Lamins impede 3D migration but also promote survival against migration-induced stresses.
Abstract
Cell migration through solid tissue often involves large contortions of the nucleus, but biological significance is largely unclear. The nucleoskeletal protein lamin-A varies both within and between cell types and was shown here to contribute to cell sorting and survival in migration through constraining micropores. Lamin-A proved rate-limiting in 3D migration of diverse human cells that ranged from glioma and adenocarcinoma lines to primary mesenchymal stem cells (MSCs). Stoichiometry of A- to B-type lamins established an activation barrier, with high lamin-A:B producing extruded nuclear shapes after migration. Because the juxtaposed A and B polymer assemblies respectively conferred viscous and elastic stiffness to the nucleus, subpopulations with different A:B levels sorted in 3D migration. However, net migration was also biphasic in lamin-A, as wild-type lamin-A levels protected against stress-induced death, whereas deep knockdown caused broad defects in stress resistance. In vivo xenografts proved consistent with A:B-based cell sorting, and intermediate A:B-enhanced tumor growth. Lamins thus impede 3D migration but also promote survival against migration-induced stresses.
Introduction
Cell motility within adult animals underlies a wide range of critical processes, including trafficking in disease and tissue repair. Cells in the periphery of a tumor, for example, crawl through surrounding tissue and ECM to divide or metastasize into narrow vessels (Weinberg, 2006). Multipotent stem cells in the bone marrow likewise transmigrate into blood capillaries or other tissues far from their niche and have potential roles in tissue regeneration (Pittenger and Martin, 2004) as well as cancer (Houghton et al., 2004; Nakamizo et al., 2005). Successful migration requires cells to survive large distortions (Fig. 1 A), but the largest single organelle in every cell is typically the nucleus—which tends to be stiff (Dahl et al., 2005; Lammerding et al., 2006; Pajerowski et al., 2007). In light of the considerable stress generated by the cytoskeleton in crawling through tissue matrix (Mierke et al., 2011), we hypothesized rate-limiting roles for nuclear mechanics in both 3D migration and post-migration survival.
Figure 1.
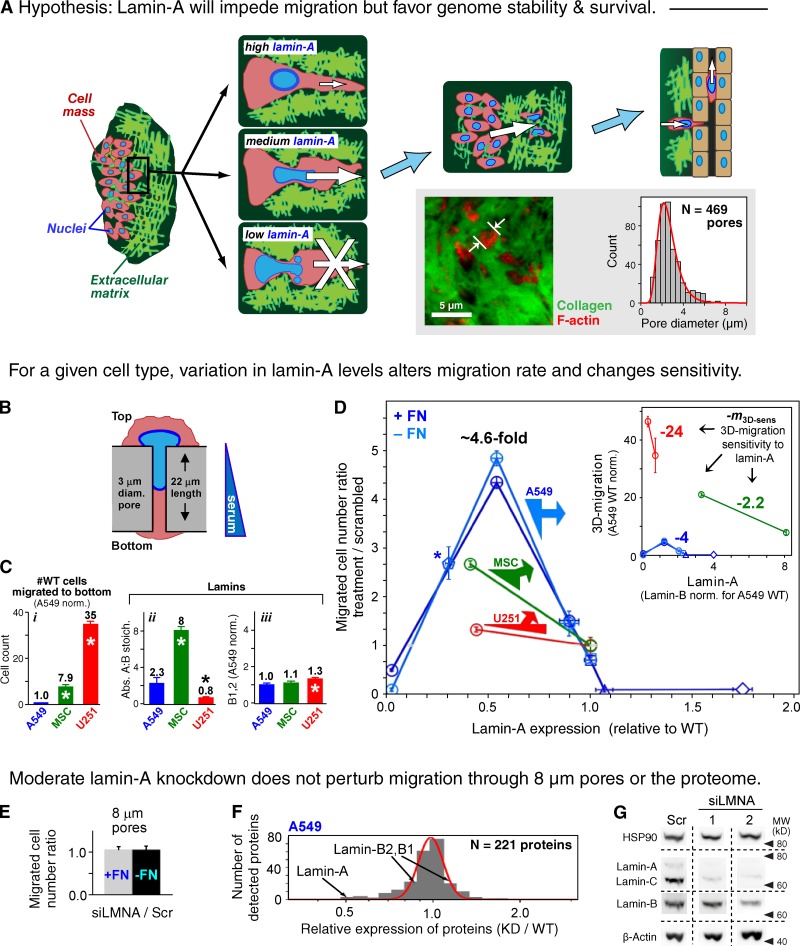
3D migration is sensitive to lamin-A levels even in the absence of major proteomic changes. (A) Hypothesis for the impact of lamin-A levels on migration. Whereas moderate expression permits migration, cells with low levels cannot withstand the stress and high levels impede migration. Inset shows confocal image of a model lung tumor in an NSG mouse and a histogram of the measured pores filled with cytoplasm. (B) Schematic of a cell passing through a filter pore. (C) Wild-type 3D migration (i) and lamina parameters (ii and iii) for human-derived cancer cells (A549, U251) and adult stem cells (MSCs). (n ≥ 3; ±SEM; *, P ≤ 0.05). (D) Lamin-A dependence of net cell migration to the filter bottom (n ≥ 3; ±SEM). Normalization is done to scrambled siRNA-treated cells, with lamin-A level determined by immunoblot. Filters were pre-coated with fibronectin (+FN) unless indicated (−FN). Circles, siLMNA-treated cells; asterisk, shLMNA-treated cells, normalized against wild type; squares, wild type; triangle and diamond, A549 cells transduced with GFP-lamin-A with low and high levels, respectively. Based on analyses of individual cells, siLMNA++ gave a monomodal, low variance population of cells and did not affect lamin-B (Fig. S2). Inset plot: net migration results rescaled to absolute lamin-A levels with migration normalized to wild-type A549 cells (Fig. 1 Ci). Slopes represent the 3D migration sensitivity to moderate lamin-A changes. (E) Net migration ratio with 8-µm pore filters shows no effects of knockdown regardless of fibronectin. (n ≥ 3; ±SEM). (F) Negligible effect on the A549 proteome after ∼50% knockdown of lamin-A, C is evident in a narrow, log-normal distribution relative to wild type. (G) Immunoblot shows little to no change in lamin-B, HSP90, and β-actin after knockdown with two different siLMNAs (1 and 2) compared with scrambled (Scr).
A- and B-type lamins are intermediate filaments that assemble with distinct membrane-binding partners into juxtaposed networks within the nuclei of nearly all adult animal cells (Moir et al., 2000; Holaska et al., 2002; Shimi et al., 2008). Yeast cells do not express lamins (Dittmer and Misteli, 2011) and do not migrate, but have rigid cell walls that physically protect their genomes. Why animal cells have two independent nucleoskeletal networks is unclear, but each lamin type might distinctly modulate gene expression (Dechat et al., 2008) and each lamin type might also stiffen and stabilize the nucleus with a distinctiveness similar to that of the many different keratin intermediate filaments in skin, nails, hair, and beaks. For migration, the polymorphonuclear leukocyte (PMN) is perhaps instructive in that it down-regulates lamins in differentiation to a cell with a multi-segmented nucleus (Olins et al., 2001) of a flexibility suitable for crawling through very small pores in endothelium and dense tissues (Chamberlain and Lichtman, 1978). However, PMNs also die within days (Pillay et al., 2010), perhaps because the chromatin is unprotected. Knockdown of lamin-A in granulocytes/monocytes derived from cultures of human hematopoietic stem/progenitor cells increases net migration of these cells by several-fold through Transwell filters with small capillary-sized micropores but not larger pores (Shin et al., 2013), with similar findings for a leukemia-derived cell line (Rowat et al., 2013). However, lamin perturbations can also impact differentiation state and cytoskeletal factors involved in migration (Ho et al., 2013; Shin et al., 2013; Swift et al., 2013b), and so any lamin knockdown or overexpression that perturbs 3D motility should in principle be shown to correspond well to perturbations of nuclear mechanics as assessed with biophysically relevant stress levels and time scales. Alternative determinants indeed seem conceivable: physical limits of cell migration might be defined, for example, not by the lamina but as recently postulated by a “non-compressible intranuclear component (e.g., chromatin)” (Wolf et al., 2013). Any systematic effects of variations in lamins on 3D migration thus remain unclear.
Average levels of lamin-A within normal solid tissues have been found recently to scale with tissue stiffness (Swift et al., 2013b), but lamin-A levels also change in disease, including cancer (Kaufmann et al., 1991; Willis et al., 2008). Low lamin-A reportedly correlates with the recurrence of colon cancer (Belt et al., 2011), whereas high lamin-A correlates with malignant carcinomas and colorectal cancer (Tilli et al., 2003; Foster et al., 2010). Although in vivo models have yet to address the roles of lamins in cancer, mouse knockouts for lamin-A (Sullivan et al., 1999) and lamin-B (Coffinier et al., 2011; Kim et al., 2011) are small and die, respectively, at 2–3 wk or at birth. Lamin-B knockouts show poor innervation migration into the diaphragm and death of neuroprogenitors migrating into the dense mid-cortex of brain. The nucleus in neuroblasts normally stretches fivefold for many minutes at a time in migration through developing cortex (Tsai et al., 2005; Pajerowski et al., 2007), and brain-slice migration of glioblastoma cells likewise shows the nucleus stretches by approximately twofold (Beadle et al., 2008). With solid tissue cells studied here, lamin-A:B stoichiometry is found to govern 3D migration in vitro, with a key role in survival and predictable effects in xenografts in vivo.
Results
3D migration is biphasic in lamin-A levels
Mesenchymal-type motility in 2D is exhibited by all three human cell types studied here. Glioblastoma-derived U251 cells migrate 35-fold faster through 3-µm pore Transwell filters (Fig. 1, B and Ci) than lung carcinoma–derived A549 cells, whereas mesenchymal stem cells (MSCs) obtained from bone marrow show an intermediate motility. Rigid, micro-porous filters with 3-µm or 8-µm pores seem appropriate for studies of cell transmigration because A549 xenografts in the flank as well as the nearby subcutaneous tissue are stiff (Swift et al., 2013b) relative to a typical nucleus that slowly flows under stress (Pajerowski et al., 2007), and also because the porosity of tumors appears to be in the range of microns (Fig. 1 A, inset) as suggested previously (Wolf et al., 2009). In 2D migration, speeds of the three cell types follow the same trend in 3D migration and vary by just twofold, with U251 > MSC > A549 (Fig. S1). These cells migrate in 2D at 10–20 µm/h, and although cell protrusions can be seen to extend through the ∼20-µm-thick filters within hours, almost no cell nuclei cross the filters in such a short time, consistent with a rate-limiting barrier to nuclear translocation.
