Nonspecific intermolecular collision between the central pair apparatus and radial spokes underlies a mechanosensing mechanism that regulates dynein activity in Chlamydomonas flagella.
Abstract
Cilia/flagella are conserved organelles that generate fluid flow in eukaryotes. The bending motion of flagella requires concerted activity of dynein motors. Although it has been reported that the central pair apparatus (CP) and radial spokes (RSs) are important for flagellar motility, the molecular mechanism underlying CP- and RS-mediated dynein regulation has not been identified. In this paper, we identified nonspecific intermolecular collision between CP and RS as one of the regulatory mechanisms for flagellar motility. By combining cryoelectron tomography and motility analyses of Chlamydomonas reinhardtii flagella, we show that binding of streptavidin to RS heads paralyzed flagella. Moreover, the motility defect in a CP projection mutant could be rescued by the addition of exogenous protein tags on RS heads. Genetic experiments demonstrated that outer dynein arms are the major downstream effectors of CP- and RS-mediated regulation of flagellar motility. These results suggest that mechanosignaling between CP and RS regulates dynein activity in eukaryotic flagella.
Introduction
Cilia and flagella are conserved motile organelles that play essential roles in cellular motility of eukaryotes and development of higher organisms by generating fluid flow (Gibbons, 1981; Hirokawa et al., 2006). In mammals, several disorders are associated with motility defects of cilia and flagella (Pazour and Rosenbaum, 2002; Zariwala et al., 2007). Axonemes are the core structure of motile cilia and flagella. Most axonemes share a 9 + 2 microtubule arrangement, composed of nine outer doublet microtubules (DMTs) and a central pair of microtubules (the central pair apparatus [CP]; Fig. 1 A).
Figure 1.
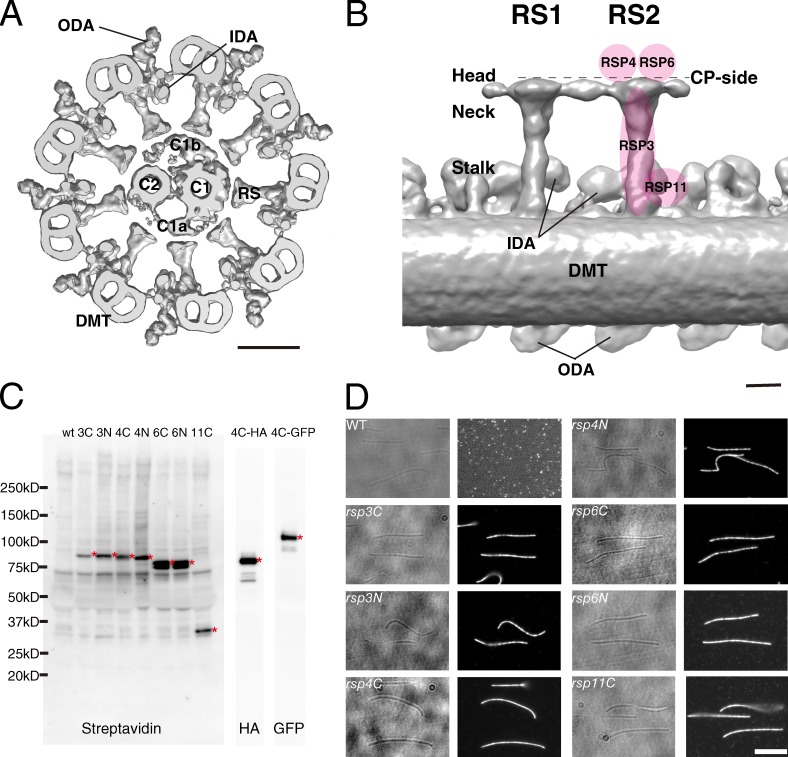
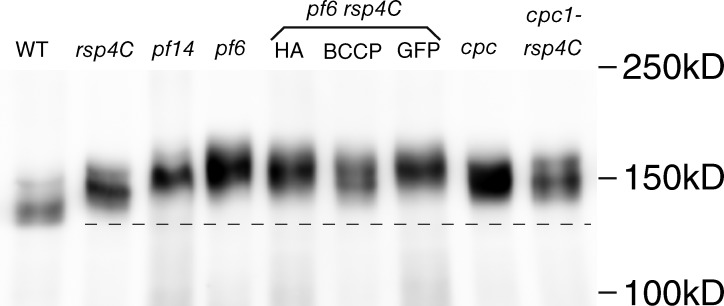
Expression and characterization of BCCP-tagged RSPs in C. reinhardtii. (A) Organization of the C. reinhardtii axoneme. A base to tip view of the 9 + 2 structure is shown. Positions of outer and inner dynein arms (ODA and IDA), outer doublet microtubules (DMT), radial spokes (RS), central pair microtubules (C1 and C2), and C1a and C1b projections are indicated. The image was generated by superimposing the averaged tomograms of the DMT and CP onto an unaveraged tomogram of the wild-type 9 + 2 structure. Bar, 50 nm. (B) Predicted positions of RSPs in RSs based on previous mutant analyses (Yang et al., 2006; Pigino et al., 2011) superimposed on a side view of the averaged tomogram of DMT. The distal end of the flagellum is to the right. We call the upper surface of the RS heads the CP side in this article (broken line). Bar, 10 nm. (C) Immunoblot of tagged RSPs. Axonemal proteins from wild-type (wt) and RSP mutant strains were separated by SDS-PAGE and probed with horseradish peroxidase–conjugated streptavidin, anti-HA, or anti-GFP antibodies. The asterisks indicate the detected bands of RSPs. 3C, rsp3C; 3N, rsp3N; 4C, rsp4C; 4N, rsp4N; 6C, rsp6C; 6N, rsp6N; 11C, rsp11C; 4C-HA, rsp4C-3HA; 4C-GFP, rsp4C-GFP. (D) Immunofluorescence of axonemes. Left, phase contrast; right, Alexa Fluor 546. The axonemes with BCCP-tagged RSPs were incubated with Alexa Fluor–conjugated streptavidin, and the signals were detected by immunofluorescence microscopy. All of the BCCP-RSPs inside the axonemes were confirmed to be accessible to streptavidin. For the fluorescence image from wild type, the contrast was enhanced to show the absence of signals. Bar, 5 µm.
To generate beating motion of flagella, only a subset of dyneins needs to be activated and the rest of the dyneins should be inactive (Sale, 1986; Kurimoto and Kamiya, 1991; Omoto et al., 1996; Wargo and Smith, 2003; Movassagh et al., 2010; Maheshwari and Ishikawa, 2012). It has been known that both CP and RS are necessary for flagellar beating in Chlamydomonas reinhardtii because mutant cells lacking one of these structures have paralyzed flagella (Witman et al., 1978; Adams et al., 1981; Smith and Lefebvre, 1997). From the A-tubules of DMT, T-shaped radial spokes (RSs) extend toward the CP (Fig. 1 B). The results of microtubule sliding assays with sea urchin sperm suggest that the CP–RS system determines which dynein arms are activated (Nakano et al., 2003; Hayashi and Shingyoji, 2008). Regulatory signals from the CP–RS system are thought to be transmitted to inner dynein arms (IDAs) through an interaction between the calmodulin- and spoke-associated complex and RS (Dymek and Smith, 2007; Dymek et al., 2011). The presence of a signaling pathway connecting the CP/RS to dyneins is also suggested by intergenic suppressor mutations that restore flagellar activity to paralyzed RS or CP mutants (Huang et al., 1982; Porter et al., 1992, 1994; Smith and Yang, 2004). Despite these extensive studies on the CP–RS-dependent regulation of flagellar motility, how CP and RS communicate with each other at a molecular level has remained to be elucidated. Although classical electron microscopic studies suggest a physical interaction between CP and RS (Warner, 1970; Warner and Satir, 1974; Goodenough and Heuser, 1985), high resolution 3D structural data and functional analyses of the CP–RS system are required to prove the presence of the molecular communication between CP and RS.
In this study, we used cryoelectron tomography, motility analysis, and genetic experiments to investigate communication between CP and RS. We demonstrate that binding of streptavidin to the biotinylated RS head proteins inhibits flagellar motility as well as that extension of RS via the addition of exogenous protein tags to RS heads restores swimming in a CP projection mutant. Furthermore, extensive genetic experiments showed that outer dynein arms (ODAs) are the major downstream effector of both the inhibition of motility by streptavidin binding and restoration of motility by RS extension. These results suggest that CP transmits signals to RSs via mechanical contacts between the CP projection and RS head and regulates dynein-driven flagellar motility.
