markers for vascular development and atherosclerosis. Circ. Res. 73:1000–1012 10.1161/01.RES.73.6.1000 [DOI] [PubMed] [Google Scholar]
- Carter M.S., Doskow J., Morris P., Li S., Nhim R.P., Sandstedt S., Wilkinson M.F. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270:28995–29003 10.1074/jbc.270.48.28995 [DOI] [PubMed] [Google Scholar]
- Chen J., Kitchen C.M., Streb J.W., Miano J.M. 2002. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell. Cardiol. 34:1345–1356 10.1006/jmcc.2002.2086 [DOI] [PubMed] [Google Scholar]
- Creemers E.E., Sutherland L.B., Oh J., Barbosa A.C., Olson E.N. 2006. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol. Cell. 23:83–96 10.1016/j.molcel.2006.05.026 [DOI] [PubMed] [Google Scholar]
- de Lanerolle P., Serebryannyy L. 2011. Nuclear actin and myosins: life without filaments. Nat. Cell Biol. 13:1282–1288 10.1038/ncb2364 [DOI] [PubMed] [Google Scholar]
- Dion V., Shimada K., Gasser S.M. 2010. Actin-related proteins in the nucleus: life beyond chromatin remodelers. Curr. Opin. Cell Biol. 22:383–391 10.1016/j.ceb.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Doi H., Iso T., Yamazaki M., Akiyama H., Kanai H., Sato H., Kawai-Kowase K., Tanaka T., Maeno T., Okamoto E., et al. 2005. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler. Thromb. Vasc. Biol. 25:2328–2334 10.1161/01.ATV.0000185829.47163.32 [DOI] [PubMed] [Google Scholar]
- Ebbert R., Birkmann A., Schüller H.J. 1999. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 32:741–751 10.1046/j.1365-2958.1999.01390.x [DOI] [PubMed] [Google Scholar]
- Fenn S., Breitsprecher D., Gerhold C.B., Witte G., Faix J., Hopfner K.P. 2011. Sturctural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. EMBO J. 30:2153–2166 10.1038/emboj.2011.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettler S., Vartiainen M.K., Miralles F., Larijani B., Treisman R. 2008. RPEL motifs link the serum response factor cofactor MAL but not myocardin to Rho signaling via actin binding. Mol. Cell. Biol. 28:732–742 10.1128/MCB.01623-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Nakamura S., Nishida W., Sobue K. 2006. Bone morphogenetic protein-induced MSX1 and MSX2 inhibit myocardin-dependent smooth muscle gene transcription. Mol. Cell. Biol. 26:9456–9470 10.1128/MCB.00759-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada K., Nishida W., Hayashi K., Ozawa K., Yamanaka Y., Saga H., Yamashita T., Tohyama M., Shimada S., Sato K., Sobue K. 1997. Coordinate expression of α-tropomyosin and caldesmon isoforms in association with phenotypic modulation of smooth muscle cells. J. Biol. Chem. 272:15396–15404 10.1074/jbc.272.24.15396 [DOI] [PubMed] [Google Scholar]
- Kervestin S., Jacobson A. 2012. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13:700–712 10.1038/nrm3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama K., Kamo M., Oma Y., Matsuda R., Uchida T., Ikura T., Tashiro S., Ohyama T., Winsor B., Harata M. 2009. The human actin-related protein hArp5: Nucleo-cytoplasmic shuttling and involvement in DNA repair. Exp. Cell Res. 315:206–217 10.1016/j.yexcr.2008.10.028 [DOI] [PubMed] [Google Scholar]
- Koteliansky V.E., Ogryzko E.P., Zhidkova N.I., Weller P.A., Critchley D.R., Vancompernolle K., Vandekerckhove J., Strasser P., Way M., Gimona M., et al. 1992. An additional exon in the human vinculin gene specifically encodes meta-vinculin-specific difference peptide. Cross-species comparison reveals variable and conserved motifs in the meta-vinculin insert. Eur. J. Biochem. 204:767–772 10.1111/j.1432-1033.1992.tb16692.x [DOI] [PubMed] [Google Scholar]
- Li S., Wang D.Z., Wang Z., Richardson J.A., Olson E.N. 2003. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. USA. 100:9366–9370 10.1073/pnas.1233635100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Chang S., Qi X., Richardson J.A., Olson E.N. 2006. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol. Cell. Biol. 26:5797–5808 10.1128/MCB.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sinha S., McDonald O.G., Shang Y., Hoofnagle M.H., Owens G.K. 2005a. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J. Biol. Chem. 280:9719–9727 10.1074/jbc.M412862200 [DOI] [PubMed] [Google Scholar]
- Liu Z.P., Wang Z., Yanagisawa H., Olson E.N. 2005b. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell. 9:261–270 10.1016/j.devcel.2005.05.017 [DOI] [PubMed] [Google Scholar]