The three different cell types possess highly distinct lamin profiles (Fig. 1 C, ii and iii) that are not predictive of differences in migration between cell types. We used mass spectrometry (MS) to measure for A549s a mean lamin-A:B stoichiometry of 2.3 (Swift et al., 2013b), which reveals lamin-A as the dominant isoform in these cells. Such a measurement of isoform stoichiometry is especially useful in calibrating results from immunoblots and immunofluorescence (Fig. S2). MSCs thus have a much higher A:B ratio while U251 cells have a much lower ratio, with lamin-A:B varying by 10-fold across the diverse cell types (MSC > A549 > U251), consistent with lamin-A level as a readout for bone being stiffer than lung and lung stiffer than brain (Swift et al., 2013b). Quantitative immunofluorescence of B-type lamins further indicates that U251s express only 20–30% more lamin-B than the other two cell types (Fig. S2), consistent with lamin-B being nearly constitutive (Jung et al., 2012). In such single-cell measurements for all cell types, the coefficient of variation for wild-type lamin-A:B levels is only ∼20%, and so the mean dominates the population variance. Whereas lamin-A:B differs considerably between cell types and shows no obvious correlation with 2D or 3D migration of wild-type cells, lamin-A:B proves surprisingly predictive of the sensitivity of 3D migration to lamin-A perturbations.
Knockdown of lamin-A by transient transfection of short interfering RNA (siLMNA) was followed by studies of migration. A549 cells showed a strongly biphasic dependence on relative lamin-A levels in net migration through 3-µm pores (Fig. 1 D), with the inset plot showing the same effect in terms of absolute lamin-A levels for comparison between cell types. Relative to scrambled (Scr) siRNA-treated cells in the same experiment, a partial knockdown (by ∼50%, denoted as siLMNA+) produced the greatest increase in 3D migration of more than fourfold, a trend confirmed with additional siLMNA (Fig. S3). Knockdown had no effect on migration through 8-µm pores (Fig. 1 E), which are approximately sevenfold larger in cross-section area than the smaller pores. 2D migration was also unaffected by knockdown (Fig. S1 A), and Scr was invariably the same as wild-type cells. Immunofluorescence showed that knockdown of lamin-A occurred in the majority of cells, with a similar variance as wild type (Fig. S2); heterogeneity thus does not dominate the average. Overexpression of lamin-A strongly impeded migration of A549 cells, and followed the same linear trend in migration established with partial knockdown: migration was suppressed by 90% with just ∼10% more lamin-A (with similar variance as wild type). Knockdown of these overexpressing cells increased the net migration as expected (Fig. S3), but further overexpression reached the same minimum level of net migration. Between partial knockdown of lamin-A and moderately overexpressing cells, the net migration of A549 cells spans a 50-fold range, indicating a strong functional dependence.
Sustained disruption of lamins can impact a broad range of cell processes (Hutchison and Worman, 2004; Swift et al., 2013b), and so we assessed broader perturbations to the proteome after lamin-A knockdown in A549 cells. Label-free mass spectrometry was used to quantify the most abundant structural proteins after ∼50% knockdown (Fig. 1 F): only 2 of 221 quantifiable proteins differed from control more than lamin-A, and neither protein is known to impact motility (importin-4 and major vault protein). Lamin-B1 and -B2 changed by less than 15% as validated by immunoblot (Fig. 1 G). No significant change was detected with β-actin, which has a critical role in motility. Transient knockdown of cells with siRNA was preferred over methods such as transduction with short hairpin RNA because the latter is difficult to titrate and control compensatory changes and selection. Lamin-A knockdown here also did not perturb proliferation (Fig. S3), consistent with past findings (Elbashir et al., 2001). Partial transient knockdown thus proved lamin-A specific and revealed only one obvious phenotype, namely enhanced migration through highly constraining micropores.
Knockdown of lamin-A by ∼50% in both U251 cells and MSCs also showed significant increases in net migration (Fig. 1 D). A proper comparison of sensitivity to lamin changes requires accounting for changes in absolute lamin levels rather than just percent change (Fig. 1 D, inset plot; Fig. S2). The slope (Δmigration/Δlamin-A) for each cell type reveals the net migration sensitivity to a lamin-A change (m3D-sens), and this cell type–specific m3D-sens is seen to vary by >10-fold. For a small change in the absolute levels of lamin-A in U251 cells, the net number of cells that migrate through constraining pores is much higher than that for the other cell types. U251 cells thus show the greatest sensitivity and MSCs show the least sensitivity (m3D-sens: U251 > A549 > MSC). This trend in sensitivity thus relates inversely to the lamin-A:B stoichiometry in wild-type cells. Knockdown did not affect nuclear volume (Fig. S2) or projected nuclear area (for U251 and A549 cells), and so the results are intrinsic to lamins.
Extruded shape is sustained by high lamin-A nuclei after cell migration through 3-µm pores
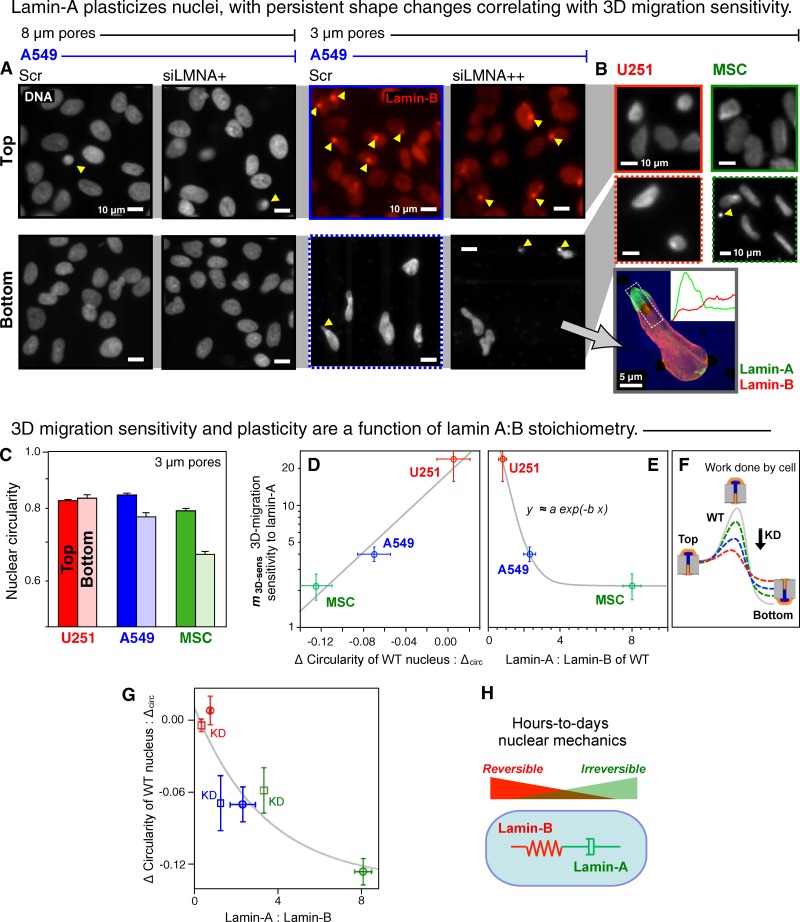
In assessing migration, we noticed that nuclear shapes on the bottom surfaces of the 3-µm Transwell filters were strikingly elongated compared with the typical elliptical shapes on the top surface (Fig. 2 A). With 8-µm Transwell filters, shapes of nuclei on the top and bottom of the filters appeared indistinguishable from healthy 2D cultures and cells migrating in 2D (Fig. 2 A; Fig. S3). For both 3- and 8-µm Transwell filters, images showed nuclei partially or fully inside pores (yellow arrowheads), but only for the 3-µm pores did we observe lamin-A segregation from lamin-B (Fig. 2 A, inset). Nuclear blebs in 2D culture have likewise been seen to have a lamin-B–deficient rim of lamin-A around inactive chromatin (Shimi et al., 2008). For A549 cells, the 3-µm pores are only 3% of the projected nuclear area versus 23% for 8-µm pores, with similar numbers for knockdown cells and U251 nuclei (Fig. S2). Sustained deformation of nearly all nuclei is apparent with 3-µm pores, regardless of lamin-A levels. However, for 8-µm pores, lamin-A knockdown had no effect on shapes of nuclei on the filter bottoms (Fig. S3), consistent with no effect on migration. These findings provided additional evidence that moderate perturbations of lamin-A do not broadly perturb migration machinery, consistent with the proteomic analyses.
Figure 2.
Lamin-A plasticizes nuclei, with persistent shape changes correlating with both 3D migration sensitivity to lamin-A and also with the lamin-A:B ratio. (A) A549 nuclei on the filters stained for either lamin-B or DNA. Arrowheads indicate a nucleus in a pore. Knockdown with siLMNA was >95% (3 µm) or 60% (8 µm). Inset: confocal image of siLMNA cell on the bottom of 3-µm pore filter showing segregation of lamins, with intensity profiles quantified from z-stacks. (B) U251 and MSC cells imaged on the top and bottom of 3-µm pore filters. (C) Circularity of wild-type nuclei on the top versus bottom of 3-µm filters (log scale). (D) Exponential correlation between 3D migration sensitivity to lamin-A levels (values in Fig. 1 C) and the difference in circularity (n > 3 repeats; ±SEM). The three datapoints were fitted to an exponential, y = α exp(−β x1) with two parameters: α = 19, β = 19 (R2 > 0.95). (E) For 3D migration sensitivity to lamin-A:B ratio, we fit a similar exponential y = a exp(−b x2) + c with similar fit parameters: a (∼α) = 80, b = 2, c << a (R2 > 0.94). (F) Scheme illustrates migration through a pore as an activated process in which an equi-mass knockdown of lamin-A suppresses the barrier and facilitates migration as U251 > A549 > MSC. (G) Migration-induced nuclear circularity change is also nonlinear in lamin-A:B for both wild-type and knockdown cells (KD, squares), with a fit based on the previous fits. (H) Schematic illustrates distinct physical roles of lamin-A in nuclear viscosity (dash pot) and lamin-B in nuclear elasticity (spring).