Results
Expression of biotinylated RS proteins (RSPs) in C. reinhardtii
To investigate possible interactions between CP and RS using the biotin-streptavidin system, we created seven new C. reinhardtii mutants, rsp3N, rsp3C, rsp4N, rsp4C, rsp6N, rsp6C, and rsp11C, which express RSPs tagged with a 9-kD fragment of biotin carboxyl carrier protein (BCCP) at the N or C terminus. Plasmids encoding BCCP-tagged RSPs were introduced into mutant strains with deficiencies in the respective RSPs: pf14 (ΔRSP3), pf1 (ΔRSP4), pf26 (ΔRSP6), and pf25 (ΔRSP11; Piperno et al., 1977; Witman et al., 1978; Huang et al., 1981; Diener et al., 1990; Yang and Yang, 2006). All of the transformants exhibited wild-type motility (Table S1), and all of the BCCP-tagged RSPs were biotinylated (Fig. 1 C). Moreover, fluorescence microscopy showed that Alexa Fluor–conjugated streptavidin molecules were able to bind BCCP-tagged RSPs inside the axoneme (Fig. 1 D).
Localization of RSPs by cryoelectron tomography
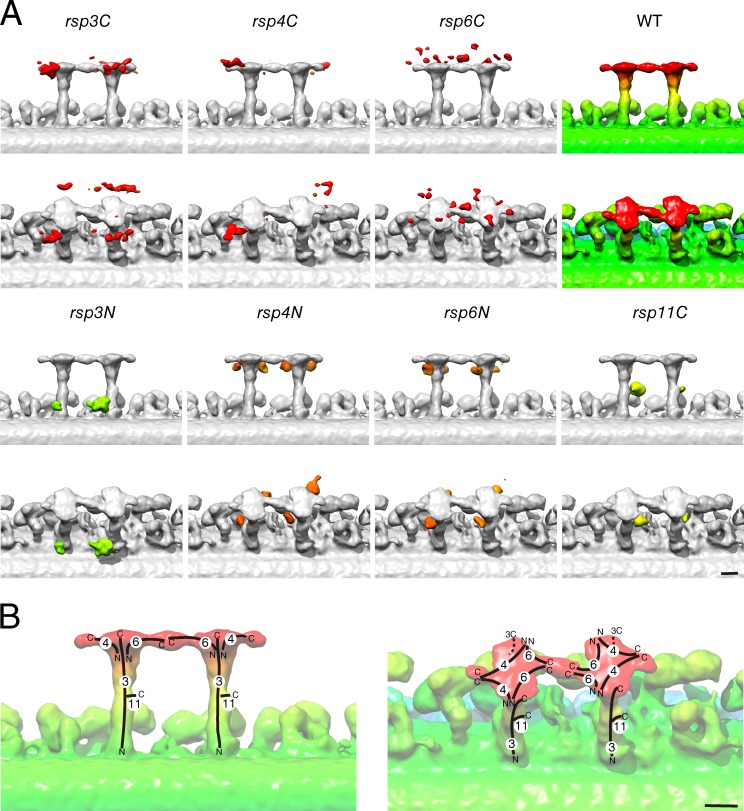
We next performed cryoelectron tomography of streptavidin-labeled axonemes to precisely locate the positions of the BCCP tags on RSs (Figs. 2 and S1). The positions of the bound streptavidin molecules can be visualized by comparison of the unlabeled and labeled 3D structures (Oda and Kikkawa, 2013). We enhanced the streptavidin signal by adding biotinylated cytochrome c, thereby increasing the total mass bound to each BCCP tag. The averaged tomograms showed that the streptavidin–cytochrome c labels bound to rsp3C, 4C, and 6C axonemes were localized on or above the RS heads, whereas rsp3N, 4N, 6N, and 11C axonemes had label densities below the RS heads (Fig. 2). Based on these results, we expected that streptavidin binding would block the proposed CP–RS communication in rsp3C, 4C, and 6C and cause impaired flagellar motility in these mutants.
Figure 2.
Architecture of RS revealed by cryoelectron tomography and enhanced streptavidin labeling. (A) Positions of the enhanced streptavidin labels were visualized by comparing the averaged subtomogram of the unlabeled wild-type DMT with those of the labeled BCCP-RSP mutant DMTs. The densities of the enhanced streptavidin labels and the wild-type structure are colored gradients of red, orange, yellow, or green, reflecting the distance from the CP–RS interface. The isosurface threshold values were t > 8.61, with a one-tailed probability of <0.05%. (B) A model of the RS architecture. Possible locations of the RSPs are shown with black lines. N and C indicate N and C termini of RSPs, respectively. 3, 4, 6, and 11 correspond to RSP3, RSP4, RSP6, and RSP11, respectively. (A and B) Distal ends are to the right. Bars, 10 nm.
Streptavidin inhibits motility of ATP-reactivated cells expressing BCCP-RSPs
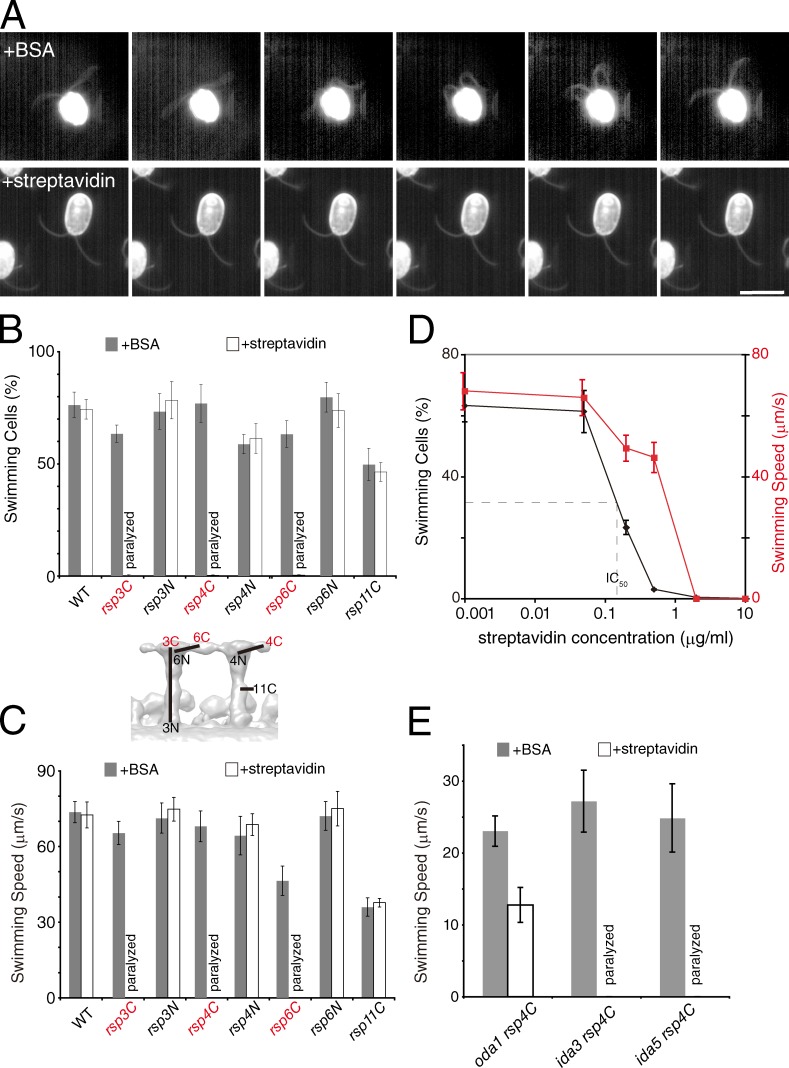
To test our prediction, we observed the motility of demembranated and ATP-reactivated cells in the presence of streptavidin or BSA (Fig. 3). As expected, rsp3C, 4C, and 6C mutants showed flagellar paralysis after the addition of streptavidin, whereas the flagellar motility of the other four mutants was unaffected (Fig. 3, A–C; and Video 1). The clear correlation between inhibition of flagellar motility and binding of streptavidin to the CP side of RS heads strongly suggests that mechanical interaction between RS and CP plays an important role.
Figure 3.
Streptavidin-dependent inhibition of motility in BCCP-RSP mutant cells. (A) Dark-field image sequences of demembranated, ATP-reactivated rsp4C cells. The interval between each frame is 1/300 s. The rsp4C cells showed normal motility in the presence of BSA, whereas in the presence of streptavidin, the cells exhibit paralyzed flagella in a slightly bent form. Bar, 10 µm. (B) Percentages of ATP-reactivated swimming cells after demembranation and addition of BSA or streptavidin. rsp3C, 4C, and 6C cells showed flagellar paralysis in response to addition of streptavidin. A smaller version of Fig. 2 B is included for reference. (C) Averaged swimming speed of ATP-reactivated swimming cells. (D) The dose-dependent percentage and speed of swimming cells relative to the concentration of streptavidin were measured. (B–D) The means ± SEM of the percentages of swimming cells and swimming speed were calculated from 10 measurements and 20 cells, respectively. (E) Mean swimming speeds of ATP-reactivated cells were measured for oda1 rsp4C, ida3 rsp4C, and ida5 rsp4C mutants. In the presence of streptavidin, oda1 rsp4C cells showed swimming with reduced speeds compared with the BSA condition, whereas ida3 rsp4C and ida5 rsp4C cells exhibited flagellar paralysis after the addition of streptavidin. The means ± SEM were calculated from 20 cells. WT, wild type.
