Abstract
Microtubules are important components of the cellular cytoskeleton that play roles in various cellular processes such as vesicular transport and spindle formation during mitosis. They are formed by an ordered organization of α-tubulin and β-tubulin hetero-polymers. Altering microtubule polymerization has been known to be the mechanism of action for a number of therapeutically important drugs including taxanes and epothilones. Traditional cell-based assays for tubulin-interacting compounds rely on their indirect effects on cell cycle and/or cell proliferation. Direct monitoring of compound effects on microtubules is required to dissect detailed mechanisms of action in a cellular setting. Here we report a high-content assay platform to monitor tubulin polymerization status by directly measuring the acute effects of drug candidates on the cellular tubulin network with the capability to dissect the mechanisms of action. This high-content analysis distinguishes in a quantitative manner between compounds that act as tubulin stabilizers versus those that are tubulin destabilizers. In addition, using a multiplex approach, we expanded this analysis to simultaneously monitor physiological cellular responses and associated cellular phenotypes.
Keywords: High-content analysis, microtubule polymerization, tubulin stabilizer, destabilizer.
INTRODUCTION
Microtubules are important structural components of the cytoskeleton. Microtubules are composed of polymerized α-tubulin and β-tubulin subunits organized into structures called protofilaments, ten to fifteen of which associate to form the hollow cylindrical polymers of microtubules [1]. The dynamics of microtubule assembly and disassembly play an important role in cellular processes such as vesicular transport [2]. Also, it has long been known that, in cells undergoing cell division, the organization of microtubules into spindle fibers allows polarized migration of duplicated chromosomes during mitosis.
A number of well characterized anti-cancer drugs have demonstrated cytotoxic effects by altering microtubule dynamics in cancer cells, in turn disrupting the formation of mitotic spindles and ultimately the progression of mitotic process. For example, paclitaxel binds to a pocket within β-tubulin that putatively counteracts the effect of GTP hydrolysis, resulting in stabilization of microtubule structure [3]. Another class of compounds, including nocodazole, vinblastine, colchicine or tubulysin, interferes with microtubule dynamics by causing tubulin depolymerization [4]. The stabilization of microtubules or disruption of tubulin polymerization by these compounds leads to cell cycle arrest and cancer cell death. As a result, monitoring drug effects on microtubule polymerization is essential to addressing anti-cancer activity. On the other hand, many other therapeutic approaches aim to avoid interference of tubulin dynamics, given that this may cause cytotoxicity. Therefore, an assay that can accurately monitor microtubule dynamics is broadly applicable to drug discovery programs seeking to optimize and progress lead compounds.
The commonly used methods to assess compounds for their microtubule activity include both biochemical and cell-based assays. The biochemical assays normally employ purified tubulin, and are based on the principle that polymerization of tubulin leads to changes in turbidity. The change in turbidity can be monitored as absorbance change due to the light scattering property of tubulin polymerization [5, 6]. A fluorescent-based version of the biochemical assay takes advantage of the ability of polymerized tubulin to bind the fluorescent molecule DAPI with higher affinity than unpolymerized tubulin [7]. Accordingly, tubulin polymerization is reflected by an increase in fluorescence intensity of DAPI, and tubulin depolymerization leads to decreased fluorescence intensity of DAPI. These assays typically require a relatively large amount of purified tubulin, resulting in an assay that is not cost-effective for screening large numbers of compounds. Cell-based assays for tubulin polymerization often utilize either fluorescent microscopy or cell cycle analysis. In fluorescent microscopic studies, either the α- or β-tubulin can be labeled directly with a tubulin antibody-conjugated fluorescent probe, or indirectly via a secondary antibody. Such studies, while straightforward, are limited in throughput, and they lack adequate methods for quantification (e.g., [8, 9]). Quantitative cell-based assays for assessing microtubule activities of compounds are often achieved by measuring the indirect effects on cell cycle which result from a disruption of microtubule networks. Tubulin stabilizers and destabilizers cause cell cycle arrest at the G2/M phase [10], and therefore measurement of the percentage of cells arrested in G2/M phase is often used as a surrogate endpoint for microtubule activity. The cell cycle analysis can be efficient due to a simple workflow and amenability to high throughput platforms; however, the method is not direct and cannot be used to discern mechanistically between tubulin stabilization and destabilization.
To address the need for a quantitative cell-based assay for direct measurement of tubulin effects, we describe here the development of a high-content platform to monitor tubulin polymerization. High-content assays have the power to simultaneously measure a variety of cell features in a highly quantitative manner, and they have been previously applied to the study of compound cytotoxic effects on tubulin, in which certain cytotoxic compounds can lower the tubulin staining intensity [11]. This approach indicates the feasibility of a quantitative high-content analysis at least in a 96-well format [11]. In the present study, we seek to address the bi-directional polymerization or de-polymerization status in a 384-well screening assay format. The biologically relevant measurement parameters were identified using a set of compounds with known microtubule activity in an unbiased parameter screen. For benchmarking, we also compared the data from the high-content assay with a biochemical assay and a cell-cycle assay. Our results demonstrate that this high-content assay platform is sensitive, reproducible and correlates well with data generated using both the biochemical and cell cycle assays. Importantly, this assay can distinguish tubulin stabilizers from destabilizers and has been applied to profile compounds for tubulin activities. Finally, we expanded this high-content platform to multiple cellular response analyses linking mechanisms of tubulin interaction to associated cellular phenotypes.