Sustained elongation of “extruded” nuclei was evident in MSCs after migration through 3-µm pores, whereas U251 nuclei appeared to be similar in shape on top and bottom (Fig. 2, B and C). Changes in circularity of nuclei after migration through 3-µm pores were therefore quantified for the three different cell types, with more distorted nuclei on the bottom producing a negative value for the characteristic “ΔCircularity of Nucleus” (ΔCirc); this structural characteristic correlates with the functional metric m3D-sens for 3D migration sensitivity to lamin-A, as do the lamin-A:B stoichiometries (Fig. 2, D and E). The latter exponential formally indicates that, relative to the wild type of any given cell type, the rate-limiting process of migrating across the microporous barrier requires less work for low lamin-A levels (Fig. 2 F).
A nonlinear relationship between ΔCirc and lamin-A:B stoichiometries across control and knockdown cell types (Fig. 2 G) further indicates that A- and B-type lamins play separate roles in nuclear shape and viscoelasticity. When lamin-B is the dominant isoform as in the U251 cells, the nucleus readily recovers after deformation through a small pore, behaving reversibly like an elastic spring (Fig. 2 H). In contrast, when lamin-A is the dominant isoform as in the A549 cells and MSCs, the nucleus deforms in a plastic manner after migration through a small pore, behaving irreversibly like a highly viscous fluid. Past photobleaching studies of GFP-lamin chimeras had shown lamin-B1 to be immobile whereas lamin-A appeared mobile (Shimi et al., 2008), consistent with the long-time elastic versus fluid contributions of lamins to nuclear shape changes. We had previously quantified the irreversible plasticity of nuclei after aspiration of a nucleus into a micropipette (Dahl et al., 2005; Pajerowski et al., 2007), and our mass spectrometry method allowed us to calibrate the lamin-A:B stoichiometries and thereby determine which isoform dominates in any given cell type. The physical plasticity shown here results from active cell migration and seems consistent with the sensitivity of 3D migration.
Sorting subpopulations: Advantages in 3D migration of more malleable nuclei
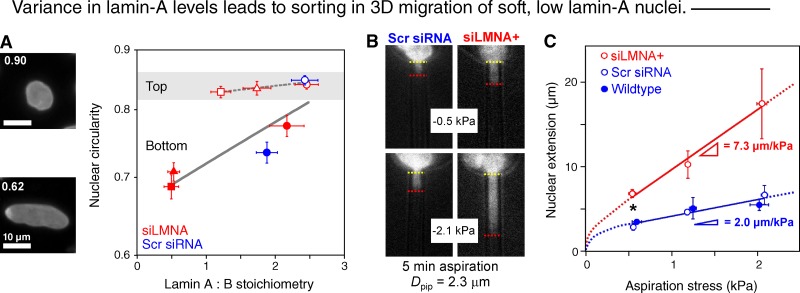
Lamin-A:B levels appear moderately variable even within wild-type cell populations, leading us to hypothesize that cell subpopulations with lower lamin-A:B levels could migrate faster through small pores. Indeed, for both wild-type A549 cells and several levels of siLMNA knockdown, nuclei on the bottoms of the 3-µm filters showed lower lamin-A:B levels in addition to appearing less circular than nuclei on top (Fig. 3 A). Similar changes were not observed with the 8-µm pores.
Figure 3.
Variations in lamin-A lead to cell sorting in 3D migration, consistent with lamin-A:B regulation of nuclear response time. (A) Nuclei on the bottom of 3-µm filters compared with the top in A549 cells show lower absolute ratio of lamin-A:B and lower nuclear circularity (n ≥ 50 cells in 3 experiments; ±SEM), consistent with segregation in 3D migration based on softness of the nucleus. Immunofluorescence intensities of lamin-A (shown) and lamin-B were rescaled to the absolute ratio of 2.3 (Fig. 1 Cii). Red circle, triangle, and square symbols correspond to high, medium, and low doses of siLMNA, respectively. Results are fit segmentally to exponentials: y = 0.81 exp(0.015 x) (top, R2 = 0.86) and y = 0.66 exp(0.083 x) (bottom, R2 = 0.78). (B) Nucleus of A549 pulled into a micropipette of similar diameter as the filters after disrupting the actin cytoskeleton. Fluorescent DNA images are at low and high aspiration pressures. (C) Plot shows nuclear extension at each pressure after 5 min (n = 4; ±SEM; *, P < 0.01). Controls are both scrambled siRNA-treated and nontreated cells fitted to single line.
Because nuclear size does not change after lamin-A knockdown, we hypothesized that the rigidity of the nucleus is rate-limiting to pore entry (Fig. 2 F). We used aspiration of individual nuclei into micropipettes of diameter similar to the Transwell pores in order to probe nuclear mechanics independent of cytoskeleton (Pajerowski et al., 2007). Lamin-A knockdown is already known to soften nuclei at short times and high stresses, but here we studied much longer time scales (Fig. 3, B and C) with aspiration rates similar to cell speeds in 3D migration (e.g., fibrosarcoma cells in a collagen lattice crawl at ∼0.75 µm/min; Wolf et al., 2003). We also used aspiration pressures similar to traction stresses exerted by migrating cells (Dembo and Wang, 1999). After 5 min of aspiration, nuclear membrane extension proves to be linear in aspiration pressure and almost fourfold steeper after partial knockdown of lamin-A compared with control cells. This is in remarkable agreement with the approximately fourfold enhancement in active migration through pores (Fig. 1 D), and also consistent with migration-based sorting of the low lamin-A subpopulation (Fig. 3 A).
In a broader examination of the lamin dependence of nuclear mechanics, nuclei in all three cell types were aspirated together with knockdown and overexpressing cells (Fig. S4). Once a constant aspiration pressure was applied, each nucleus extended to a given length (e.g., 5 µm) over a time, τ. For the 10-fold range in lamin-A:B ratio between wild-type cells, τ increased more than 100-fold, which effectively stiffens nuclei against rapid shape changes. Similarly strong effects for lamin-A knockdown and overexpression in A549 cells were likewise systematic. We have also recently shown that lamin-B1 knockdown in U251 cells increased the relaxation time τ for an intermediate level of nuclear extension from ∼10 s in wild-type nuclei to ∼300 s after ∼50% knockdown, consistent with lamin-B’s role as a “spring” (Shin et al., 2013). The range of τ across all nuclei is thus similar to timescale differences obtainable in aspirating water versus honey! The results are consistent with lamin-A conferring nuclear viscosity and impeding 3D migration even in the absence of perturbations either to migration speed in 2D culture (Fig. S1) or to other abundant cellular proteins (Fig. 1 F).
Lamin-A protects against 3D migration–induced apoptosis
In addition to observing sustained, plastic deformation of nuclei after migration through small pores, we noticed that the total number of cells on the 3-µm filters after 24 h was very low with deep lamin-A knockdown (siLMNA++) and generally in proportion to knockdown level (Fig. S3). No such changes were observed with 8-µm filters. For net migration through the 3-µm filters (Fig. 1 D), we confirmed the relative decrease for deep knockdown cells compared with ∼50% partial knockdown cells by using shLMNA to generate an A549 line with “partial-deep” knockdown to ∼30% of wild-type levels, which gave the expected intermediate migration result (Fig. 1 D). The various observations prompted the hypothesis that mechanical stresses of migration could compromise viability. Protective roles for lamin-A are reasonable because enhanced apoptosis and necrosis is reported both in vivo with laminopathies and in in vitro studies (Dahl et al., 2008).
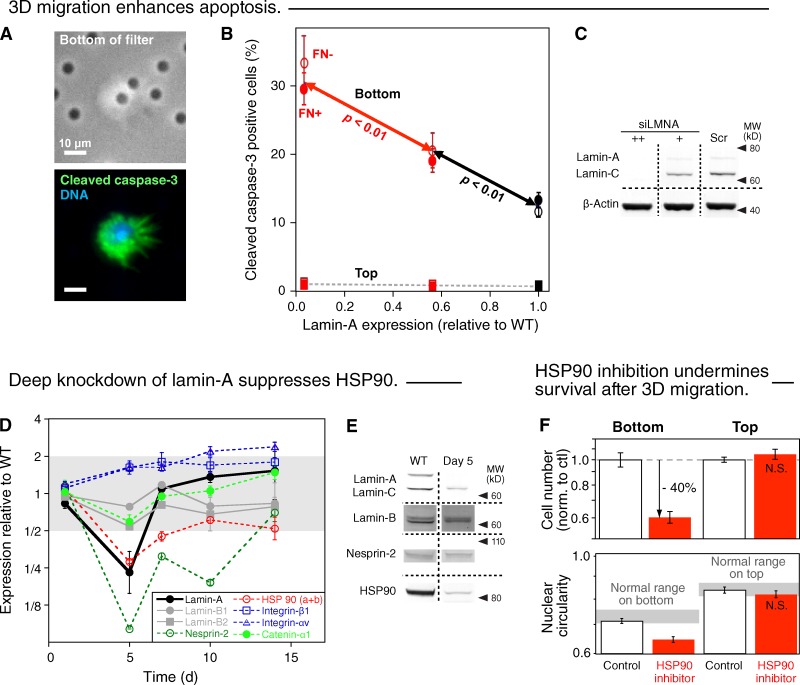
Cells on both sides of the filters were therefore immunostained for the apoptosis marker, cleaved caspase-3, which is downstream of stress pathways that include mechanical stress (Shive et al., 2002; Cheng et al., 2009) and that can be activated within a few hours of insult (e.g., staurosporine; Zhang et al., 2004). This is a minimum time scale for cells to migrate across the 22-µm thickness of the pores (Fig. S1). Imaging indeed showed apoptotic cells with their nuclei still in the pore (Fig. 4 A), which suggests the coincidence of apoptosis and 3D motility. With scrambled siRNA-treated cells, only ∼1% or fewer cells on top of the filter were apoptotic compared with ∼12% of cells that had migrated to the bottom of the filter (Fig. 4, B and C). Fibronectin had no statistically significant effect on apoptosis or net migration throughout these studies, which indicates that such adsorbed matrix or a lack of it is not a factor. The results thus reveal a significant wild-type level of 3D migration enhanced apoptosis.