The half-maximal inhibitory concentration (IC50) of streptavidin-dependent inhibition of flagellar motility was 0.15 µg/ml and the Hill coefficient was 2.37 (Fig. 3 D). At the IC50 concentration, the amount of streptavidin bound to BCCP tags was ∼5% of the saturating levels (Fig. S2 A), suggesting that the inhibitory effect of a streptavidin molecule bound to RS propagates along the axoneme. It is also noteworthy that the speed of the swimming cells did not show steep drop even when the concentration of streptavidin was high enough to inhibit the motility in 98% of the cells (Fig. 3 D, red). This all-or-none behavior suggests that inhibition of motility occurs when the amount of streptavidin bound to one axoneme is above a certain threshold.
To identify the axonemal dynein that is the downstream effector of the streptavidin-dependent inhibition of motility in rsp4C mutant, we created the strains oda1 rsp4C (lacking ODAs), ida3 rsp4C (lacking IDA subspecies f), and ida5 rsp4C (lacking IDA subspecies a, c, d, and e) and examined the flagellar motility of ATP-reactivated cells of each genotype. Interestingly, oda1 rsp4C cells swam slowly even in the presence of streptavidin, whereas ida3 rsp4C and ida5 rsp4C cells exhibited paralysis of flagella (Fig. 3 E and Video 1). These results suggest that streptavidin molecules bound to RSP4C cause inhibition of motility mainly via alteration in ODA activity.
We next examined the effects of streptavidin bound to RSPs on axonemal ATPase and microtubule sliding activities (Table S2 and Fig. S2), which mainly depend on ODA activity (Kagami and Kamiya, 1990, 1991). In wild-type axonemes, microtubule sliding occurs only on one or two DMTs in the presence of 1 mM ATP, whereas almost all the DMTs slide out in the presence of 50 µM ATP as a result of nonspecific activation of ODA caused by low concentration of ATP (Kurimoto and Kamiya, 1991; Omoto et al., 1996). In most mutant axonemes, the addition of streptavidin did not change ATPase or microtubule sliding activities, suggesting that if present, any alterations in ODA activity in rsp4C are below the range of what can be detected with this method (see Discussion). However, microtubule sliding in rsp3C axonemes was almost completely inhibited by streptavidin, even in the presence of 50 µM ATP, whereas rsp4C and rsp6C axonemes showed normal microtubule sliding even in the presence of streptavidin (Table S2 and Fig. S2 C). These results suggest that the mechanism of the inhibition of motility in the rsp3C strain is different from that in rsp4C and rsp6C. One possible explanation for the unique response of rsp3C to streptavidin is that the streptavidin molecules bound to the RS of rsp3C axonemes might serve as mechanical obstacles to microtubule sliding because the BCCP tags of rsp3C locate at the circumference edges of the RS heads and the bound streptavidin would extend toward the neighboring RS heads on the adjacent DMT (Figs. 2 and S1 C). In contrast, the reason for the flagellar paralysis in rsp4C and rsp6C cells appears to be related to the regulation of beating, as microtubule sliding itself was unaffected by the addition of streptavidin in these mutant strains (see Discussion; Table S2 and Fig. S2 C).
Tags on RS heads rescue a CP mutant, pf6
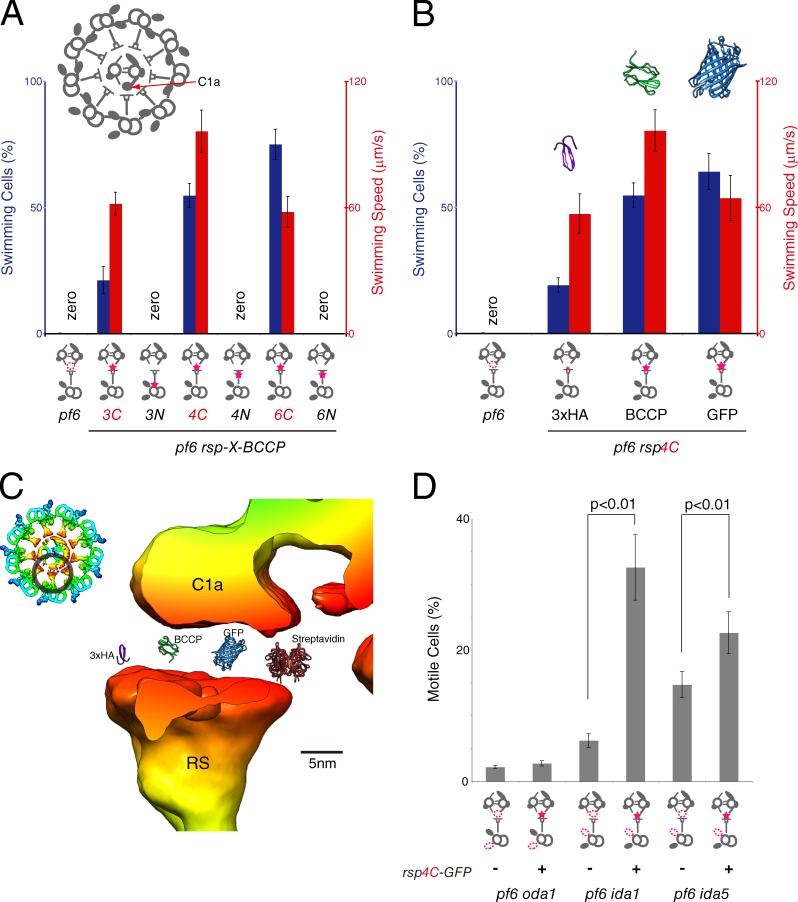
If communication between CP and RS is mediated by mechanical contacts and does not require specific protein–protein interaction, exogenous proteins should be able to transmit mechanical signals from CP to RS. To test this hypothesis, we crossed rsp3C, 3N, 4C, 4N, 6C, and 6N with pf6, which lacks C1a projections (Fig. 4 A; Dutcher et al., 1984; Mitchell and Sale, 1999). We expected that exogenous protein tags on top of the RS heads would extend the RS head and rescue the flagellar motility by restoring the interaction between CP and RS. As expected, pf6 rsp3C, pf6 rsp4C, and pf6 rsp6C mutant cells showed recovery of motility, whereas pf6 cells cannot swim and instead jiggle on the bottom of the culture dish (Fig. 4 A and Video 2). The jagged swimming paths of pf6 rsp4C cells (Fig. S3) suggest that coordination between the two flagella was not restored despite recovery of swimming ability. In contrast, pf6 rsp3N, pf6 rsp4N, and pf6 rsp6N cells exhibited pf6-like motility defects, demonstrating that location of the tag is important for recovery (Fig. 4 A).
Figure 4.
Rescue of a CP projection mutant pf6 in rsp tag mutant backgrounds. (A) Motility of pf6 rsp mutants. pf6 rsp3C, 4C, and 6C mutants showed recovery of motility. (B) Motility of pf6 rsp4C mutants having different tags. There is a positive correlation between recovery of motility and the size of the tags on RSP4. (A and B) Diagrams below the charts indicate the positions of tags (red stars) on the RS. Because rescued strains made frequent turns after short straight swimming, swimming speed was measured by taking the mean speed between one turn and the next. (B) The crystal structures of 3×HA (violet, predicted structure), BCCP (green, Protein Data Bank accession no. 1BOD), and GFP (blue, Protein Data Bank accession no. 1EMA) are shown for size comparison. The tertiary structure of 3×HA was predicted using the PROTINFO tool (Hung and Samudrala, 2003). (C) Comparison of the CP–RS gap and the size of tags fused to RSP4. The crystal structures of 3×HA (violet), BCCP (green), GFP (blue), and streptavidin (red; Protein Data Bank accession no. 4JO6) were superimposed above the RS head. Images of averaged subtomogram were colored based on the distance from the CP–RS interface. (D) Motility of pf6 oda/ida rsp4C-GFP mutants. As forward swimming was not observed in any of the mutant strains, percentages of motile cells were calculated by counting rotating or jiggling cells. The p-values were calculated using Student’s t test. (A, B, and D) Means ± SEM for mean percentages of swimming/motile cells and mean swimming speeds were calculated from 10 measurements and 20 cells, respectively.
Because our hypothesis predicts that the size of the tags on RS heads would affect recovery of motility, we compared the motility of three different pf6 rsp4C mutants, which have tags of varying size (3–27 kD) at the C terminus of RSP4. Interestingly, we observed a positive correlation between the size of the tag and the percentage of swimming cells (Fig. 4 B). In addition, pf6 rsp3C, pf6 rsp4C, and pf6 rsp6C cells showed streptavidin-dependent inhibition of motility if they were demembranated (Table S1), indicating that BCCP + streptavidin (9 + 50 kD) was too large to restore proper interactions between CP and RS. These results clearly demonstrate that the physical interaction between CP and RS, which is disrupted in pf6 mutant cells, can be restored by artificial extension of the RS head (Fig. 4 C).