- Meagher R.B., Kandasamy M.K., McKinney E.C., Roy E. 2009. Chapter 5. Nuclear actin-related proteins in epigenetic control. Int. Rev. Cell Mol. Biol. 277:157–215 10.1016/S1937-6448(09)77005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., Treisman R. 2009. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11:257–268 10.1038/ncb1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.I., Treisman R. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 113:329–342 10.1016/S0092-8674(03)00278-2 [DOI] [PubMed] [Google Scholar]
- Morita T., Mayanagi T., Sobue K. 2007a. Reorganization of the actin cytoskeleton via transcriptional regulation of cytoskeletal/focal adhesion genes by myocardin-related transcription factors (MRTFs/MAL/MKLs). Exp. Cell Res. 313:3432–3445 10.1016/j.yexcr.2007.07.008 [DOI] [PubMed] [Google Scholar]
- Morita T., Mayanagi T., Sobue K. 2007b. Dual roles of myocardin-related transcription factors in epithelial–mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 179:1027–1042 10.1083/jcb.200708174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouilleron S., Guettler S., Langer C.A., Treisman R., McDonald N.Q. 2008. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 27:3198–3208 10.1038/emboj.2008.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Hayashi K., Iwasaki K., Fujioka T., Egusa H., Yatani H., Sobue K. 2010. Nuclear import mechanism for myocardin family members and their correlation with vascular smooth muscle cell phenotype. J. Biol. Chem. 285:37314–37323 10.1074/jbc.M110.180786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann F.R., Dion V., Gehlen L.R., Tsai-Pflugfelder M., Schmid R., Taddei A., Gasser S.M. 2012. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 26:369–383 10.1101/gad.176156.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Wang Z., Wang D.Z., Lien C.L., Xing W., Olson E.N. 2004. Target gene-specific modulation of myocardin activity by GATA transcription factors. Mol. Cell. Biol. 24:8519–8528 10.1128/MCB.24.19.8519-8528.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Richardson J.A., Olson E.N. 2005. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc. Natl. Acad. Sci. USA. 102:15122–15127 10.1073/pnas.0507346102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G., Sotiropoulos A., Treisman R. 2002. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell. 13:4167–4178 10.1091/mbc.02-05-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan M., Wuerges J., Bose D., McCormack E.A., Cook N.J., Zhang X., Wigley D.B. 2012. Interactions between the nucleosome histone core and Arp8 in the INO80 chromatin remodeling complex. Proc. Natl. Acad. Sci. USA. 109:20883–20888 10.1073/pnas.1214735109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Mizuguchi G., Hamiche A., Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 406:541–544 10.1038/35020123 [DOI] [PubMed] [Google Scholar]
- Shen X., Ranallo R., Choi E., Wu C. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 12:147–155 10.1016/S1097-2765(03)00264-8 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Sato H., Doi H., Yoshida C.A., Shimizu T., Matsui H., Yamazaki M., Akiyama H., Kawai-Kowase K., Iso T., et al. 2008. Runx2 represses myocardin-mediated differentiation and facilitates osteogenic conversion of vascular smooth muscle cells. Mol. Cell. Biol. 28:1147–1160 10.1128/MCB.01771-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen M.K., Guettler S., Larijani B., Treisman R. 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 316:1749–1752 10.1126/science.1141084 [DOI] [PubMed] [Google Scholar]
- Wang D., Chang P.S., Wang Z., Sutherland L., Richardson J.A., Small E., Krieg P.A., Olson E.N. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 105:851–862 10.1016/S0092-8674(01)00404-4 [DOI] [PubMed] [Google Scholar]
- Weitz D., Ficek N., Kremmer E., Bauer P.J., Kaupp U.B. 2002. Subunit stoichiometry of the CNG channel of rod photoreceptors. Neuron. 36:881–889 10.1016/S0896-6273(02)01098-X [DOI] [PubMed] [Google Scholar]
- Yin H., Jiang Y., Li H., Li J., Gui Y., Zheng X.L. 2011. Proteasomal degradation of myocardin is required for its transcriptional activity in vascular smooth muscle cells. J. Cell. Physiol. 226:1897–1906 10.1002/jcp.22519 [DOI] [PubMed] [Google Scholar]
- You K.T., Li L.S., Kim N.G., Kang H.J., Koh K.H., Chwae Y.J., Kim K.M., Kim Y.K., Park S.M., Jang S.K., Kim H. 2007. Selective translational repression of truncated proteins from frameshift mutation-derived mRNAs in tumors. PLoS Biol. 5:e109 10.1371/journal.pbio.0050109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Herring B.P. 2005. Mechanisms responsible for the promoter-specific effects of myocardin. J. Biol. Chem. 280:10861–10869 10.1074/jbc.M411586200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.