Figure 4.
3D migration enhances apoptosis, and ablation of lamin-A compromises HSP90-dependent stress protection. (A) Images of A549 cell partially in a 3-µm pore (top) and staining positive for the apoptosis marker cleaved caspase-3 (green, bottom; blue, DNA Hoechst 33342). (B) Increased frequency of apoptotic A549 cells on the bottom of 3-µm filter after lamin-A knockdown. Cells on top show a negligible dependence of apoptosis on lamin-A. (n = 3; ±SEM). (C) Measurements were made by immunoblotting. (D) Changes in the expression levels of proteins of interest, detected by quantitative mass spectrometry, after transient lamin-A knockdown and recovery over 14 d (n = 3; ±SEM). Although up-regulation of the fibronectin receptor integrin α5:β1 in epithelial cells can favor mesenchymal-like motility (Friedl et al., 1997), correlations of levels with extents of cell behaviors remain controversial (Desgrosellier and Cheresh, 2010) and, more importantly, FN coating of filters had no effect in our studies (Fig. 1, D and E). Likewise, while decreases in α-catenin (by > 25%) also associate with an epithelial-mesenchymal transition (EMT; Yang et al., 2004), deep lamin-A knockdown suppresses net 3D migration (Fig. 1 D). (E) Changes in lamin-A, lamin-B, nesprin-2, and HSP90 were confirmed by immunoblot for wild-type and 5-d post-knockdown cells. (F) A549 cells pretreated for 24 h with HSP90 inhibitor (17-AAG, 90 nM in DMSO) or the same volume of DMSO (control) were plated in the same solutions on the top of 3-µm filters and allowed to migrate for 24 h. Cells were fixed, immunostained for lamin-A, imaged, and measured (n = 3; ±SEM); drug gave fewer cells on bottom and nuclei were less circular (P < 0.01; log scales).
Partial knockdown of lamin-A led to a significant increase in apoptosis on the bottom of the filter, which is consistent with the decrease of total cell number after migration by 10% or more (Fig. S3). Because partial reduction of lamin-A in A549 cells showed negligible perturbation to the proteome, the results support the hypothesis that the nuclear lamina protects physically against stress. In this case, “stress” is the mechanical stress generated by each cell during its migration. Of course, despite these stresses and an increase in percentage of apoptotic cells, partial knockdown also produced a fourfold increase in the net number of migrated cells, which indicates that the flexibility gains are considerably greater than the compromised stress response. Deep knockdown of lamin-A to <10% of wild-type A549 cells showed an average of 30% apoptotic cells that had migrated through the pores (Fig. 4 B). Decreased lamin-A had no significant effect on the minimal apoptosis of cells in standard 2D culture or on the top of the 3-µm filters, but apoptosis in 3D migration is consistent with the large decrease in total cell number after migration (Fig. S3) and with suppressed migration of these cells (Fig. 1 D).
Deep knockdown of lamin-A also changed the levels of many additional proteins quantifiable by MS (Fig. 4 D), which is unlike moderate knockdown (Fig. 1 F) and which could contribute to the dramatic decrease in migrated cell numbers (Fig. 1, C and D). Among several stress-response proteins that showed large and intriguing decreases with deep knockdown of lamin-A was HSP90, as validated by immunoblot (Fig. 4 E). HSP90 accounts for ∼2% of protein mass in many cell types. As a molecular chaperone, HSP90 interacts with >200 client proteins and contributes to repair of DNA damage (Stecklein et al., 2012), and although greatly understudied, mechanical stress can “induce” DNA damage (Mayr et al., 2002). A protective role for HSP90 in 3D migration stresses seemed a pharmacologically testable hypothesis. Wild-type A549 cells were treated for one day with the HSP90 inhibitor 17-AAG (Maloney et al., 2007) and were then allowed to migrate through 3-µm pores in the presence of drug. Inhibition of HSP90 consistently led to 40% fewer cells on the filter bottoms, consistent with measurements of fourfold greater apoptosis with drug even though there was no significant difference in cell numbers on the filter tops (Fig. 4 F). Of the cells that remained on the filter bottoms, nuclei were also more distended even though lamin-A levels appeared similar; greater distension is consistent with migration-coupled unfolding of structural proteins such as lamins that might need chaperones to refold in shape recovery. HSP90 is normally a highly abundant protein but can be down-regulated when lamin-A is repressed, with suppressed function of HSP90 being a major impediment to survival in 3D migration.
Lamin-A is low in the periphery of xenografts, and lamin-A moderation enhances growth
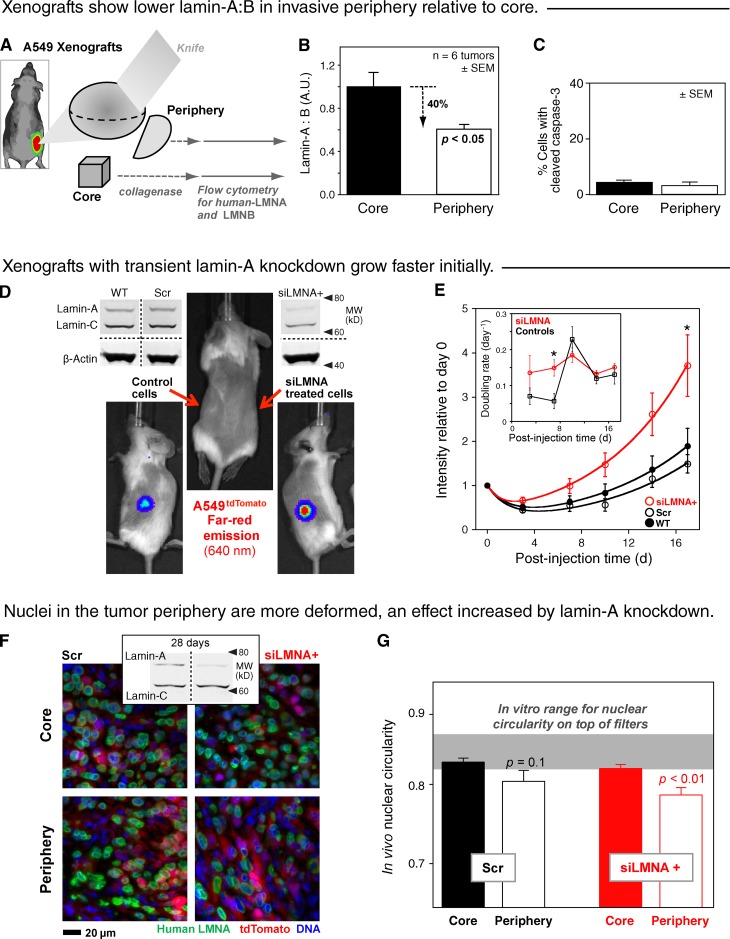
For insights into lamin effects in vivo, A549 cells were injected into the flanks of immunodeficient NSG mice, and after 4–8 wk, xenografts were sufficiently large to be harvested and separated into periphery and core sections for lamin analysis (Fig. 5 A). Collagenase treatment was followed by immunostaining of suspended A549 cells for human-specific lamin-A and lamin-B. Higher lamin-A:B in the tumor cores compared with the peripheries was consistently measured by flow cytometry (Fig. 5 B). Cleaved caspase-3 showed very few apoptotic cells compared with in vitro studies (Fig. 4 B and Fig. 5 C), but in vivo processes (e.g., phagocytosis by macrophages) could lead to rapid removal of apoptotic cells. The lamin-A:B results seem consistent with segregation and invasion of the periphery by cells with softer nuclei—similar to in vitro sorting across the Transwell filters (Fig. 3 A).
Figure 5.
Cells in the tumor periphery show low lamin-A:B in more deformed nuclei, consistent with enhanced tumor growth after moderate knockdown of lamin-A. (A) Immunostaining of lamins in A549 cells isolated from core and periphery tumor sections (of relative width: ∼1/4, ∼1/2, ∼1/4) from wild-type xenografts followed by flow cytometry. (B) A549 cells were distinguished from mouse cells based on positive staining with human specific lamin-A primary antibody. lamin-A:B intensity ratio showed a statistically significant difference between core and periphery (n = 6 mice; ±SEM). (C) Frequencies of cleaved caspase-3–positive tumor-derived A549 cells were obtained by imaging cells positively stained for human lamin-A (n = 3; ±SEM). (D) In situ imaging of A549 xenografts that were stably expressing tdTomato. Each mouse received siLMNA-treated cells in one side and control cells on the opposite side: wild-type (WT) or scrambled-RNA (Scr). Knockdown was confirmed by immunoblot. (E) Tumor propagation measured by tdTomato signal from tumors up to 17 d post-engraftment, with double exponential fits (n = 10 for siLMNA+, n = 5 for NT and Scr; ±SEM). Inset: growth rates as reciprocal doubling times calculated from consecutive time points (n = 10; ±SEM; P < 0.05). (F) Representative fluorescence images of cells in core and periphery regions of tumor tissue sections, stained for DNA (Hoechst 33342) and anti–(human lamin-A) with red fluorescence from tdTomato. Inset: immunoblots of tumors with anti–(human lamin-A) and constant total protein show lamin-A:C has recovered after moderate knockdown. (G) Circularity of A549 nuclei in control or lamin-A knockdown tumor slices (n ≥ 25 cells; ±SEM; log scale). Cell populations were again divided into core or periphery (n = 4 tumors).