To identify the axonemal dyneins that is the downstream effector of the rsp4C-dependent motility restoration in pf6, we compared the motility of three pairs of mutants (Fig. 4 D and Video 3): pf6 oda1 and pf6 oda1 rsp4C-GFP (lacking ODAs), pf6 ida1 and pf6 ida1 rsp4C-GFP (lacking IDA subspecies f), and pf6 ida5 and pf6 ida5 rsp4C-GFP (lacking IDA subspecies a, c, d, and e). Interestingly, both pf6 oda1 and pf6 oda1 rsp4C-GFP mutants exhibited paralyzed flagella, whereas pf6 ida1 rsp4C-GFP and pf6 ida5 rsp4C-GFP mutants showed improved motility compared with pf6 ida1 and pf6 ida5, respectively. These results indicate that ODAs are the major downstream effector of the restoration of motility in pf6 with rsp4C mutant background.
We also examined the phosphorylation state of IC138 of IDA subspecies f because IC138 is reported to be hyperphosphorylated in RS and CP mutants and involved in the regulation of flagellar motility (Hendrickson et al., 2004; VanderWaal et al., 2011). We expected that recovery of motility in pf6 by rsp4C would reduce phosphorylation of IC138 because kinase inhibitors have been shown to increase microtubule sliding activity in C. reinhardtii axonemes (Howard et al., 1994; Gokhale et al., 2009). However, the degree of hyperphosphorylation of IC138 in pf6 rsp4C mutants was almost the same as that in pf6 (Fig. 5). Thus, recovery of pf6 motility by rsp4C appears to be independent of the phosphorylation state of IC138. In addition, moderate IC138 phosphorylation was observed in rsp4C, but motility of rsp4C was normal, suggesting that this level of phosphorylation seems to be unrelated to motility. Phosphorylation of IC138 in rsp4C probably results from the temporal absence of RSs at the beginning of the flagella formation caused by the relatively low expression of RSP4 and delayed assembly of RSs.
Figure 5.
Phosphorylation of intermediate chain IC138 of IDA subspecies f. TCA-fixed axonemal proteins were separated on a 5% acrylamide gel and transferred to a polyvinylidene difluoride membrane. The blot was probed with IC138 antiserum. Hyperphosphorylation of IC138 was observed in pf14, pf6, pf6 rsp4CHA, pf6 rsp4C, pf6 rsp4CGFP, cpc1, and cpc1 RSP4C, and intermediate phosphorylation was observed in rsp4C. The broken line shows the height of the wild-type (WT) band.
Discussion
In this study, we investigated communication between CP and RS by expressing biotin (BCCP)-tagged RSPs in C. reinhardtii flagella. We applied cryoelectron tomography to streptavidin-labeled axonemes and identified the precise 3D locations of the termini of RSPs. We next showed that addition of streptavidin to C. reinhardtii mutants expressing BCCP-tagged RSP inhibited motility of demembranated cells. The results of motility analyses, along with the results of cryoelectron tomography, indicate that covering the CP-facing surfaces of RS heads using exogenous proteins inhibits flagellar motility via alteration in ODA activity. We further confirmed the involvement of CP in RS-mediated regulation of motility by crossing a CP projection–missing mutant pf6 with various types of rsp tag strains. The pf6 rsp3C, pf6 rsp4C, and pf6 rsp6C strains showed recovery of motility, suggesting that the CP–RS interaction disrupted by the absence of a CP projection was restored by extension of the RS head with artificial tags. Immotility of pf6 oda1 rsp4C-GFP mutant indicates that the recovery of flagellar motility requires ODAs. These results strongly suggest that the signals required for flagellar motility are transmitted from CP to RS via mechanical contacts between CP projections and RS heads, modulating activities of ODAs as the effector of the signal.
Collision-based mechanosignaling between CP and RS
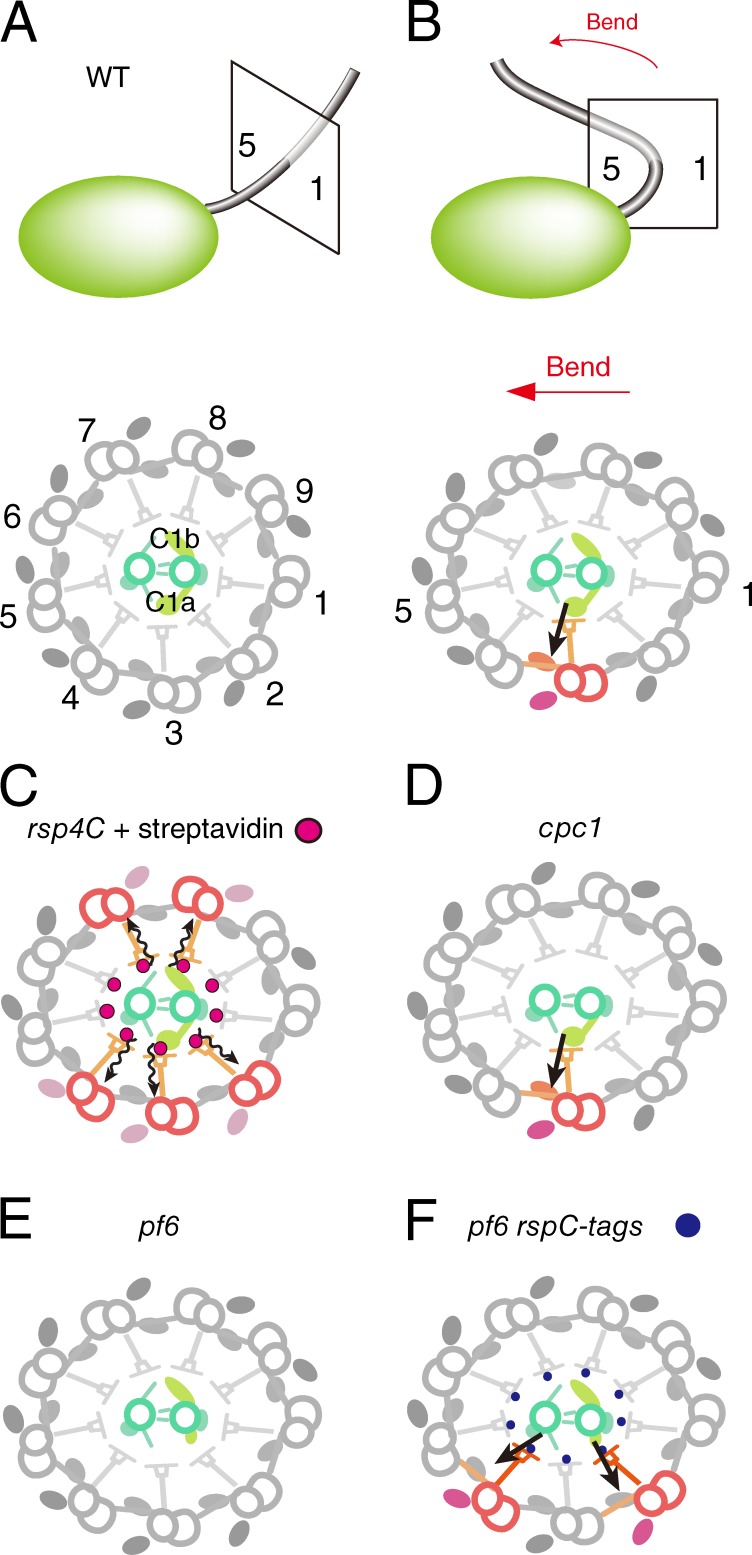
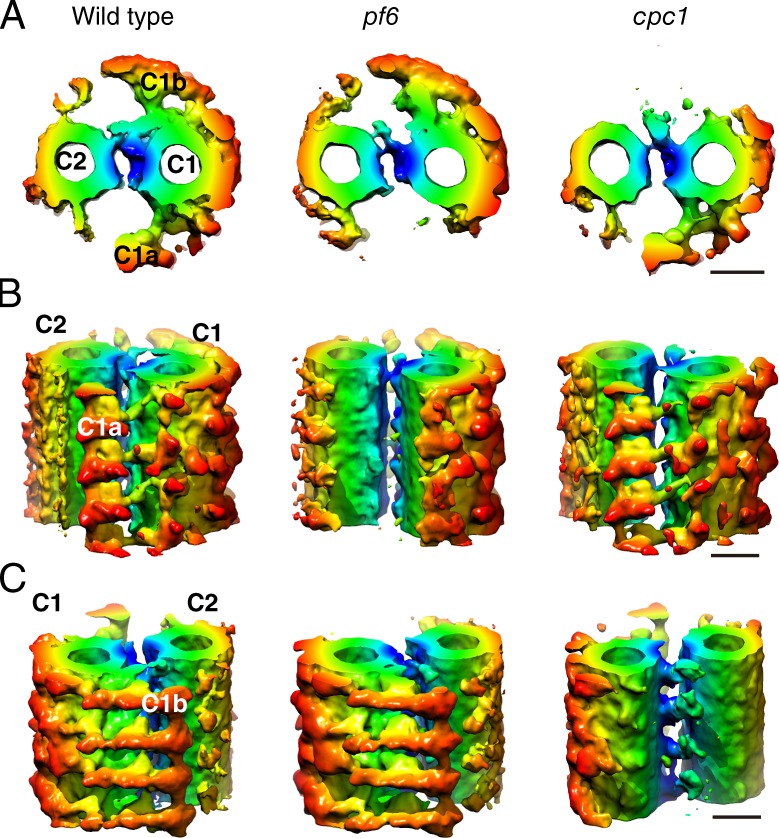
Rescue of the motility defect in pf6 via the addition of tags to the RS heads (Fig. 4) indicates that communication between CP and RS does not require specific protein–protein interactions; rather, an artificial interaction between the remnant of the C1a projection and an exogenous protein can transmit signals that regulate motility. This result strongly suggests that CP–RS communication is mediated, in part, by collision-based mechanosignaling (Fig. 6). In the straight axoneme, which we used for cryoelectron tomography, there are gaps of ∼5 nm between CP projections and RS heads (Figs. 4 C and 6 A). In the bending region of flagella (Fig. 6 B), the CP is oriented parallel to the bend plane, with its C1a projection pointing toward DMT 2, 3, and 4 (Mitchell, 2003), which are thought to be the active sites of dyneins that generate the principal bend (Sale, 1986; Wargo and Smith, 2003). Lindemann and Mitchell (2007) reported that bending of flagella distorts the circular arrangement of the nine DMTs and increases the diameter parallel to the bending plane, which may in turn decrease the diameter of the axoneme perpendicular to the bending plane, bringing the RS heads closer to the C1a projection (Fig. 6 B). We propose that collision-based mechanosignaling between the C1a projection and RS head modulates dyneins, mainly ODA, on DMT 2, 3, and 4. The shrinking in diameter appears to bring the C1b projection in contact with the RS heads of DMT 6, 7, and 8 as well, but this possible contact between the C1b and RS does not seem to activate dynein activity, as sliding of DMT 6, 7, and 8 would antagonize that of DMT 2, 3, and 4. The idea that the C1a projection, not the C1b projection, is essential for generating the bending motion is supported by the difference in the severity of motility defects in pf6 (missing C1a) versus cpc1 (missing C1b): pf6 cells cannot swim, whereas cpc1 cells can swim with a wild-type waveform and reduced beat frequency (Dutcher et al., 1984; Mitchell and Sale, 1999; Rupp et al., 2001; Goduti and Smith, 2012). The difference in the 3D structures of C1a and C1b projections might explain their roles in CP–RS mechanosignaling (Fig. 6, D and E; Carbajal-González et al., 2013). The C1a projection looks like a fist extending from C1 (Fig. 7, A and B) and creates bumpy surfaces against the RS heads, whereas the C1b is like a curved blade and forms a smooth surface (Fig. 7 C). The undulating surface of the C1a projection might be important to “hit” the RS heads and transmit signals to dyneins (Wargo and Smith, 2003). Our hypothesis that bending of flagella causes activation or inactivation of specific dyneins also agrees well with a previous study on sea urchin sperm flagella showing that mechanical stress created by flagellar bending regulates activation and inactivation of dynein arms (Hayashi and Shingyoji, 2008).