MATERIALS AND METHODS
Reagents
Nocodazole was purchased from Sigma-Aldrich (St. Louis, MO). The anti-tubulin antibody (anti-α-Tubulin, CBL270) from rats was obtained from Millipore (Billerica, MA), and the anti-rat secondary antibody conjugated to Alexa488 was obtained from Life Technology (Carlsbad, CA). Other high-content reagents used in washing and blocking were obtained from ThermoFisher Scientific (Pittsburg, PA). Cell culture reagents were purchased from Life Technologies or Sigma.
Cell Culture
A549 and HCT116 cells from ATCC were grown in RPMI 1460 medium (Gibco) Life Technology (Carlsbad, CA) supplemented with 10% FBS (Sigma, St Louis, MO) and Penicillin-Streptomycin (Gibco). One day prior to assay, cells were harvested from tissue culture flasks and resuspended in growth medium in the absence of antibiotic and antifungal agents. Cells were seeded in 384-well clear bottom black-wall poly-D-lysine coated microplates (BD Falcon) at a density of 2,000 cells per well. The microplates were maintained at 37 °C in a CO2 incubator overnight until assay.
Tubulin High-Content Assay
The effect on cellular microtubule structure was assayed by treating the cells with increasing concentrations of compounds at 37 °C for either 3, 6 or 18 h. At the end of treatment, cells were fixed at room temperature with 4% formaldehyde freshly prepared in DPBS containing Ca2+ and Mg2+ (DPBS+/+) for 30 min. Fixed cells were washed twice with DPBS+/+, followed by a permeabilization step for 20 min using Permeabilization Buffer (ThermoFisher), and washing for two times in DPBS+/+. Cytoplasmic tubulin structures were stained using standard fluorescent staining technique. Blocking was performed for 1 h with 1X Blocking Buffer (ThermoFisher). The anti-α-tubulin rat primary antibody was diluted 1:125 in 1X Blocking Buffer and incubated overnight at 4 °C. The anti-rat secondary antibody conjugated to Alexa488 was used at 1:500 diluted in 1X Blocking Buffer for 3 h at room temperature. Hoechst33342 (Life Technology) was used for nuclear counter staining at 4 μg/mL and was included in the secondary antibody incubation step. Cells were washed twice with DPBS+/+ after each antibody incubation step. Fluorescent images were acquired on GE InCell 2000 high-content platform using a 20X objective.
Analysis of high-content images was carried via in GE Analysis Workstation using multi-target analysis. Segmentation was performed for nuclei, cytoplasmic and organelle structures. Tubulin staining was measured as general cytoplasmic staining or from identified organelle structures. In order to determine the proper measurements, all available parameters were measured and exported into Spotfire (Tibco) for comparison. Concentration-response curves were analyzed using GraphPad Prism v5 (La Jolla, CA), or by in-house data analysis software.
Tubulin Polymerization Fluorescence Assay
The direct effect of compounds on tubulin polymerization was determined in a biochemical fluorescent-based assay (Cytoskeleton, Denver, CO), essentially according to manufacturer’s protocol. Briefly, purified tubulin from porcine brain (2 mg/mL) in assay buffer (80 mM PIPES pH 6.9, 2.0 mM MgCl2, 0.5 mM EGTA) was allowed to polymerize in the presence of 1 mM GTP, along with 10% glycerol as a polymerization enhancer, as well as 6.3 μM DAPI to follow the extent of polymerization fluorimetrically. The reaction mixtures were incubated with increasing concentrations of compound in 384-well black wall microplates with a non-binding surface (Corning, Tewksbury, MA). Fluorescence changes were monitored kinetically in the Spectramax Gemini fluorescence plate reader (Molecular Devices, Sunnyvale, CA) at 37 °C for 1 h, using excitation at 360 nm and emission at 420 nm. Kinetic measurements were calculated by the area under the curve (AUC) using Softmax Pro v5.4.1 (Molecular Devices, Sunnyvale, CA). Data were analyzed using GraphPad Prism, or by in-house data analysis software.
Cell Cycle Assay
Cell cycle assays with HCT116 cells were carried out by treatment with compounds at 37 °C for 24 h. Cells were then fixed with 4% paraformaldehyde prepared in DPBS+/+ for 15 min, followed by washing two times in DPBS+/+ and blocking for 1 h at room temperature. Cells were permeabilized and labeled with anti-Phospho-Histone H3 (Ser) antibody (Cell Signaling, Danvers, MA) diluted 1:200 in blocking buffer for 1 h at RT and then anti-goat secondary antibody Alexa488 (Life Technology) at 1:1000 dilution along with DAPI at 5 μg/ml for nuclear staining.