Lamin-A:B differences in a tumor might reflect population-wide mechanisms that require time (e.g., adaptation), and so a controlled study was done in which we assessed the impact on in vivo tumor growth of an initial partial knockdown of lamin-A. In preparation for in vivo study, lung tumor-derived A549 cells were transduced with lentivirus for expression of the deep red fluorescent protein tdTomato (A549tdTomato), thus enabling noninvasive monitoring of tumors at wavelengths where tissue autofluorescence is low (Winnard et al., 2006). A549tdTomato cells were treated as in Fig. 1 D with siLMNA+ to achieve transient knockdown to one-third of wild-type lamin-A levels (without major changes to the proteome; Fig. 1 F), and these were then injected into the flanks of immunodeficient NSG mice (Fig. 5 D). Similar numbers of control cells were injected into the contralateral sites achieving a 10-fold range in day-0 xenograft intensity. Unlike in the membrane migration assay, it was important to consider the potential for cells to modify the surrounding matrix, thus altering pore dimensions within the tumor. However, we found no variation in matrix metalloproteinase (MMP) levels with changes in lamin-A in cell model systems (Fig. S5). Compared with wild-type and scrambled siRNA-treated control cells, tumors with partial LMNA knockdown cells grew more rapidly (Fig. 5 E). Importantly, the rates at which tumors doubled in size were significantly higher within the first week (up to threefold) for knockdown tumors compared with controls (Fig. 5 E, inset). Recall that the in vitro measurements of partially knocked down cells had shown, relative to wild type, approximately fourfold softer nuclei on minute timescales (Fig. 3 C) and an approximately fivefold faster 3D migration on a 1-d timescale (Fig. 1 D). In vivo growth rates of knockdown and wild-type A549 grafts become similar with time, consistent with recovery from the knockdown (Fig. 4 D). Indeed, the timescale for recovery of expression after knockdown in vitro (Fig. S2) was similar to that for the loss of difference in tumor growth rates (Fig. 5 E, inset), and immunoblotting of late tumors confirmed similarity of human lamin-A levels across tumors (Fig. 5 F, blots). Although a transient knockdown produced a transient bulk tumor response, the in vivo results provide a measure of the advantage in tumor propagation for more flexible nuclei.
Tumor sections immunostained for human lamin-A revealed a general colocalization of positively stained nuclei with tdTomato fluorescence and also showed a range of A549 nuclear shapes (Fig. 5 F, microscope images). Importantly, wherever we saw tdTomato, we saw lamin-A, which indicates that lamin-A expression is not lost completely or sustainably in vivo even though it can decrease. Indeed, in the periphery of these dense tissues, nuclei appeared more elongated than in the core within the same slice, consistent with in vitro observations of plastically elongated nuclei after migration through micropores (Fig. 2, A and B; and Fig. 3 A). Nuclear circularity was always statistically smaller at the periphery for knockdown nuclei relative to the core (P < 0.01); control nuclei showed an intermediate circularity compared with knockdowns, consistent perhaps with slower growth of controls (Fig. 5 G). A mean circularity of knockdown and control nuclei (∼0.82–0.83) in the core also compares well with the in vitro results for wild-type nuclei on the top of the micro-filter (0.83–0.84 in Fig. 3 A), and knockdown nuclei in the periphery showed a decreased circularity (ΔCirc ∼ −0.04) similar to that seen in vitro only for small pores (Fig. 2, C and D) and not large pores (Fig. 2 A and Fig. S3).
Discussion
Although lamin-A varies more widely than lamin-Bs in normal and cancerous solid tissue cells (Fig. 1 C), the migratory cells here with the lowest levels of lamin-A (U251 cells) show the greatest increase in 3D migration after lamin-A perturbation (Fig. 1 D, inset). A549s express an intermediate level of lamin-A:B, and moderate knockdown has an intermediate effect on their 3D migration. Importantly, 2D migration was unaffected for any cell type, and at least for the A549 cells migration was also unaffected through 8-µm pores that require much less distortion of the nucleus compared to 3-µm pores (i.e., by a factor of (8/3)2 = 7.1-fold less). One recent study of deep lamin-A knockdown (with shRNA) in prostate tumor cell lines (Kong et al., 2012) reported decreased cell growth (24–72 h), decreased cell migration in 2D (72 h), and decreased migration through 8-µm pore filters (24 h), and all of these processes also increased about twofold with lamin-A overexpression (to more than ∼50% wild type). The cells underwent selection for weeks or more and clearly changed phenotype in ways not seen here with transient knockdown (Fig. 1 E and Fig. S2). Past work had also demonstrated a dependence of 3D but not 2D migration on cytoskeletal contractility with inhibition of myosin-II compromising migration of U251 cells through 3-µm pores but not through 8-µm pores or on surfaces (Beadle et al., 2008), and very similar effects of myosin-II inhibition could also be demonstrated here with A549 cells independent of lamin-A level (Fig. S4). While cytoskeletal forces actively push and pull a nucleus through a constraining pore, scaling concepts from polymer physics largely explain the lamin-A:B dependence of biphasic motility that underlies tumor growth and migration-induced death.
Polymer physics of the nuclear lamina and distinct mechanical roles for lamin isoforms
Many cytoskeletal and ECM polymer assemblies have been mathematically modeled in physicochemical terms over the last decade with successful predictions of nonlinear responses (Gardel et al., 2004; Storm et al., 2005). Similar models might also apply here for the juxtaposed assemblies of lamin-A and lamin-B to predict shape changes of nuclei and response times (Figs. 2 and 3). Because polymer theories focus on averages such as mean concentration and mean molecular weight despite the many molecular states in polymer systems (biological or synthetic), of secondary importance here are post-transcriptional heterogeneities of lamins that range from lamin-A spliceoform ratios to nucleoplasmic partitioning. Moreover, an object’s properties are dictated foremost by the material rather than its arrangement: consider the bouncing behavior of a ping-pong ball with a steel shell versus a solid rubber ball. Previous findings that lamin-A is mobile or “fluid” whereas lamin-B is immobile (Shimi et al., 2008) are thus critical and qualitatively consistent with the physical plasticity evident here in 3D migration (Fig. 2). However, lamin-A’s strongly nonlinear contribution to viscosity in nuclear mechanics relative to lamin-B’s solid-like elasticity are predictable from the physics of large interacting polymers (Swift et al., 2013b) and also key to understanding 3D migration.
For the physical plasticity model of Fig. 2, standard analyses of elongation yield a response time τ = (viscosity/elasticity) (Barnes, 2000). Our recent studies of nuclei aspirated in micropipettes yield steeply positive power laws τ ∼ [lamin-A:B]2.5 that therefore reveal the dominating contribution of lamin-A to nuclear viscosity relative to lamin-B’s contribution to nuclear elasticity (Swift et al., 2013b). Viscosity exponents of ∼3 are typical of large interacting polymers (Swift et al., 2013b) and explain why the very high lamin-A:B ratio in MSCs produces the most elongated, extruded nuclei after hours-long migration through small pores (Fig. 2 B); it also explains why lamin-B knockdown stiffens the nucleus (Fig. S4). The evidence here that lamin-B is key to nuclear elasticity contrasts with past conclusions from observing cells in 2D culture that lamin-B does not regulate nuclear mechanics (Lammerding et al., 2006). Lamina physics can now be used to predict the key observations here that moderate changes in lamin-A have the greatest effect on cells with the lowest levels of lamin-A. The nucleus appears rate-limiting in 3D migration, and given the stochastic nature of motility, an activated model (Fig. 2 F) would have the rate k of filter crossing impeded by the work ΔG required for the cell to deform its own nucleus,
with ko and β assumed constant across cell types. Because lamin-B is almost constant, higher lamin-A:B gives an effectively stiffer nucleus that requires more work (ΔG). A linear dependence,
is close to the geometric mean for the 1/2-scaling of the effective stiffness E with A:B (Fig. S4) and the 2.5-scaling of apparent viscosity with A:B (Swift et al., 2013b). The “3D migration sensitivity to lamin-A” m3D-sens (Fig. 1 D, inset) is simply the derivative of the rate:
The exponential result fits Fig. 2 E. Based on this fit and also the exponential relation between m3D-sens and the change in circularity of the nucleus (ΔCirc; Fig. 2 D), ΔCirc should also depend nonlinearly on lamin-A:B as found in Fig. 2 G.
An optimal lamina: Low lamins limit survival under stress but high lamins anchor cells in place
Stiffer materials typically withstand more stress before they break: they are stronger. Because wild-type cells generate sufficient stress in migration through small pores to apoptose and die (∼12% of cells per Fig. 4 B; Fig. S3), softening of nuclei by approximately three- to fourfold even with ∼50% knockdown of lamin-A (Fig. 2, B and C; and Fig. S4) is readily predicted to increase the nuclear stress in proportion so that increases in apoptotic cells and cell loss by approximately two- to threefold are understandable. Although modest knockdown of lamin-A perturbs the proteome to a minimal extent (Fig. 1 F), the relevant molecular stresses are hinted at by the major proteomic changes caused by deep knockdown (Fig. 4 D). While lamin-Bs change minimally, as expected for independent regulation, detected integrins increase about twofold, suggesting that adhesion is more than adequate. However, the protein that is most obviously indicative of compromised stress resistance is HSP90, which was suppressed almost as much as lamin-A (fourfold or more). Lamin-A regulates multiple transcription factors (Ho et al., 2013; Swift et al., 2013b), and we hypothesize that these can influence the HSP pathway (Fig. S5). Importantly, HSPs stabilize and/or refold proteins under stress conditions, which can include mechanical unfolding of proteins within contractile cells (Johnson et al., 2007). In mechanically stressed nuclei, even lamin-A’s immunoglobulin domain unfolds as measured by stress-enhanced labeling of sequestered cysteines (Swift et al., 2013b). HSP90 also has roles in chromatin repair (Dote et al., 2006), and a failure to repair DNA damage in 3D migration could promote cell death. Here, an HSP90 inhibitor 17-AAG indeed suppressed cell numbers that migrated across the filter (Fig. 4 F), and so the anti-cancer efficacy of this drug (Goetz et al., 2005) might reflect or even require stresses in 3D migration.