Figure 6.
Models of the regulation of dynein activity via the CP–RS mechanosignaling. (A and B, top) Schematic diagrams of C. reinhardtii cells and flagella showing the orientation of cross sections of axonemes relative to the cell and bending direction. The 9 + 2 structures are viewed from the base. DMT numbering was adapted from Hoops and Witman (1983). (A, bottom) In a straight flagellum, CP projections are not in contact with RS heads, and all dyneins are in the resting state. (B, bottom) When a flagellum makes a principal bend (red arrows), the circular arrangement of the nine DMTs is distorted (Lindemann and Mitchell, 2007), and C1 is located on the outer side (closest to DMT 1) of the curved flagellum (Mitchell, 2003). As a result of this distortion, C1a projections collide with RS heads and modulate the dynein activity on DMT 3 (or DMTs 2–4). (C) When streptavidin molecules bind to RS heads, nonphysiological CP–RS contacts occur on many DMTs, and irregular activation or inactivation of ODA causes flagellar paralysis. (D) In a cpc1 flagellum, the absence of C1b projections does not cause a serious motility defect (Mitchell and Sale, 1999). The remaining C1a projection alone can modulate dynein activity through a collision with RS heads at bent regions of the flagellum. (E) The absence of C1a projections in pf6 severely impairs motility (Dutcher et al., 1984; Rupp et al., 2001). Loss of the CP–RS contact by C1a projection would result in failure of dynein regulation. (F) Addition of tags to RS heads restores CP–RS contacts in bent regions of the flagellum, and dyneins (mainly ODAs) are modulated through collisions between a remnant of the C1a projection or other CP projections and the tagged RS heads. WT, wild type.
Figure 7.
3D structures of CP. (A) Top views from the bases of flagella. (B) Side views from the C1a side. The fist and arm–like structure of C1a is absent in pf6. (C) Side views from the C1b side. The bladelike structure of the C1b is absent in cpc1. (B and C) The distal end of flagellum is to the bottom. Bars, 20 nm.
Streptavidin-dependent inhibition of motility in demembranated rsp4C and rsp6C cells is likely to be related to our model of CP–RS mechanosignaling (Fig. 6 C). The streptavidin bound to RS heads might transmit aberrant signals and cause irregular modulation of dynein activities, especially ODA (Fig. 3 E). However, the results of our ATPase and sliding disintegration assays did not reveal elevation or reduction in dynein activities in response to the addition of streptavidin (Table S2 and Fig. S2). We suggest that irregular activation (or inactivation) of ODA by streptavidin occurs only in a limited region in bending flagella and that ATPase and sliding disintegration assays cannot detect such local changes in dynein activities.
Architecture of RS
The results with streptavidin-labeled RS structures provide clues on how RSPs are arranged in 3D (Fig. 2 B). The rsp3C, 4N, and 6N structures reveal that each spoke contains at least two RSP3, 4, and 6 molecules and furthermore, indicate a partial twofold rotational symmetry in the RS head around the RS stalk (Pigino et al., 2011). However, the densities of the labels for rsp4C and 6C suggest that the C termini of RSP4 and 6 break the rotational symmetry. Although the signals for rsp6C appear to be dispersed on the RS heads, the strongest density is on the junction between the two RSs (Fig. S1 A). As there is a high amino acid sequence similarity between RSP4 and 6 (Curry et al., 1992), the two proteins may create pseudorotational symmetry, in which the C termini of RSP6 form the RS1–2 junction, and the C termini of RSP4 extend in opposite directions, away from the junction, forming the longitudinal edges of RS (Fig. 2 B). Note that there remains an ambiguity in the determination of the C termini of RSP6 because of the dispersed appearance of the label signals.
In agreement with the previous studies (Diener et al., 1993; Patel-King et al., 2004; Sivadas et al., 2012), RSP3 appears to be localized all the way from the RS head to the root of the RS stalk, and one spoke contains at least two copies of RSP3 (Figs. 2 and S1; Wirschell et al., 2008; Diener et al., 2011). RSP3 may form the core of the RS stalk, as the RSP3-missing mutant pf14 lacks the whole RS (Luck et al., 1977; Piperno et al., 1981). The position of the N termini of RSP3, i.e., near the root of the RS stalk, agrees with previous studies indicating that RSP3 interacts with the calmodulin- and spoke-associated complex, which mediates regulatory signals between RS and IDA (Dymek and Smith, 2007; Dymek et al., 2011).
RSP11 is located in the middle of the RS stalk (Fig. 2), a distribution that is unique among RSPs and is consistent with the previous deletion mutant analysis of RSP3 (Sivadas et al., 2012). RSP11 has sequence similarity to the regulatory subunit (RIIa) of cAMP-dependent protein kinase (PKA), and it has been proposed to regulate flagellar beating through interaction with RSP3 (Yang and Yang, 2006). As RSP11 is localized to the middle of the RS stalk, it might reinforce the stalk and facilitate effective transmission of mechanosignaling through the extended RSP3.
Future directions
The combination of cell biological, biochemical, and genetic approaches with cryoelectron tomography we applied in this study has revealed evidence for mechanosignaling between CP and RS. A remaining question is how the mechanosignal is transduced, becoming a regulatory signal that controls dynein activity. One possibility is that the physical contact between RS and CP slightly displaces the DMT outward and applies tensions to the nexin–dynein regulatory complex, outer–inner dynein linkers, and dynein stalks (Porter et al., 1994; Heuser et al., 2009; Oda et al., 2013). Another possibility is that mechanosignals transmit from RS to ODA via physical connections among IDA subspecies f, MIA (modifier of inner arms) complex, and the outer–inner dynein linker (Dymek and Smith, 2007; Yamamoto et al., 2013). We will pursue the signaling pathway downstream of CP–RS mechanosignaling by tagging of axonemal proteins and cryoelectron tomography.