RESULTS
Establishment of a High-Content Platform to Monitor Tubulin Activity
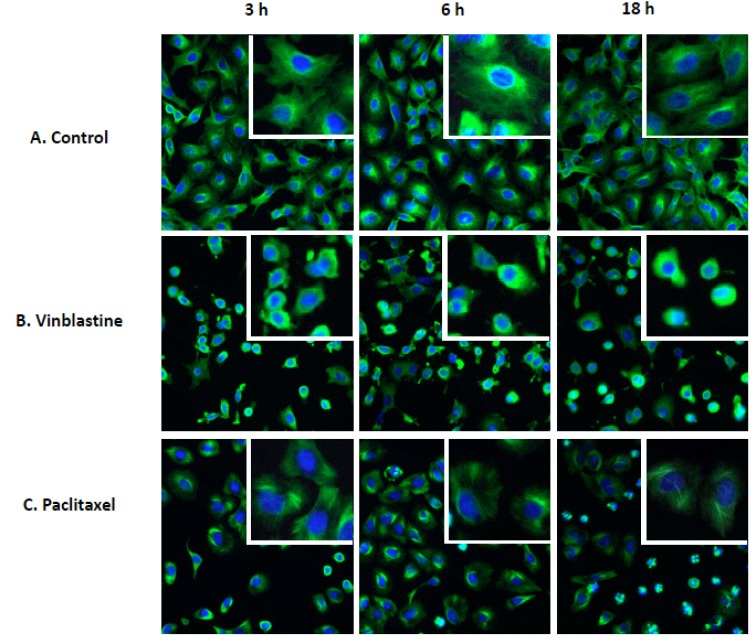
To establish a high throughput cellular based assay to quantify tubulin stabilization and destabilization, high content screening (HCS) was explored since the microtubule network in our model cell line A549 can be visualized by fluorescence staining with anti α-tubulin antibody (Fig. 1). Compounds known to affect microtubule assembly were used as tool molecules to develop this tubulin activity assay. These tool molecules include colchicine, vinblastine, nocodazole and tubulysin, all of which disrupt microtubule assembly, as well as paclitaxel, which is known to stabilize microtubule assembly [4]. We also included camptothecin, a DNA topoisomerase I inhibitor [12], and a compound known not to interact with tubulin. These latter two molecules were used as negative controls to assess the specificity of the assay. The HCS method was then optimized by adjusting cell number, fluorescent labeling conditions and exposure time during image acquisition. Fluorescent images of the tubulin network were acquired on the GE InCell 2000 high-content platform as in Fig. (1). As shown in the control, very fine microtubule strands that resemble the cytoskeletal network covering the entire cytoplasmic area were clearly observable in A549 cells (Fig. 1A). In agreement with its known effects on tubulin activity, the destabilizer, vinblastine, caused diffuse tubulin staining due to disruption of the microtubule network that occurred as soon as 3 h after treatment at 2 μM, and this effect could sustain up to 18 h (Fig. 1B). In terms of the cell morphology, cells treated with vinblastine became round and with visibly smaller cytoplasmic area. In contrast, 2 μM paclitaxel treatment led to thickening of the microtubule fibers that were observable as early as 3 h and as late as 18 h. Morphologically, cells became round after a 3-h treatment with paclitaxel, and at 18 h, a large number of cells showed condensed nulcei, suggesting apoptosis could be taking place (Fig. 1C). Moreover, those cells showing condensed nuclei appeared to have the tubulin structure completely collapsed, suggesting that these cells have died from severe toxicity. Overall, the results clearly indicate qualitatively distinguishable tubulin effects from different treatments and tubulin staining can be used to assess the activities of compounds on tubulin. To enable precise detection of time-dependent effect and concentration-dependent effect, there is a need to develop a method that quantifies the images in a way that capture the characteristic changes.
Fig. (1).
High-content images of fluorescently-labeled α-tubulin in A549 cells treated with tubulin-active compounds. A549 cells were treated with DMSO control, 2 µM vinblastine or paclitaxel for 3 h, 6 h or overnight (18 h). Microtubule structure (green) was visualized by fluorescent labeling using anti-α-tubulin rat primary antibody followed by anti-rat-Alexa488, as described in the “Materials and Methods”. Nuclei are counterstained using Hoescht33342 (blue). Microtubule network making up the cytoskeleton was visible in the control cells. Vinblastine caused disruption of the microtubule network, leading to a diffused staining of α-tubulin, that was also visible after 3 h and lasted up to 18 h. Treatment with paclitaxel induced a thickening of the microtubule fiber that was most prominent at 18 h. Enlarged images for 3 h treatment are shown in the inserts to further visualize the changes in microtubule network upon the different treatments.