Poor survival with low lamins and susceptibility to stress is likely to dominate the advantage in 3D migration of a low lamin-A nucleus, which is easier to push or pull through a narrow constraint. PMNs have very low lamins (Olins et al., 2001) and can invade any tissue during an infection, but these cells apoptose in days (Pillay et al., 2010). In the opposite extreme, the nucleus can act as an anchor that physically immobilizes a cell if the nucleus is simply too stiff to be pushed or pulled through a small opening by a given cell type (with its characteristic cytoskeleton), as seen with lamin-A overexpression in A549 cells here. Our recent studies of various human hematopoietic cells indeed show that the cell types with the highest lamin levels predominate in marrow, seeming less capable of migrating through endothelial micropores that separate marrow from blood; the marrow “niche” is thus defined in part by differential nuclear mechanics (Shin et al., 2013). Nonetheless, large decreases in net migration from moderate to deep knockdown of lamin-A could reflect additional defects here, especially in cytoskeleton structures and/or their folding (if not levels) because inhibition of myosin-II also impedes 3D migration regardless of lamin-A knockdown (Fig. S4). Nesprin-2 is substantially lower with deep lamin-A knockdown, and although direct knockdown of this linker between the cytoskeleton and nucleus impedes 2D migration (Luxton et al., 2010), any links to apoptosis are as yet unreported.
Malignancies associated with increased levels of lamin-A (Tilli et al., 2003; Foster et al., 2010) could therefore reflect the pro-survival function of lamin-A in stressful migration of these cells in 3D. Xenograft studies here indeed show that lamin-A levels vary within a tumor, and lamin-A levels can also impact tumor growth. A549 cells have more lamin-A than lamin-B, but there is also sufficient variation within any cell population to sort in 3D migration. In vivo studies here demonstrated lamin-A levels are lower in the growing periphery of an established wild-type tumor than in the core, correlating with nuclear shape in vivo. Although the timescale for low lamin-A cells to dominate at the periphery is unknown, if that timescale is a few days or longer, then tumors generated with transient lamin-A knockdown cells will initially grow faster than the wild type before the latter catch up—consistent with the observed dynamics. Such findings seem especially pertinent to low lamin-A levels that correlate with colon cancer recurrence (Belt et al., 2011), although increases as well as decreases of lamin-A in tumorigenesis and other processes are likely rooted in survival limits that underlie the biphasic motility of cells.
Conclusion
Nuclear lamins are of great interest in aging, development, and differentiation, in addition to cancer. Specific roles of lamin isoforms seem controversial, with recent lamin-B knockouts appearing surprisingly viable until birth when apoptosis in migration through the brain is enhanced (Kim et al., 2011). Because lamin-A is low in the brain and does not compensate, the fundamental insight here into the polymeric basis for lamina stabilization of the nucleus in 3D migration offers an explanation for the phenotype. Further understanding of the roles for both lamin isoforms in cell migration and viability could be useful in fine-tuning the behaviors of cells such as stem cells in vivo, thereby contributing to therapy as well as diagnosis.
Materials and methods
Cells and mice
Human lung carcinoma A549 and mesenchymal stem cells were purchased from ATCC and Lonza, respectively. Glioblastoma multiforme U251 cells were a gift from J. Dorsey (University of Pennsylvania, Philadelphia, PA). Cells were cultured in accordance with the supplier’s instructions. Lentiviral particles encapsulating tdTomato-encoding plasmid were a gift from C. Carpenito (University of Pennsylvania, Philadelphia, PA). 105 A549 cells were incubated with lentivirus with MOI of ∼7.5 for 24 h. Cells were expanded after selection of tdTomato-positive cells, using a cloning cylinder (Bel-Art Products). Non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice with null expression of interleukin-2 receptor gamma chain (NSG mice) were maintained by the Stem Cell and Xenograft Core Facility at the University of Pennsylvania, using breeders obtained from The Jackson Laboratory. All animal experiments were planned and performed according to IACUC protocols.
Lamin-A knockdown and characterization
All siRNAs used in this study were from Thermo Fisher Scientific. A549 cells were passaged 24 h before transfection. A complex of siRNA (30 nM; siLMNA 1, 5′-GGUGGUGACGAUCUGGGCU-3′; and siLMNA 2, 5′-AACUGGACUUCCAGAAGAACAUC-3′ [Elbashir et al., 2001]) and 1 µg/ml Lipofectamine 2000 was prepared per manufacturer’s instructions and incubated for 24 h per dose for up to three doses (in high glucose DMEM with 10% FBS for A549 and U251 and in low glucose DMEM with 10% FBS for MSC). A single 30 nM dose is denoted in this study as “siLMNA+,” and three daily doses of 30 nM to achieve deep knockdown as “siLMNA++.” Results were compared with those using the same dose of scrambled siRNA. For wild-type controls, an equal volume of opti-MEM (Invitrogen) was used. For shRNA treatment, A549 cells were infected with lentiviral supernatants targeting lamin-A:C (TRCN000061833; Sigma-Aldrich) at a multiplicity of infection (MOI) of 10 in the presence of 80 µg/ml polybrene (Sigma-Aldrich), and cultured for 24 h. The cells were then selected by 2 µg/ml puromycin (Sigma-Aldrich) for 30 d. Knockdown efficiency was determined by Western blot following standard methods. Antibodies against lamin-A (mouse, sc-7292) and β-actin (mouse, sc-47778) were from Santa Cruz Biotechnology, Inc. The effect of knockdown on proliferation was measured with thiazolyl blue tetrazolium (Sigma-Aldrich), following the standard assay protocol. The effect of lamin level on MMPs was determined by zymogram assay, conducted according to the manufacturer’s protocol (Novex, Life Technologies).
Lamin-A overexpression and characterization
After transduction with lentivirus constructs for GFP-lamin-A, positive cells were seeded on 6-well plates at very low density of less than one cell per field observed with a 4×/0.13 NA objective lens. Colonies of GFP-lamin-A–expressing cells were isolated using a cloning cylinder (Bel-Art Products). Trypsinized cells from isolated colonies were further expanded for experiments. Relative standard deviation values of cells used for Transwell migration assay were 55% and 66% for “high” and “low” GFP populations, respectively, per immunofluorescence.
2D and 3D migration assays
2D (wound-healing assay).
105 A549 cells were seeded on 24-well plates. After 24 h, cells were treated with 1 dose of 30 nM siRNA (lamin knockdown or scrambled) as described above. After 5 d, a scratch was made in the monolayer of cells and the width of the scratch measured in three places at 0, 3, 6, and 9 h. Lamin-A expression in migrating cells was measured by immunostaining cells at the wound frontiers.
3D (Transwell migration assay).
Multi-well inserts with 3-µm or 8-µm pore filters were purchased from BD. Fibronectin coating was achieved by incubation with 50 µg/ml human fibronectin solution (BD) for 2 h at 37°C (Meng et al., 2009). 105 cells were seeded in serum-free medium in the top well and DMEM supplemented with 20% FBS was added to the bottom well to establish a nutrient gradient across the filter. For drug treatment during migration assay, 2 µM of (−)-blebbistatin or 60–90 nM of 17-AAG (LC Laboratories) was added to both top and bottom wells. In the case of 17-AAG, cells were pretreated with the drug for 24 h before starting the assay. After incubation at 37°C with 5% CO2 (24 h for A549 and MSC, and 6 h for U251 cells), cells were fixed with 3.7% formaldehyde (Thermo Fisher Scientific). Cells were stained for either DNA with Hoechst 33342, lamin-A (mouse, sc-7292), or lamin-B (goat, sc-6217) from Santa Cruz Biotechnology, Inc.; or cleaved caspase-3 (rabbit, AB3623; EMD Millipore), with either Alexa Fluor 488– or Alexa Fluor 546–conjugated secondary antibodies (Invitrogen). After staining, filters were cut out from the insert and mounted on coverslips using Fluoro-Gel (Electron Microscopy Sciences). 20×/0.4 NA and 40×/0.6 NA objectives were used for nucleus counting and caspase-3 imaging, respectively. For confocal microscopy (model TCS SP8; Leica), unless otherwise stated, a 63×/1.4 NA objective was used.
Micropipette aspiration
Just before aspiration experiments, cells were treated as we have done previously (Lammerding et al., 2006; Pajerowski et al., 2007) with 0.2 µg/ml of latrunculin-A (Sigma-Aldrich) for 1 h at 37°C, detached with trypsin/EDTA, centrifuged, and resuspended in aspiration buffer (135 mM NaCl, 5 mM KCl, 5 mM Hepes, 1.8 mM CaCl2, 2 mM MgCl2, 2% BSA, and 1:3,000 propidium iodide [Molecular Probes]). Cell nuclei were stained with Hoechst 33342. Nuclear compliance was measured as membrane extension under negative pressure inside a micropipette needle. Epifluorescence imaging was done with an inverted microscope (model TE300; Nikon) coupled with a digital CCD camera (Roper Scientific), using a 60×/1.25 NA oil immersion objective at RT. Harvested A549 tumor tissues, pretreated with collagenase (Sigma-Aldrich) or blebbistatin (EMD Millipore) if necessary, were soaked in Ham’s F12 media (Invitrogen) during aspiration, visualized with a 20×/0.4 NA objective.
Quantitative proteomic profiling
Protein content was quantified by BCA Protein Assay (Thermo Fisher Scientific). Samples were prepared for gel electrophoresis by addition of 25% NuPAGE LDS sample buffer (4x; Invitrogen) and 1% β-mercaptoethanol, followed by heating to 80°C for 10 min. SDS-PAGE gels (NuPAGE 4–12% Bis-Tris; Invitrogen) were loaded with constant total protein. Gel electrophoresis was run for 10 min at 100 V and 25 min at 160 V. Gel sections were excised with the MW range 60–80 kD, washed (50% 0.2 M ammonium bicarbonate [AB] solution and 50% acetonitrile [ACN] for 30 min at 37°C), dried by lyophilization, incubated with a reducing agent (20 mM tris(2-carboxyethyl)phosphine [TCEP] in 25 mM AB solution at pH 8.0 for 15 min at 37°C), and alkylated (40 mM iodoacetamide [IAM] in 25 mM AB solution at pH 8.0 for 30 min at 37°C). The gel sections were dried by lyophilization before in-gel trypsinization (20 µg/ml sequencing grade modified trypsin in buffer as described in the manufacturer’s protocol [Promega] for 18 h at 37°C with gentle shaking). The resulting solutions of tryptic peptides were acidified by addition of 50% digest dilution buffer (60 mM AM solution with 3% formic acid).