Materials and methods
Strains and reagents
C. reinhardtii strains 137c, cpc1, oda1 (Kamiya, 1988; Takada et al., 2002), ida1, ida3 (Kamiya et al., 1991), ida5 (Kato et al., 1993), pf1, pf6, pf14, pf18 (Adams et al., 1981), pf25, and pf26 were provided by R. Kamiya (Gakushuin University, Tokyo, Japan) and M. Hirono (The University of Tokyo, Tokyo, Japan). Details of the aforementioned mutant strains are described in Table S1. The cells were grown in TAP (Tris-acetate-phosphate) medium. The triple and quadruple mutants were constructed using standard methods (Harris et al., 2009). For the screening of transformants, cells were grown on TAP agar supplemented with 10 ng/ml paromomycin (Sigma-Aldrich). The IC138 antiserum, raised against the N-terminal part of IC138 in rabbit, was provided by W.S. Sale (Emory University, Atlanta, GA).
Construction and expression of BCCP-tagged RSPs
A fragment spanning from ∼500 bp upstream noncoding sequence to immediately before the stop codon for the genes encoding RSP3 (2.8 kbp), RSP4 (3.2 kbp), RSP6 (3.3 kbp), and RSP11 (3.0 kbp) were amplified by genomic PCR using genomic DNA from the wild-type strain 137c and then inserted into the pGenD plasmid (Fischer and Rochaix, 2001; Nakazawa et al., 2007). After the last codon of each RSP gene, we ligated the 3′UTR of the LC8 (FLA14) gene. To insert a BCCP tag, both the tag sequence corresponding to amino acids 141–228 (for N terminus tagging) or 151–228 (for C terminus tagging) of C. reinhardtii acetyl–coenzyme A carboxylase BCCP and pGenD-RSPs were amplified by PCR, and the BCCP tag was fused immediately before the start or stop codon of RSP genes using a cloning kit (In-Fusion HD; Takara Bio Inc.). To add 3×HA or GFP, the RSP genes were subcloned into pGenD plasmids containing a codon-optimized sequence encoding 3×HA or GFP.
Preparation of axonemes
C. reinhardtii cells were deflagellated with dibucaine-HCl (Wako Chemicals USA), and axonemes were collected by centrifugation (Witman et al., 1978). Flagella were demembranated with 1% Nonidet P-40 in HMDENa buffer or HMDEK buffer comprised of 30 mM Hepes-NaOH, pH 7.2, 5 mM MgCl2, 1 mM DTT, 1 mM EGTA, 50 mM NaCl, or 50 mM CH3COOK, and protease inhibitor cocktail (Nacalai Tesque).
To observe the phosphorylation state of IC138 protein, flagella were first prepared using TCA fixation method (Wakabayashi and King, 2006). C. reinhardtii cells were deflagellated with acetic acid, and then, TCA (final concentration of 5% vol/vol) was immediately added to fix the phosphorylation state of the axonemal proteins. Cell bodies were removed by differential centrifugation, and then, flagella were pelleted and washed three times with acetone. Dried flagella were resuspended in urea-SDS sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 8 M urea, and 5% 2-mercaptoethanol) and boiled for 10 min.
Electrophoresis and immunoblotting
RSPs and other axonemal proteins were resolved by SDS-PAGE on 5–20% polyacrylamide gradient gels (Nacalai Tesque) and blotted onto polyvinylidene difluoride membranes. The blots were probed with streptavidin conjugated with horseradish peroxidase (Thermo Fisher Scientific), anti-HA (mouse; Wako Chemicals USA), or anti-GFP (rat; Nacalai Tesque) antibodies.
Fluorescence microscopy detection of axonemes
Demembranated axonemes were attached to glass slides and blocked with 1 mg/ml BSA in HMDEK buffer. Axonemes were incubated with 1 µg/ml Alexa Fluor 546–conjugated streptavidin (Invitrogen) for 1 min. Labeled axonemes were washed three times with HMDEK buffer and observed using a fluorescence microscope (IX70; Olympus). Images were recorded using a charge-coupled device (CCD) camera (ORCA-R2; Hamamatsu Photonics). Signal intensity was measured using ImageJ (National Institutes of Health) as follows: the background was subtracted from each image of Alexa Fluor–streptavidin-labeled axonemes, and the signal intensity was defined as the sum of the pixel values of one axoneme divided by the length of the axoneme.
Measurements of swimming velocity and beat frequency
The swimming velocity of C. reinhardtii cells was recorded using an inverted microscope (IX70) at a total magnification of 100×. A red filter with a cutoff wavelength of 630 nm was inserted before the condenser lens to suppress the cellular response to light. The beat frequency of flagella was measured as described previously (Kamiya, 2000). In brief, fluctuations in the intensity of microscopic images of swimming cells were analyzed using a photo detector, and spectra were generated using fast Fourier transform. The mean value was considered the position of a peak, and the standard deviation was obtained from the shape of the peak, fitted with a Gaussian curve. In a typical experiment at a total magnification of 100×, ∼500–1,000 cells contributed to one fast Fourier transform spectrum.
Biotinylation of cytochrome c
To biotinylate cytochrome c, 5 mg/ml equine heart cytochrome c (Sigma-Aldrich) was incubated with 2 mg/ml biotin-(AC5)2-OSu (Dojindo Molecular Technologies) in 10 mM Hepes-NaOH buffer, pH 7.2, for 2 h at room temperature. Unreacted biotin was quenched with 0.1 M glycine (Wako Chemicals USA) and removed by three rounds of desalting using PD MiniTrap G-25 columns (GE Healthcare).
Cryosample preparation
Demembranated axonemes were incubated with 0.05 mg/ml streptavidin (Wako Chemicals USA) for 15 min at 4°C in HMDEK buffer. Axonemes were then washed five times with HMDEK buffer and incubated with 0.05 mg/ml biotinylated cytochrome c for 15 min at 4°C in the presence of 1 mg/ml BSA and 0.1 mg/ml of unlabeled cytochrome c. Next, axonemes were again washed five times with HMDEK and incubated with 0.05 mg/ml streptavidin for 15 min at 4°C. Labeled axonemes were separated from unbound streptavidin by centrifugation, resuspended in HMDEK buffer at a concentration of 0.02 mg/ml, and mixed with an equal amount of 15-nm colloidal gold suspension conjugated with BSA (Aurion). Homemade holey carbon grids were glow discharged and coated with 20-nm colloidal gold (BBInternational). Suspended axonemes plus colloidal gold (5 µl) were loaded onto the grids and plunge frozen in liquid ethane at −180°C with an automated plunge-freezing device (EM GP; Leica).
Image acquisition
Grids were transferred into a transmission electron microscope (JEM-3100FEF; JEOL) with a high-tilt liquid nitrogen cryotransfer holder (914; Gatan, Inc.). Tilt series images were recorded at −180°C using a 4,096 × 4,096–pixel complementary metal-oxide semiconductor camera (TemCam-F416; Tietz Video and Image Processing Systems) and automated acquisition using Recorder software (System in Frontier, Inc.). The angular range of the tilt series was from −65 to 65° with 2.0° increments. The total electron dose was limited to ∼100 e−/Å2. Images were recorded at 300 keV, with 6–12-µm defocus, at a magnification of 25,700× and a pixel size of 6 Å. An in-column Ω energy filter was used to enhance image contrast in the zero-loss mode with a slit width of 23 eV.
Image processing
The image processing for subtomogram averaging of DMT structures was performed as described previously (Oda and Kikkawa, 2013). In brief, tilt series images were aligned and back projected to reconstruct 3D tomograms using Composer software (System in Frontier, Inc.). Tomograms of intact axonemes with a high signal-to-noise ratio were selected and used for subtomogram averaging of the 96-nm repeats of the outer DMTs. Alignment and averaging of subtomograms were conducted using custom Ruby-Helix scripts (Metlagel et al., 2007) and the PEET (Particle Estimation for Electron Tomography) software suite (Nicastro et al., 2006). For subtomogram averaging of CP structures, we cut out the subvolumes of 220 × 220 × 220 pixels, including CP microtubules at the center of the volume, every 16 nm along the center of the axoneme. The initial values for the rotational angle were determined based on the orientation of the C1 microtubule. The numbers of DMT subtomograms averaged were as follows: 644 for wild type, 604 for rsp3C, 680 for rsp3N, 512 for rsp4C, 700 for rsp4N, 580 for rsp6C, 502 for rsp6N, and 524 for rsp11C. The numbers of CP subtomograms averaged were as follows: 779 for wild type, 574 for pf6, and 695 for cpc1. The effective resolutions determined by Fourier shell correlation with a cutoff value of 0.5 were as follows: 6.8 nm for wild-type DMT, 6.9 nm for rsp3C, 6.9 nm for rsp3N, 7.0 nm for rsp4C, 6.9 nm for rsp4N, 7.0 nm for rsp6C, 7.1 nm for rsp6N, 6.9 nm for rsp11C, 7.1 nm for wild-type CP, 7.2 nm for pf6 CP, and 7.1 nm for cpc1 CP (Fig. S1, D and E). Surface renderings were generated using University of California, San Francisco Chimera (Pettersen et al., 2004).