Development of a Quantification Method to Distinguish the Effects of Tubulin Stabilization and Destabilization
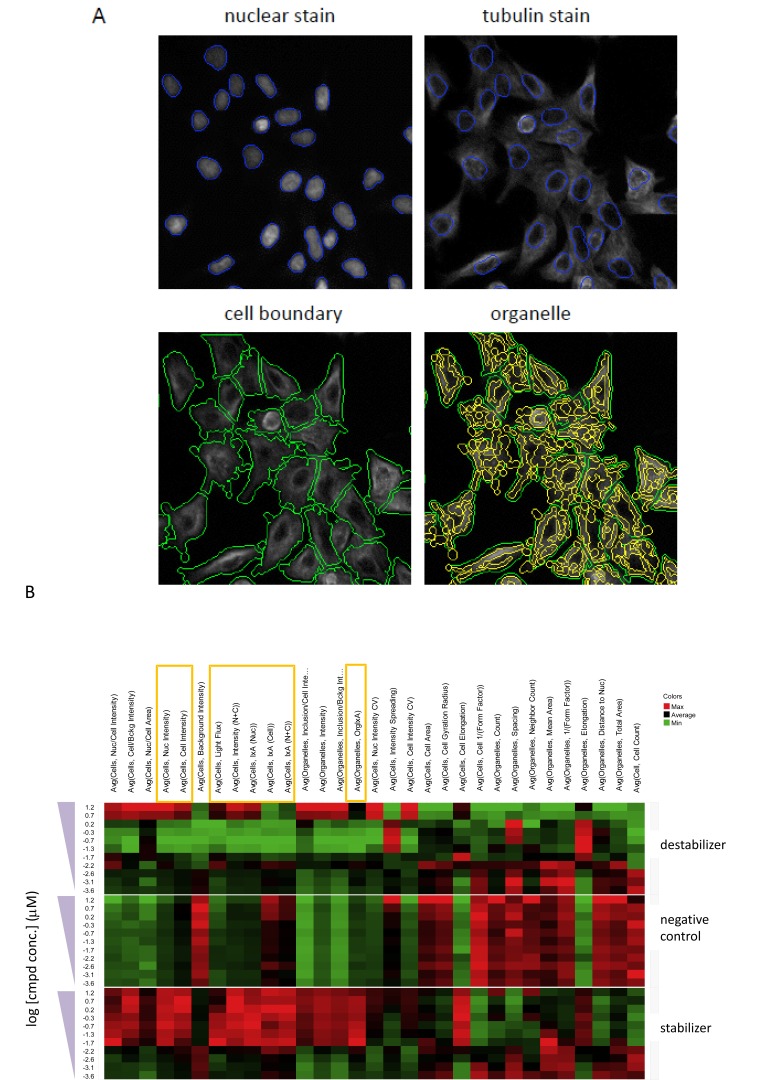
A challenge for developing a high-content assay for tubulin activity assessment was to determine the appropriate algorithm that can capture the opposing tubulin effects of stabilizers and destabilizers and thus reflect the observed effects on microtubule networks. To develop a quantification method, vinblastine, paclitaxel and a negative control were utilized in a concentration-response study with 6-h treatment. Image analysis was subsequently performed with the goal to identify response measurements that can robustly distinguish the opposing effect of microtubule stabilizers and destabilizers. Segmentation of the nuclei was achieved by the use of Hoechst nuclear stain, while segmentation of cellular boundary was achieved by means of the α-tubulin fluorescent staining. Furthermore, the tubulin structures in each cell are segmented as intracellular “organelle” objects (Fig. 2A). Quantification of the tubulin staining was performed by taking the tubulin structures as single cell objects or by prior segmentation of the structures as cytoplasmic “organelles”. Fig. (2B) shows a set of heat maps generated from such a concentration-response study with the test set, in which a number of different response measurements were taken for both the cell objects and organelles. These measurements can be grouped broadly into three categories: 1) measurements of intensity, 2) measurements of intensity distribution, and 3) morphological measurements (Table 1). Measurements of intensity include those that measure average intensity and total intensity (also called integrated intensity), whereas measurements of intensity distribution include those that provide a statistical description on the intensity signals. The morphological measurements are the object area, gyration, elongation and form factor (Table 1). As shown in Fig. (2B), the characteristic opposite effects of stabilizers and destabilizers are reflected by intensity measurements (category 1). Specifically, compared with the negative control, increasing concentrations of paclitaxel can result in increasing magnitude of the intensity measurements either in the cellular objects or in the identified organelle, while increasing concentrations of vinblastine lead to decreasing magnitude of the measurements, with exceptions at the two highest concentrations in which severe cytotoxicity caused the tubulin network to become collapsed and condensed (data not shown). In contrast, the opposing effects between stabilizers and destabilizers are not discernible with measurements from categories 2 and 3.
Fig. (2).
Object segmentation in high-content algorithm and determination of biologically relevant high-content measurements using heat maps.
(A) Object segmentation was carried out using Multi-target analysis in GE Analysis Workstation to identify the nuclei (blue) and cell boundary (green). Tubulin structures were further identified as intracellular “organelle” objects (yellow). (B) A549 cells treated for 6 h with decreasing concentrations of paclitaxel, vinblastine or a known tubulin-inactive compound (negative control). The microtubule structure was visualized by fluorescent labeling of α-tubulin and high-content imaging. The images were quantified using different measurement parameters as shown in the figure. The measured values from the different treatments and concentrations for each parameter were organized into heat maps to determine the parameters that can capture opposite effects of microtubule stabilizer and destabilizer. Red color in the heat maps indicates the highest value, whereas green color indicates the lowest value, and average values are shown in black. The heat maps reveal that several cell or organelle intensity measurements (highlighted in rectangles) may capture biologically relevant changes.
Table 1.