Peptide separations (5 µl injection volume) were performed on a 15-cm PicoFrit column (75 µm inner diameter; New Objective) packed with Magic 5 µm C18 reversed-phase resin (Michrom Bioresources) using a nanoflow high-pressure liquid chromatography system (Eksigent Technologies), which was coupled online to a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ion source. Chromatography was performed with Solvent A (Milli-Q water with 0.1% formic acid) and Solvent B (acetonitrile with 0.1% formic acid). Peptides were eluted at 200 nL/min for 3–28% B over 42 min, 28–50% B over 26 min, 50–80% B over 5 min, 80% B for 4.5 min before returning to 3% B over 0.5 min. To minimize sample carryover, a fast blank gradient was run between each sample. The LTQ-Orbitrap XL was operated in the data-dependent mode to automatically switch between full-scan MS (m/z = 350–2,000 in the orbitrap analyzer [with resolution of 60,000 at m/z 400]) and the fragmentation of the six most intense ions by collision-induced dissociation in the ion trap mass analyzer.
Raw mass spectroscopy data were processed using Elucidator (version 3.3; Rosetta Biosoftware). The software was set up to align peaks in data from samples derived from corresponding molecular weight regions of the 1D gels. Peptide and protein annotations were made using SEQUEST (version 28; Thermo Fisher Scientific) with full tryptic digestion and up to two missed cleavage sites. Peptide masses were selected between 800 and 4,500 amu with peptide mass tolerance of 1.1 amu and fragment ion mass tolerance of 1.0 amu. The peptide database was modified to search for alkylated cysteine residues (monoisotopic mass change, Δ = +57.021 D) and oxidized methionine (Δ = +15.995 D). Further analysis was performed with Mathematica (Wolfram Research; Swift et al., 2013a,b).
Establishment of A549 tumors in mice
tdTomato-expressing A549 cells were detached with trypsin, washed with PBS, and suspended in PBS containing 25% Matrigel (BD) at a concentration of 2 × 107 cells/ml. Each mouse received two injections of one million cells at two flank sites, siLMNA-treated cells on one side and control cells on the other. Tumor progression was evaluated by tdTomato fluorescence emission from tumors using an IVIS Spectrum Imaging System (Caliper Life Sciences), imaged with 535/640-nm excitation/emission. Image analysis was done with Living Image Software (Caliper Life Sciences). Tumors were grown for 28 d before harvesting. Tumor slices (10 µm thick) were fixed with 3.7% formaldehyde, permeabilized with 0.25% Triton X-100, and blocked with BSA. Tissue was stained for human lamin-A and nuclei were counterstained with Hoechst 33342, and then observed with a 40×/0.6 NA objective. Alternatively, for pore size visualization, the tissue was stained for F-actin and collagen-1 (phalloidin and mouse, C2456, respectively, from Sigma-Aldrich), and then imaged by confocal microscopy (63×/1.4 NA). The cross-section area of pores that were occupied by the cytoplasm (F-actin) were measured and used to derive the pore diameter. Tissue remaining after cryo-sectioning was lysed in RIPA buffer and prepared for MS and Western blotting following standard protocol found elsewhere. For tumor tissue decellularization, tissue was first soaked in 1% SDS (Bio-Rad Laboratories) solution and incubated at RT overnight with one solution exchange. Then tissue was washed with 1% Triton X-100 solution for 30 min at RT, followed by equilibration with PBS.
Intracellular staining of lamins in tumor-derived A549 cells
A549 tumors were separated into core and peripheral portions, and further cut into smaller pieces of approximately a few mm3. Tumor tissue pieces were incubated in F12 growth medium containing 0.2 mg/ml collagenase (Sigma-Aldrich) at 37°C with 5% CO2 for 0.5–1 h. Isolated cells were collected and fixed/permeabilized using flow cytometry fixation/permeabilization buffer (R&D Systems), following the manufacturer’s instructions. For cultured A549 cells, fixation and permeabilization was done using 3.7% formaldehyde and 0.25% Triton X-100 (MP Biomedicals) in PBS, respectively. Cells were stained for lamin-A (mouse, sc-7292) and B (goat, sc-6217) from Santa Cruz Biotechnology, Inc.) with Alexa Fluor 488– and Alexa Fluor 647–conjugated secondary antibodies (all samples were treated with antibodies of fixed concentration ratio), respectively, and intensities were measured with a FACSCalibur system (BD). Apoptotic cells were detected based on positive signal for both lamin-A (mouse, sc-7292 [Santa Cruz Biotechnology, Inc.] with Alexa Fluor 488 secondary antibody) and cleaved capsase-3 (rabbit, AB3623 [EMD Millipore] with Alexa Fluor 647) by microscopic observation with a 20×/0.4 NA objective lens.
Fixation, immunostaining, and microscopy
Cells were fixed with 3.7% formaldehyde (Sigma-Aldrich) in PBS for 10 min at RT followed by PBS washing 2× for 5 min. Blocking and antibody staining was performed in 1% BSA in PBS. Antibodies were used at ∼1:100, and all primary antibodies were incubated at RT for 1 h or overnight at 4°C. All donkey secondary antibodies (Alexa Fluor dyes 488, 564, and 647) were stained for 1 h at RT at 1:300 dilution in PBS in 1% BSA. Hoechst 33342 (Invitrogen) was used to stain DNA at a concentration of 1 µg/ml for 10 min at RT. Unless otherwise stated, epifluorescence imaging was performed using an inverted microscope (model IX-71; Olympus) with a CCD camera (512B, Cascade; Photometrics), and image acquisition was performed with Image-Pro Plus software (Media Cybernetics). For quantification purposes, a 40×/0.6 NA objective was used and image analysis was performed using ImageJ (National Institutes of Health) according to JCB guidelines. For quantitation, samples that were to be directly compared were imaged at the same sitting, and the same gain and exposure time were used.
Online supplemental material
Fig. S1 shows that moderate changes in lamin-A do not affect 2D migration, but 2D migration always correlates positively with 3D migration. Fig. S2 shows that isoform ratios can be calibrated by mass spectrometry for comparisons between cell types, and that single cell measurements of nuclear properties indicate no change in nuclear volume with knockdown. Fig. S3 shows that siLMNAs with distinct targets consistently increase 3D migration without affecting viability in 2D or else viability and nuclear shape after large pore migration, but also that cell numbers are reduced with small pores and with knockdown. Fig. S4 used micropipette aspiration to reveal that lamin-A prolongs relaxation times and effectively stiffens nuclei, while myosin-II drives migration through small pores. Fig. S5 shows that lamin-A does not influence MMP levels, and also because deep knockdown of lamin-A suppresses HSP90, lamin-A regulation of transcription factors could also affect the HSP90 pathway. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308029/DC1.
Supplementary Material
Acknowledgments
We thank many colleagues in the field for help with this manuscript. We thank the Proteomics Facility at The Wistar Institute (Philadelphia, PA) for analysis.
This work was supported by the National Institutes of Health (P01DK032094; R01HL062352; R01EB007049), the National Science Foundation (1200834), and the Human Frontier Science Program (D.E. Discher).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- MMP
- matrix metalloproteinase
- MS
- mass spectrometry
- MSC
- mesenchymal stem cell
- PMN
- polymorphonuclear leukocyte
- siLMNA
- short interfering LMNA
References
- Barnes H.A. 2000. Handbook of Elementary Rheology. University of Wales, Institute of Non-Newtonian Fluid Mechanics, Cardiff, UK: 204 pp [Google Scholar]
- Beadle C., Assanah M.C., Monzo P., Vallee R., Rosenfeld S.S., Canoll P. 2008. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell. 19:3357–3368 10.1091/mbc.E08-03-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belt E.J., Fijneman R.J., van den Berg E.G., Bril H., Delis-van Diemen P.M., Tijssen M., van Essen H.F., de Lange-de Klerk E.S., Beliën J.A., Stockmann H.B., et al. 2011. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur. J. Cancer. 47:1837–1845 10.1016/j.ejca.2011.04.025 [DOI] [PubMed] [Google Scholar]
- Chamberlain J.K., Lichtman M.A. 1978. Marrow cell egress: specificity of site if penetration into sinus. Blood. 52:959–968 [PubMed] [Google Scholar]
- Cheng G., Tse J., Jain R.K., Munn L.L. 2009. Micro-environmental mechanical stress controls tumor spheroid size and morphology by suppressing proliferation and inducing apoptosis in cancer cells. PLoS ONE. 4:e4632 10.1371/journal.pone.0004632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C., Jung H.J., Nobumori C., Chang S., Tu Y.P., Barnes R.H., II, Yoshinaga Y., de Jong P.J., Vergnes L., Reue K., et al. 2011. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell. 22:4683–4693 10.1091/mbc.E11-06-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K.N., Engler A.J., Pajerowski J.D., Discher D.E. 2005. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 89:2855–2864 10.1529/biophysj.105.062554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K.N., Ribeiro A.J.S., Lammerding J. 2008. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102:1307–1318 10.1161/CIRCRESAHA.108.173989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D.K., Solimando L., Goldman R.D. 2008. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22:832–853 10.1101/gad.1652708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M., Wang Y.L. 1999. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76:2307–2316 10.1016/S0006-3495(99)77386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier J.S., Cheresh D.A. 2010. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 10:9–22 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer T.A., Misteli T. 2011. The lamin protein family. Genome Biol. 12:222 10.1186/gb-2011-12-5-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dote H., Burgan W.E., Camphausen K., Tofilon P.J. 2006. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer Res. 66:9211–9220 10.1158/0008-5472.CAN-06-2181 [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- Foster C.R., Przyborski S.A., Wilson R.G., Hutchison C.J. 2010. Lamins as cancer biomarkers. Biochem. Soc. Trans. 38:297–300 10.1042/BST0380297 [DOI] [PubMed] [Google Scholar]