Statistical analysis
To identify statistically significant differences, we applied the Student’s t test to compare wild-type and streptavidin-labeled axonemes as described previously (Oda and Kikkawa, 2013). First, wild-type and streptavidin-labeled subtomograms were randomly divided into three datasets. Subtomograms for each dataset were aligned and averaged, and a total of six averaged subtomograms were created. We calculated the t-value for each voxel and presented them as a single t-value map. The isosurface threshold values were t > 8.61, with a one-tailed probability of <0.05%.
Motility analysis of demembranated C. reinhardtii cells
C. reinhardtii cells were washed three times with HMES buffer (10 mM Hepes-NaOH, pH 7.2, 5 mM MgCl2, 1 mM EGTA, and 2% sucrose) and gently resuspended in HMDEK plus 1% polyethylene glycol and 0.2% NP-40. The demembranated cells were reactivated with 1 mM ATP and motility was analyzed after a 1-min incubation with BSA or streptavidin using a dark-field microscope (BX51; Olympus) equipped with a 20× and 40× oil immersion objective lens and a 100-W mercury lamp. The image sequences were recorded using a progressive scan–type CCD camera (MC681SPD; Texas Instruments) or a high speed CCD camera (MC1362; Mikrotron).
ATPase assay
The rate of phosphate release by axonemes was measured using Biomol green reagent (Enzo Life Sciences). 0.1 mg/ml axonemes were incubated for 5 min in HMDEK buffer in the presence of 1 mM ATP. The concentrations of released phosphate were calculated based on changes to the absorbance at 620 nm.
Sliding disintegration of the axoneme
To measure sliding velocity, suspensions of flagella were sonicated using a sonicator (S-4000; Qsonica, LLC). Sonication was repeated until, on average, flagella were sheared in half lengthwise. The flagella were then demembranated with 1% Nonidet P-40 in HMDEK buffer and centrifuged. Axonemes were resuspended in HMDEK buffer without protease inhibitors and absorbed onto a glass slide. The solution was covered with a coverslip, and two sides of the coverslip were sealed with Vaseline. Axonemes were activated with HMDEK buffer with 1 mM ATP. Sliding disintegration was then initiated by washing the flow chamber with HMDEK buffer with 1 mM ATP and 2 µg/ml trypsin. To measure the degree of sliding disintegration, intact flagella were demembranated and absorbed onto a glass slide. Sliding disintegration was initiated with HMDEK buffer with 1 mM ATP and 0.3 µg/ml nagarse (Sigma-Aldrich). Sliding of DMTs was observed using a dark-field microscope (BX51) equipped with a 40× oil immersion objective lens and a 100-W mercury lamp. The image sequences were recorded using a progressive scan–type CCD camera (MC681SPD).
Accession numbers
The EM maps of averaged DMT and CP tomograms are available at the EMDataBank under the following accession numbers: wild-type DMT, EMD-5845; rsp3N DMT, EMD-5846; rsp3C DMT, EMD-5847; rsp4N DMT, EMD-5848; rsp4C DMT, EMD-5849; rsp6N DMT, EMD-5850; rsp6C DMT, EMD-5851; rsp11C DMT, EMD-5852; wild-type CP, EMD-5853; pf6 CP, EMD-5854; and cpc1 CP, EMD-5855.
Online supplemental material
Fig. S1 is related to Fig. 2 and shows averaged tomograms of DMT used for Student’s t test. Fig. S2 is related to Fig. 3 and shows the results of sliding disintegration assays of wild-type and mutant axonemes. Fig. S3 is related to Fig. 4 and shows the swimming paths of wild-type and pf6 rsp4C cells. Table S1 describes details of the mutant strains used in this study. Table S2 is related to Fig. 3 and shows the results of ATPase and sliding disintegration assays of wild-type and mutant axonemes. Video 1 is related to Fig. 3 and shows swimming of demembranated, ATP-reactivated mutant cells. Video 2 is related to Fig. 4 and shows impaired motility of pf6 cells and the recovered motility of pf6 rsp4C tags live cells. Video 3 is related to Fig. 4 and shows motility of pf6 oda/ida double mutants and pf6 oda/ida rsp4C-GFP quadruple mutants. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201312014/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Winfield S. Sale for the gift of IC138 antiserum.
This work was supported by the Funding Program for Next Generation World-Leading Researchers, the Uehara Memorial Foundation, and the Takeda Science Foundation (to M. Kikkawa).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- BCCP
- biotin carboxyl carrier protein
- CCD
- charge-coupled device
- CP
- central pair apparatus
- DMT
- doublet microtubule
- fps
- frames per second
- IC50
- half-maximal inhibitory concentration
- IDA
- inner dynein arm
- ODA
- outer dynein arm
- RS
- radial spoke
- RSP
- radial spoke protein
References
- Adams G.M.W., Huang B., Piperno G., Luck D.J.L. 1981. Central-pair microtubular complex of Chlamydomonas flagella: polypeptide composition as revealed by analysis of mutants. J. Cell Biol. 91:69–76 10.1083/jcb.91.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajal-González B.I., Heuser T., Fu X., Lin J., Smith B.W., Mitchell D.R., Nicastro D. 2013. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken). 70:101–120 10.1002/cm.21094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry A.M., Williams B.D., Rosenbaum J.L. 1992. Sequence analysis reveals homology between two proteins of the flagellar radial spoke. Mol. Cell. Biol. 12:3967–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D.R., Curry A.M., Johnson K.A., Williams B.D., Lefebvre P.A., Kindle K.L., Rosenbaum J.L. 1990. Rescue of a paralyzed-flagella mutant of Chlamydomonas by transformation. Proc. Natl. Acad. Sci. USA. 87:5739–5743 10.1073/pnas.87.15.5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D.R., Ang L.H., Rosenbaum J.L. 1993. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J. Cell Biol. 123:183–190 10.1083/jcb.123.1.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D.R., Yang P., Geimer S., Cole D.G., Sale W.S., Rosenbaum J.L. 2011. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken). 68:389–400 10.1002/cm.20520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K., Huang B., Luck D.J. 1984. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98:229–236 10.1083/jcb.98.1.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek E.E., Smith E.F. 2007. A conserved CaM- and radial spoke–associated complex mediates regulation of flagellar dynein activity. J. Cell Biol. 179:515–526 10.1083/jcb.200703107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek E.E., Heuser T., Nicastro D., Smith E.F. 2011. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol. Biol. Cell. 22:2520–2531 10.1091/mbc.E11-03-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N., Rochaix J.D. 2001. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics. 265:888–894 10.1007/s004380100485 [DOI] [PubMed] [Google Scholar]
- Gibbons I.R. 1981. Cilia and flagella of eukaryotes. J. Cell Biol. 91:107s–124s 10.1083/jcb.91.3.107s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goduti D.J., Smith E.F. 2012. Analyses of functional domains within the PF6 protein of the central apparatus reveal a role for PF6 sub-complex members in regulating flagellar beat frequency. Cytoskeleton (Hoboken). 69:179–194 10.1002/cm.21010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A., Wirschell M., Sale W.S. 2009. Regulation of dynein-driven microtubule sliding by the axonemal protein kinase CK1 in Chlamydomonas flagella. J. Cell Biol. 186:817–824 10.1083/jcb.200906168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Heuser J.E. 1985. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100:2008–2018 10.1083/jcb.100.6.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H., Stern D.B., Witman G.B. 2009. The Chlamydomonas Sourcebook. Second edition Academic Press, Oxford, England, UK: 2,000 pp [Google Scholar]
- Hayashi S., Shingyoji C. 2008. Mechanism of flagellar oscillation-bending-induced switching of dynein activity in elastase-treated axonemes of sea urchin sperm. J. Cell Sci. 121:2833–2843 10.1242/jcs.031195 [DOI] [PubMed] [Google Scholar]
- Hendrickson T.W., Perrone C.A., Griffin P., Wuichet K., Mueller J., Yang P., Porter M.E., Sale W.S. 2004. IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol. Biol. Cell. 15:5431–5442 10.1091/mbc.E04-08-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T., Raytchev M., Krell J., Porter M.E., Nicastro D. 2009. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 187:921–933 10.1083/jcb.200908067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Tanaka Y., Okada Y., Takeda S. 2006. Nodal flow and the generation of left-right asymmetry. Cell. 125:33–45 10.1016/j.cell.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Hoops H.J., Witman G.B. 1983. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol. 97:902–908 10.1083/jcb.97.3.902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard D.R., Habermacher G., Glass D.B., Smith E.F., Sale W.S. 1994. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J. Cell Biol. 127:1683–1692 10.1083/jcb.127.6.1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Piperno G., Ramanis Z., Luck D.J. 1981. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88:80–88 10.1083/jcb.88.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Ramanis Z., Luck D.J. 1982. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for Flagellar function. Cell. 28:115–124 10.1016/0092-8674(82)90381-6 [DOI] [PubMed] [Google Scholar]