Categories of response measurements used in quantification.
| Category 1 — Intensity measurements | |
|---|---|
| Nuc/Cell Intensity | Intensity from nuclear region / intensity from cytoplasmic region |
| Cell/Bckg Intensity | Intensity from cytoplasmic region / background intensity |
| Nuc Intensity | Intensity from nuclear region per unit area |
| Cell Intensity | Cytoplasmic intensity per unit area |
| Background Intensity | Average intensity in the local background |
| Light Flux | Total intensity in per nuclear region / cytoplasmic intensity per unit area |
| Intensity (N+C) | Intensity in whole cell per unit area |
| IxA (Nuc) | Total nuclear intensity per cell |
| IxA (Cell) | Total cytoplasmic intensity per cell |
| IxA (N+C) | Total intensity from whole cell per cell |
| Organelle Inclusion/Cell Intensity | Ratio of the organelle intensity inclusions to cell intensity |
| Organelle Inclusion/Bckg | Intensity of organelle objects / local background intensity |
| Organelle Intensity | Intensity of pixels within inclusions per unit area |
| OrgIxA | Total organelle intensity per cell |
| Category 2 — Measurements of intensity distribution | |
| Nuc Intensity CV | Coefficient of variation for intensity in the nuclear region |
| Intensity Spreading | Cellular intensity distribution |
| Cell Intensity CV | Coefficient of variation of cell intensity |
| Category 3 —Morphological measurements | |
| Cell Area | Cell area |
| Nuc/Cell Area | Nuclear area / cell area |
| Cell Gyration Radius | Average radius of cells |
| Cell Elongation Mean | Ratio of the short axis to long axis of the cell |
| Cell 1/(Form Factor) | Cell roundness index |
| Organelle Count | Number of identified organelle objects |
| Organelle Spacing | Distance between organelle objects |
| Organelle Neighbor Count | Number of neighboring organelle |
| Organelle Mean Area | Mean area of identified organelle objects |
| Organelle 1/(Form Factor) | Roundness index of organelle |
| Organelle Elongation | Mean ratio of the short axis of the organelle to the long axis |
| Organelle Distance to Nuc | Distance from the center of gravity of organelle to center of gravity of nucleus |
| Organelle Total Area | Total area of organelle |
| Cell Count | Number of cells |
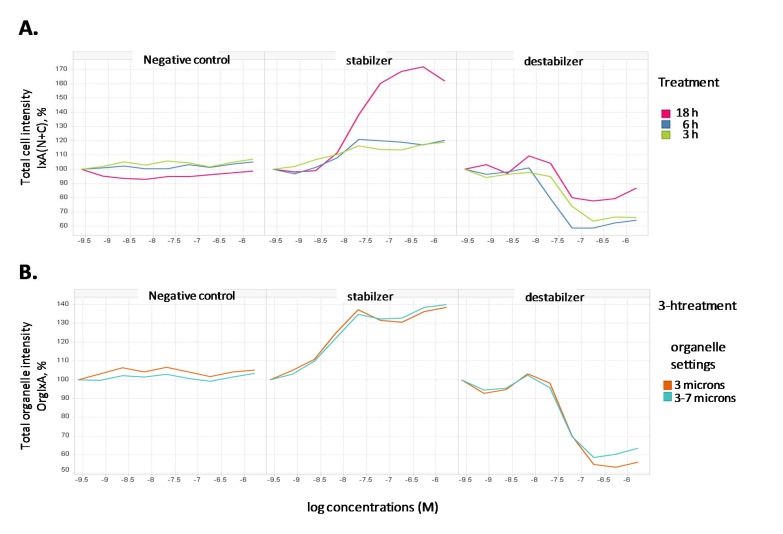
To further validate the quantification method, the response measurements in category 1 under different durations of compound treatment were analyzed (Fig. 3). Total cell intensity [IxA(N+C)] measurements demonstrated significant differences comparing paclitaxel, vinblastine and the negative control compound (Fig. 3A). No significant change for the total cell intensity was found with the negative control at all time points. While paclitaxel showed dramatic increase of total cell intensity at 18 h, the increase could be due to the results of complete collapse of tubulin structure in those apoptotic cells which we have observed in Fig. (1C). The increase at 3 h and 6 h by paclitaxel, although moderate, was concentration-dependent and the similarity at both time points was consistent with the observations in Fig. (1). With vinblastine, clear decrease of total cell intensity was shown with at all three time points with the 18 h having the least effect, most likely due to cellular toxicity. To minimize the interference of cellular toxicity and dead cells, we focused on shorter treatment time and aim at identifying a readout in category 1 that significantly differentiates among negative control, vinblastine and paclitaxel. As shown in Fig. (3B), measurements of the organelle objects can yield good assay windows at 3h for both vinblastine and paclitaxel, with opposite trends. In addition, we compared the quantification for two different sizes of organelles identified during object segmentation. With either size, the resultant measurement is found to give a reasonable assay window for both paclitaxel and vinblastine. The average change with the stabilizer was 50±18% (n=5) and with the destabilizer was 57±7% (n=3), indicating consistency of the measurement. Therefore, we finalized the measurements of the organelle objects as our high content assay quantification method.
Fig. (3).
Quantification of cell and organelle intensity from concentration-response data.
Cells from treatment with a negative control compound, paclitaxel or vinblastine for 3, 6 and 18 h were stained for microtubule structures as described in “Materials and Methods”. (A) Tubulin staining was quantified as total cell intensity (integrated intensity N+C), which is the average light intensity measured from the whole cell multiplied by the average area of cell. (B) Tubulin staining from 3-h treatment was quantified as total organelle intensity (integrated organelle intensity), which is the average light intensity measured from the identified organelle structures multiplied by the average area of cell. Microtubule structures were identified using two different parameters (3 microns or 3-7 microns) to compare the assay window. Either parameter is found to produce reasonable windows for both paclitaxel and vinblastine. The EC50 values for the stabilizer were determined as 4.2 nM (3 microns) and 4.6 nM (3-7 microns) and IC50 values for the destablizer as 58 nM (3 microns) and 45 nM (3-7 microns).