- Friedl P., Maaser K., Klein C.E., Niggemann B., Krohne G., Zänker K.S. 1997. Migration of highly aggressive MV3 melanoma cells in 3-dimensional collagen lattices results in local matrix reorganization and shedding of alpha2 and beta1 integrins and CD44. Cancer Res. 57:2061–2070 [PubMed] [Google Scholar]
- Gardel M.L., Shin J.H., MacKintosh F.C., Mahadevan L., Matsudaira P., Weitz D.A. 2004. Elastic behavior of cross-linked and bundled actin networks. Science. 304:1301–1305 10.1126/science.1095087 [DOI] [PubMed] [Google Scholar]
- Goetz M.P., Toft D., Reid J., Ames M., Stensgard B., Safgren S., Adjei A.A., Sloan J., Atherton P., Vasile V., et al. 2005. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J. Clin. Oncol. 23:1078–1087 10.1200/JCO.2005.09.119 [DOI] [PubMed] [Google Scholar]
- Ho C.Y., Jaalouk D.E., Vartiainen M.K., Lammerding J. 2013. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 497:507–511 10.1038/nature12105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska J.M., Wilson K.L., Mansharamani M. 2002. The nuclear envelope, lamins and nuclear assembly. Curr. Opin. Cell Biol. 14:357–364 10.1016/S0955-0674(02)00329-0 [DOI] [PubMed] [Google Scholar]
- Houghton J., Stoicov C., Nomura S., Rogers A.B., Carlson J., Li H.C., Cai X., Fox J.G., Goldenring J.R., Wang T.C. 2004. Gastric cancer originating from bone marrow-derived cells. Science. 306:1568–1571 10.1126/science.1099513 [DOI] [PubMed] [Google Scholar]
- Hutchison C.J., Worman H.J. 2004. A-type lamins: guardians of the soma? Nat. Cell Biol. 6:1062–1067 10.1038/ncb1104-1062 [DOI] [PubMed] [Google Scholar]
- Johnson C.P., Tang H.Y., Carag C., Speicher D.W., Discher D.E. 2007. Forced unfolding of proteins within cells. Science. 317:663–666 10.1126/science.1139857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.J., Coffinier C., Choe Y., Beigneux A.P., Davies B.S.J., Yang S.H., Barnes R.H., II, Hong J., Sun T., Pleasure S.J., et al. 2012. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. USA. 109:E423–E431 10.1073/pnas.1111780109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann S.H., Mabry M., Jasti R., Shaper J.H. 1991. Differential expression of nuclear envelope lamins A and C in human lung cancer cell lines. Cancer Res. 51:581–586 [PubMed] [Google Scholar]
- Kim Y., Sharov A.A., McDole K., Cheng M., Hao H., Fan C.M., Gaiano N., Ko M.S.H., Zheng Y. 2011. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 334:1706–1710 10.1126/science.1211222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Schäfer G., Bu H.J., Zhang Y., Zhang Y.X., Klocker H. 2012. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 33:751–759 10.1093/carcin/bgs022 [DOI] [PubMed] [Google Scholar]
- Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G., Lee R.T. 2006. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281:25768–25780 10.1074/jbc.M513511200 [DOI] [PubMed] [Google Scholar]
- Lloyd D.J., Trembath R.C., Shackleton S. 2002. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum. Mol. Genet. 11:769–777 10.1093/hmg/11.7.769 [DOI] [PubMed] [Google Scholar]
- Luxton G.W.G., Gomes E.R., Folker E.S., Vintinner E., Gundersen G.G. 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 329:956–959 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney A., Clarke P.A., Naaby-Hansen S., Stein R., Koopman J.O., Akpan A., Yang A., Zvelebil M., Cramer R., Stimson L., et al. 2007. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 67:3239–3253 10.1158/0008-5472.CAN-06-2968 [DOI] [PubMed] [Google Scholar]
- Mayr M., Hu Y.H., Hainaut H., Xu Q.B. 2002. Mechanical stress-induced DNA damage and rac-p38MAPK signal pathways mediate p53-dependent apoptosis in vascular smooth muscle cells. FASEB J. 16:1423–1425 [DOI] [PubMed] [Google Scholar]
- Meng X.N., Jin Y., Yu Y., Bai J., Liu G.Y., Zhu J., Zhao Y.Z., Wang Z., Chen F., Lee K.Y., Fu S.B. 2009. Characterisation of fibronectin-mediated FAK signalling pathways in lung cancer cell migration and invasion. Br. J. Cancer. 101:327–334 10.1038/sj.bjc.6605154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke C.T., Frey B., Fellner M., Herrmann M., Fabry B. 2011. Integrin α5β1 facilitates cancer cell invasion through enhanced contractile forces. J. Cell Sci. 124:369–383 10.1242/jcs.071985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir R.D., Yoon M., Khuon S., Goldman R.D. 2000. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151:1155–1168 10.1083/jcb.151.6.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamizo A., Marini F., Amano T., Khan A., Studeny M., Gumin J., Chen J., Hentschel S., Vecil G., Dembinski J., et al. 2005. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 65:3307–3318 [DOI] [PubMed] [Google Scholar]
- Olins A.L., Herrmann H., Lichter P., Kratzmeier M., Doenecke D., Olins D.E. 2001. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp. Cell Res. 268:115–127 10.1006/excr.2001.5269 [DOI] [PubMed] [Google Scholar]
- Pajerowski J.D., Dahl K.N., Zhong F.L., Sammak P.J., Discher D.E. 2007. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 104:15619–15624 10.1073/pnas.0702576104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay J., den Braber I., Vrisekoop N., Kwast L.M., de Boer R.J., Borghans J.A.M., Tesselaar K., Koenderman L. 2010. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 116:625–627 10.1182/blood-2010-01-259028 [DOI] [PubMed] [Google Scholar]
- Pittenger M.F., Martin B.J. 2004. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ. Res. 95:9–20 10.1161/01.RES.0000135902.99383.6f [DOI] [PubMed] [Google Scholar]
- Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R.H., Heitner S.G., et al. 2013. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41(D1, Database issue):D56–D63 10.1093/nar/gks1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat A.C., Jaalouk D.E., Zwerger M., Ung W.L., Eydelnant I.A., Olins D.E., Olins A.L., Herrmann H., Weitz D.A., Lammerding J. 2013. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 288:8610–8618 10.1074/jbc.M112.441535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Pfleghaar K., Kojima S., Pack C.G., Solovei I., Goldman A.E., Adam S.A., Shumaker D.K., Kinjo M., Cremer T., Goldman R.D. 2008. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22:3409–3421 10.1101/gad.1735208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.-W., Spinler K.R., Swift J., Chasis J.A., Mohandas N., Discher D.E. 2013. Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc. Natl. Acad. Sci. USA. 110:18892–18897 10.1073/pnas.1304996110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shive M.S., Brodbeck W.G., Anderson J.M. 2002. Activation of caspase 3 during shear stress-induced neutrophil apoptosis on biomaterials. J. Biomed. Mater. Res. 62:163–168 10.1002/jbm.10225 [DOI] [PubMed] [Google Scholar]
- Stecklein S.R., Kumaraswamy E., Behbod F., Wang W.J., Chaguturu V., Harlan-Williams L.M., Jensen R.A. 2012. BRCA1 and HSP90 cooperate in homologous and non-homologous DNA double-strand-break repair and G2/M checkpoint activation. Proc. Natl. Acad. Sci. USA. 109:13650–13655 10.1073/pnas.1203326109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm C., Pastore J.J., MacKintosh F.C., Lubensky T.C., Janmey P.A. 2005. Nonlinear elasticity in biological gels. Nature. 435:191–194 10.1038/nature03521 [DOI] [PubMed] [Google Scholar]
- Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147:913–920 10.1083/jcb.147.5.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Harada T., Buxboim A., Shin J.-W., Tang H.-Y., Speicher D.W., Discher D.E. 2013a. Label-free mass spectrometry exploits dozens of detected peptides to quantify lamins in wildtype and knockdown cells. Nucleus. 4:7–6 10.4161/nucl.27413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C.D.P., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.-W., Tewari M., et al. 2013b. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341:1240104 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilli C.M., Ramaekers F.C.S., Broers J.L.V., Hutchison C.J., Neumann H.A.M. 2003. Lamin expression in normal human skin, actinic keratosis, squamous cell carcinoma and basal cell carcinoma. Br. J. Dermatol. 148:102–109 10.1046/j.1365-2133.2003.05026.x [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Chen Y., Kriegstein A.R., Vallee R.B. 2005. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 170:935–945 10.1083/jcb.200505166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R.A. 2006. The biology of cancer. First edition Garland Science, New York: 850 pp [Google Scholar]
- Willis N.D., Wilson R.G., Hutchison C.J. 2008. Lamin A: a putative colonic epithelial stem cell biomarker which identifies colorectal tumours with a more aggressive phenotype. Biochem. Soc. Trans. 36:1350–1353 10.1042/BST0361350 [DOI] [PubMed] [Google Scholar]
- Winnard P.T., Jr, Kluth J.B., Raman V. 2006. Noninvasive optical tracking of red fluorescent protein-expressing cancer cells in a model of metastatic breast cancer. Neoplasia. 8:796–806 10.1593/neo.06304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Mazo I., Leung H., Engelke K., von Andrian U.H., Deryugina E.I., Strongin A.Y., Bröcker E.B., Friedl P. 2003. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160:267–277 10.1083/jcb.200209006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Alexander S., Schacht V., Coussens L.M., von Andrian U.H., van Rheenen J., Deryugina E., Friedl P. 2009. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 20:931–941 10.1016/j.semcdb.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A.L., Hoffman R.M., Figdor C.G., Weiss S.J., Friedl P. 2013. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201:1069–1084 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Mani S.A., Donaher J.L., Ramaswamy S., Itzykson R.A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R.A. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117:927–939 10.1016/j.cell.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Zhang X.D., Gillespie S.K., Hersey P. 2004. Staurosporine induces apoptosis of melanoma by both caspase-dependent and -independent apoptotic pathways. Mol. Cancer Ther. 3:187–197 [PubMed] [Google Scholar]
- Zou J.Y., Guo Y.L., Guettouche T., Smith D.F., Voellmy R. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 94:471–480 10.1016/S0092-8674(00)81588-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.