- Hung L.-H., Samudrala R. 2003. PROTINFO: Secondary and tertiary protein structure prediction. Nucleic Acids Res. 31:3296–3299 10.1093/nar/gkg541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami O., Kamiya R. 1990. Strikingly low ATPase activities in flagellar axonemes of a Chlamydomonas mutant missing outer dynein arms. Eur. J. Biochem. 189:441–446 10.1111/j.1432-1033.1990.tb15508.x [DOI] [PubMed] [Google Scholar]
- Kamiya R. 1988. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J. Cell Biol. 107:2253–2258 10.1083/jcb.107.6.2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R. 2000. Analysis of cell vibration for assessing axonemal motility in Chlamydomonas. Methods. 22:383–387 10.1006/meth.2000.1090 [DOI] [PubMed] [Google Scholar]
- Kamiya R., Kurimoto E., Muto E. 1991. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 112:441–447 10.1083/jcb.112.3.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Kagami O., Yagi T., Kamiya R. 1993. Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct. Funct. 18:371–377 10.1247/csf.18.371 [DOI] [PubMed] [Google Scholar]
- Kurimoto E., Kamiya R. 1991. Microtubule sliding in flagellar axonemes of Chlamydomonas mutants missing inner- or outer-arm dynein: velocity measurements on new types of mutants by an improved method. Cell Motil. Cytoskeleton. 19:275–281 10.1002/cm.970190406 [DOI] [PubMed] [Google Scholar]
- Lindemann C.B., Mitchell D.R. 2007. Evidence for axonemal distortion during the flagellar beat of Chlamydomonas. Cell Motil. Cytoskeleton. 64:580–589 10.1002/cm.20205 [DOI] [PubMed] [Google Scholar]
- Luck D., Piperno G., Ramanis Z., Huang B. 1977. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc. Natl. Acad. Sci. USA. 74:3456–3460 10.1073/pnas.74.8.3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A., Ishikawa T. 2012. Heterogeneity of dynein structure implies coordinated suppression of dynein motor activity in the axoneme. J. Struct. Biol. 179:235–241 10.1016/j.jsb.2012.04.018 [DOI] [PubMed] [Google Scholar]
- Metlagel Z., Kikkawa Y.S., Kikkawa M. 2007. Ruby-Helix: an implementation of helical image processing based on object-oriented scripting language. J. Struct. Biol. 157:95–105 10.1016/j.jsb.2006.07.015 [DOI] [PubMed] [Google Scholar]
- Mitchell D.R. 2003. Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil. Cytoskeleton. 56:120–129 10.1002/cm.10142 [DOI] [PubMed] [Google Scholar]
- Mitchell D.R., Sale W.S. 1999. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144:293–304 10.1083/jcb.144.2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movassagh T., Bui K.H., Sakakibara H., Oiwa K., Ishikawa T. 2010. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat. Struct. Mol. Biol. 17:761–767 10.1038/nsmb.1832 [DOI] [PubMed] [Google Scholar]
- Nakano I., Kobayashi T., Yoshimura M., Shingyoji C. 2003. Central-pair-linked regulation of microtubule sliding by calcium in flagellar axonemes. J. Cell Sci. 116:1627–1636 10.1242/jcs.00336 [DOI] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. 2007. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17:2169–2174 10.1016/j.cub.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Nicastro D., Schwartz C., Pierson J., Gaudette R., Porter M.E., McIntosh J.R. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 313:944–948 10.1126/science.1128618 [DOI] [PubMed] [Google Scholar]
- Oda T., Kikkawa M. 2013. Novel structural labeling method using cryo-electron tomography and biotin-streptavidin system. J. Struct. Biol. 183:305–311 10.1016/j.jsb.2013.07.003 [DOI] [PubMed] [Google Scholar]
- Oda T., Yagi T., Yanagisawa H., Kikkawa M. 2013. Identification of the outer-inner dynein linker as a hub controller for axonemal dynein activities. Curr. Biol. 23:656–664 10.1016/j.cub.2013.03.028 [DOI] [PubMed] [Google Scholar]
- Omoto C.K., Yagi T., Kurimoto E., Kamiya R. 1996. Ability of paralyzed flagella mutants of Chlamydomonas to move. Cell Motil. Cytoskeleton. 33:88–94 [DOI] [PubMed] [Google Scholar]
- Patel-King R.S., Gorbatyuk O., Takebe S., King S.M. 2004. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol. Biol. Cell. 15:3891–3902 10.1091/mbc.E04-04-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Rosenbaum J.L. 2002. Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 12:551–555 10.1016/S0962-8924(02)02410-8 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pigino G., Bui K.H., Maheshwari A., Lupetti P., Diener D., Ishikawa T. 2011. Cryoelectron tomography of radial spokes in cilia and flagella. J. Cell Biol. 195:673–687 10.1083/jcb.201106125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Huang B., Luck D.J. 1977. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 74:1600–1604 10.1073/pnas.74.4.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Huang B., Ramanis Z., Luck D.J. 1981. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88:73–79 10.1083/jcb.88.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M.E., Power J., Dutcher S.K. 1992. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J. Cell Biol. 118:1163–1176 10.1083/jcb.118.5.1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M.E., Knott J.A., Gardner L.C., Mitchell D.R., Dutcher S.K. 1994. Mutations in the SUP-PF-1 locus of Chlamydomonas reinhardtii identify a regulatory domain in the β-dynein heavy chain. J. Cell Biol. 126:1495–1507 10.1083/jcb.126.6.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G., O’Toole E., Porter M.E. 2001. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell. 12:739–751 10.1091/mbc.12.3.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale W.S. 1986. The axonemal axis and Ca2+-induced asymmetry of active microtubule sliding in sea urchin sperm tails. J. Cell Biol. 102:2042–2052 10.1083/jcb.102.6.2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadas P., Dienes J.M., St. Maurice M., Meek W.D., Yang P. 2012. A flagellar A-kinase anchoring protein with two amphipathic helices forms a structural scaffold in the radial spoke complex. J. Cell Biol. 199:639–651 10.1083/jcb.201111042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.F., Lefebvre P.A. 1997. The role of central apparatus components in flagellar motility and microtubule assembly. Cell Motil. Cytoskeleton. 38:1–8 [DOI] [PubMed] [Google Scholar]
- Smith E.F., Yang P. 2004. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton. 57:8–17 10.1002/cm.10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Wilkerson C.G., Wakabayashi K., Kamiya R., Witman G.B. 2002. The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol. Biol. Cell. 13:1015–1029 10.1091/mbc.01-04-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWaal K.E., Yamamoto R., Wakabayashi K., Fox L., Kamiya R., Dutcher S.K., Bayly P.V., Sale W.S., Porter M.E. 2011. bop5 mutations reveal new roles for the IC138 phosphoprotein in the regulation of flagellar motility and asymmetric waveforms. Mol. Biol. Cell. 22:2862–2874 10.1091/mbc.E11-03-0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K., King S.M. 2006. Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J. Cell Biol. 173:743–754 10.1083/jcb.200603019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo M.J., Smith E.F. 2003. Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 100:137–142 10.1073/pnas.0135800100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner F.D. 1970. New observations on flagellar fine structure. The relationship between matrix structure and the microtubule component of the axoneme. J. Cell Biol. 47:159–182 10.1083/jcb.47.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner F.D., Satir P. 1974. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J. Cell Biol. 63:35–63 10.1083/jcb.63.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M., Zhao F., Yang C., Yang P., Diener D., Gaillard A., Rosenbaum J.L., Sale W.S. 2008. Building a radial spoke: flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil. Cytoskeleton. 65:238–248 10.1002/cm.20257 [DOI] [PubMed] [Google Scholar]
- Witman G.B., Plummer J., Sander G. 1978. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76:729–747 10.1083/jcb.76.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R., Song K., Yanagisawa H.-A., Fox L., Yagi T., Wirschell M., Hirono M., Kamiya R., Nicastro D., Sale W.S. 2013. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J. Cell Biol. 201:263–278 10.1083/jcb.201211048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Yang P. 2006. The flagellar motility of Chlamydomonas pf25 mutant lacking an AKAP-binding protein is overtly sensitive to medium conditions. Mol. Biol. Cell. 17:227–238 10.1091/mbc.E05-07-0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M., Agrin N.S., King S.M., Sale W.S., Kamiya R., et al. 2006. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119:1165–1174 10.1242/jcs.02811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala M.A., Knowles M.R., Omran H. 2007. Genetic defects in ciliary structure and function. Annu. Rev. Physiol. 69:423–450 10.1146/annurev.physiol.69.040705.141301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.