Validation of High-Content Analysis Assay
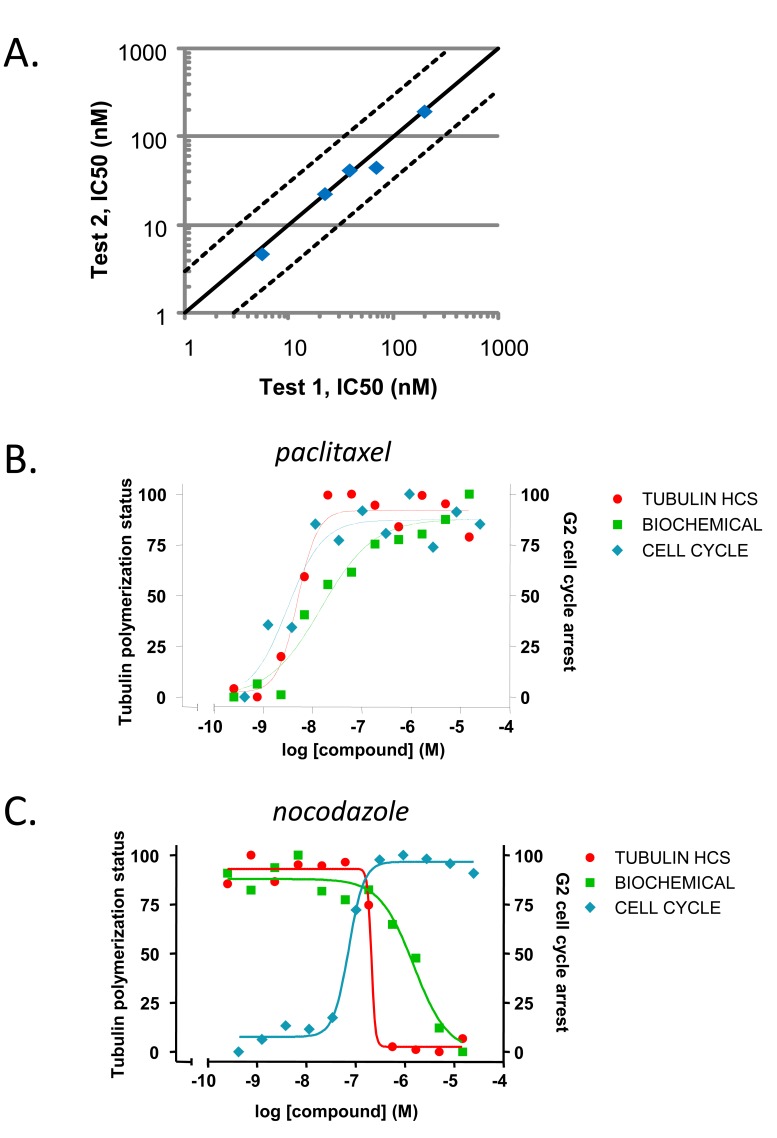
With the quantification method identified, we then examined the data reproducibility by comparing the estimated potency values from two experiments. Five compounds including both stabilizers and destabilizers were tested in two independent experiments. The measured potency values for each compound from the two experiments were plotted as individual symbols (Fig. 4A). Regression analysis of the compound potency values yielded a slope of 0.96 and r2 = 0.98, suggesting tight correlation and therefore good assay reproducibility for this high-content approach.
Fig. (4).
Assay final validation and comparison among different assays.
(A) Correlation plot to demonstrate potency reproducibility of tubulin high-content assay. A set of five compounds including both stabilizers and destabilizers were assayed in two independent experiments. The solid diagonal line illustrates theoretical 1:1 correlation. The two dash lines mark the limits for 3-fold difference from the theoretical line. (B–C) The activities of the following compounds were then compared in high content analysis, cell cycle and biochemical assays: (B) tubulin stabilizer paclitaxel and (C) destabilizer nocodazole. For tubulin high-content assay, data were analyzed by measuring total organelle intensity. For cell-cycle assay and biochemical tubulin polymerization assay, data were analyzed as described in “Materials and Methods”. All data were normalized by taking the maximum values as 100%. For paclitaxel, the potency values estimated in the three assays are 4 nM (high-content), 10 nM (biochemical) and 2 nM (cell-cycle). With nocodazole, the potency values estimated in the three assays are 244 nM (high-content), 2.292 µM (biochemical) and 72 nM (cell-cycle).
To further validate the assay, we compared the measurements from the present high-content tubulin assay with biochemical and cell cycle assays (Fig. 4B, 4C and Table 2). As expected, paclitaxel demonstrated similar activities in high content analysis as in the cell cycle assay and the biochemical assay (Fig. 4B). The activities of the tubulin destabilizer nocodazole are comparable in the high-content analysis and the cell cycle assay, while its potency in the biochemical assay might be slightly weaker (Fig. 4C). The higher sensitivity observed when interrogating compounds in a cellular context can most likely be related to the availability of a milieu of associated proteins involved in regulating microtubule dynamics. Most importantly, the high-content assay enables us to distinguish the opposite effects of a stabilizer and a destabilizer similar to biochemical assays. In contrast, the cell cycle assay does not provide such differentiation. Taken together, these results highlight the value derived from an HCS approach to monitoring tubulin dynamics. All those results indicate that the established high-content analysis not only distinguishes tubulin stabilizers from destabilizers as biochemical assays, but also reflects cellular environment.
Table 2.
Comparison of potency values across three assay platforms.
| Compound | Platform | Properties | EC50 / IC50 (µmM) | log EC50±S.E. / log IC50±S.E. (M) a | N b |
|---|---|---|---|---|---|
| Paclitaxel | Tubulin high-content | Increased tubulin polymerization | 0.004 | –8.4 ± 0.1 | 2 |
| Biochemical | Increased tubulin polymerization | 0.010 | –8.0 ± 0.3 | 1 | |
| Cell-cycle | Increased G2 arrest | 0.002 | –8.6 ± 0.4 | 2 | |
| Nocodazole | Tubulin high-content | Decreased tubulin polymerization | 0.244 | –6.6 ± 0.1 | 2 |
| Biochemical | Decreased tubulin polymerization | 2.292 | –5.6 ± 0.3 | 1 | |
| Cell-cycle | Increased G2 arrest | 0.072 | –7.1 ± 0.1 | 2 |
S.E. denotes standard error of the estimated values from non-linear dose-response regression analyses in a single curve or in a global fitting of data when there are two experiments
N = number of experiments
Expansion of High-Content Assay to Multiple Response Analysis
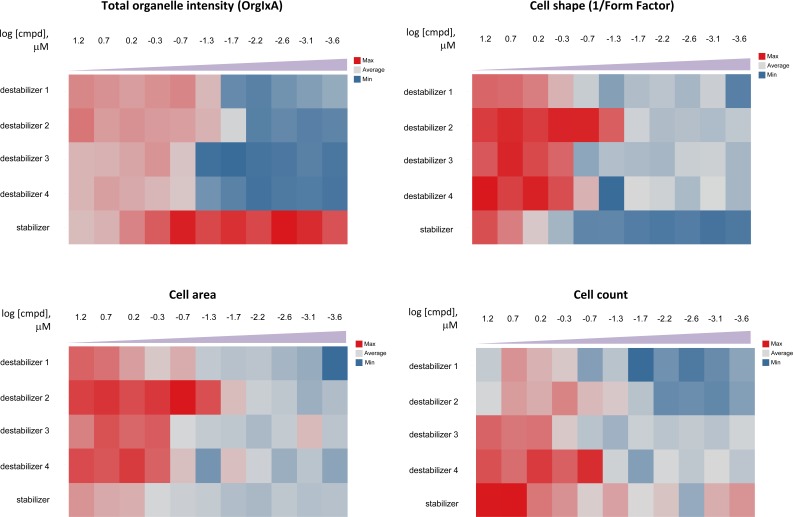
An added advantage of the high-content assay is the ability to measure multiple response parameters. Hence, we examine the different measurements and noted a concentration-dependent effect of both stabilizers and destabilizers on cell shapes, cell areas and cell counts (Fig. 5). This observation is in line with the actions of these compounds on cytoskeleton; such effects are especially reflected by cell shape measurement. Consequently, these additional phenotypic measurements could be useful to verify the mechanisms of action of these compounds.
Fig. (5).
Heat maps from concentration-response data with tubulin activity, measured as total organelle intensity, compared to other phenotypic measurements. Stabilizers and destabilizers are shown to cause an associated change in cell shape and area and a concomitant decrease in cell count. The phenotypic effects provide additional lines of evidence for their tubulin activities, which affect cytosekeleton.
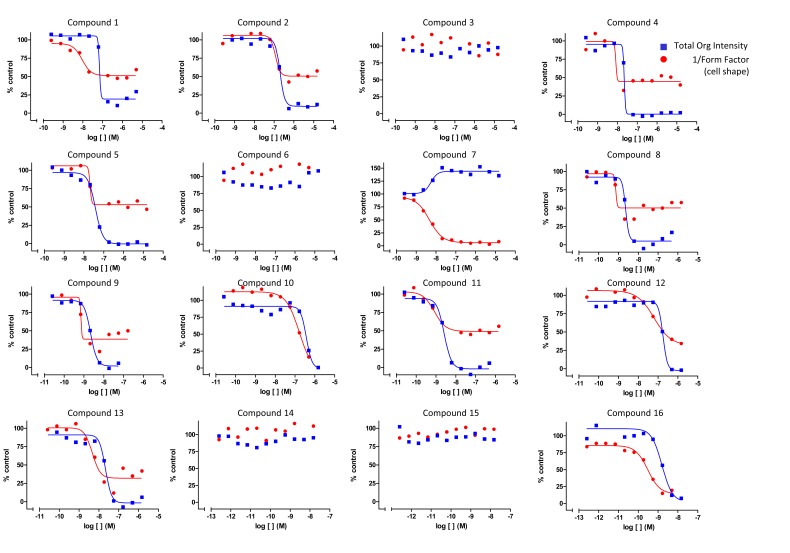
Applying this multiple response analysis approach, we performed a dose-response study for a small set of compounds to monitor both tubulin activity and cell shape. The results from this study are shown in Fig. (6). It can be seen that compound 7 is a stabilizer and induces a change in cell shape, whereas compounds 3, 6, 14 and 15 lack tubulin activity and do not cause any associated morphological change. The remaining compounds appear to be tubulin destabilizers with relatively similar efficacies on tubulin. However, their specific effect on cell shape can be different from each other. For example, compounds 10 and 16 have similar activities in effecting changes in cell shape, while compounds 1, 2, 4, 5, 8, 9, 11, 12 and 13 differ from compounds 10 and 16 in their efficacy at inducing changes in cell shape. While the reason for this is not understood, these different profiles suggest distinct underlying mechanisms of action for these compounds. Such profiling data could potentially provide evidence for differentiation of compounds.
Fig. (6).
Simultaneous profiling of compounds for tubulin activity and phenotypic effects. A set of compounds was screened for their tubulin activities and effects on cell shape. The screening set contains compounds that are destabilizers, stabilizers and inactive compounds. Only tubulin active compounds are found to affect changes in cell shape, whereas inactive compounds demonstrated no change in cell morphology.
DISCUSSION
We have established a high-content based platform that directly measures the effects of small molecules on tubulin status in cells. This high-content platform not only demonstrates the same trend of tubulin stabilizing and destabilizing activities as the biochemical methods do, but it also offers a cell-based environment to further dissect the abilities of compounds to enter the cells and to affect tubulin in the associated cellular protein complex. Furthermore, this cell-based high-content analysis provides a more sensitive readout for tubulin polymerization as reflected by increased compound potencies compared to corresponding biochemical assay results. Therefore, this high-content analysis could be employed as a primary cell-based assay to determine the compound tubulin activity for structure-activity relationship studies. Additionally, it can be applied as a liability assay when tubulin effect might be undesirable. Hence, the assay can find broad utility in oncology and other types of drug discovery programs. Compared with other cellular assays used to evaluate tubulin agents in the past, such as cell cycle analysis, our direct tubulin assay allows tubulin stabilizers to be distinguished from destabilizers; Also, we optimized our assay for compound treatment at 3 hours to avoid the complication of compound cytotoxicity from longer time treatments. Moreover, this analysis can be coupled to the evaluation of various phenotypic cellular endpoints to further characterize the cellular effects by the compounds. In our study, we further demonstrated that multi-response analysis could help to differentiate compounds that may otherwise all appear to have similar effects on tubulin. The differentiation of compound cellular activities could provide mechanistic insight and play a critical role in the final selection of drug candidates. The positive compounds identified from this study could be followed up in proliferation assays to assess their effects on cell growth in a panel of cell lines. On the other hand, biochemical analyses can be performed to further address their mechanisms of action. Tubulin-active compounds have been shown to bind to various distinct sites on tubulin [4]. Competitive radioligand binding assays can be performed to determine the sites of interaction. Recently, competitive mass spectrometry binding assay has been applied to determine compound binding to colchicine-, vinblastine- and or paclitaxel-binding sites in tubulin [13].
Although this high content platform confirmed certain advantages as we have highlighted here, it still faces some challenges. While high-content assays can be implemented on an automated platform and are amenable to a high throughput setting, the data acquisition is still relatively time consuming. One general disadvantage about high-content assays compared with the majority of other types of cell-based assays is that the size and number of image files produced by the platform can be considerably large, requiring a database solution. Fortunately, data storage nowadays is more affordable than in the past, and multiple database solutions are currently available. Another challenge of developing a high-content assay is to establish an optimal quantification algorithm that accurately reflects the biological response. We approached these issues systematically by first assessing the different response measurements in a heat map, from which we then determined a number of possible response measurements for further validation. Additionally, we examined the assay window given by the selected measurements to assess their sensitivity under the desirable assay conditions. We also addressed the reproducibility of the measurements and compared the data across different assay platforms, as would be the case for assay development for any other platform. This systematic approach can be applied generally to facilitate the development of any high content assay.
ACKNOWLEDGEMENTS
We would like to acknowledge the cell culture support from Cellular Resource Group, especially, Zia Lozewski. We also thank Mike Lenard, Normand Cloutier and Mike Thibeault for their efforts in the establishment of high-content platform at Hopewell site at Bristol-Myers Squibb Company.
CONFLICT OF INTEREST
The authors are employees of Bristol-Myers Squibb Company.
REFERENCES
- 1.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Hamm-Alvarez SF, Sheetz MP. Microtubule-dependent vesicle transport: modulation of channel and transporter activity in liver and kidney. Physiol Rev. 1998;78:1109–29. doi: 10.1152/physrev.1998.78.4.1109. [DOI] [PubMed] [Google Scholar]
- 3.Amos LA, Lowe J. How Taxol stabilises microtubule structure. Chem Biol. 1999;6:R65–9. doi: 10.1016/s1074-5521(99)89002-4. [DOI] [PubMed] [Google Scholar]
- 4.Jordan A, Hadfield JA, Lawrence NJ, McGown AT. Tubulin as a target for anticancer drugs: agents which interact with the mitotic spindle. Med Res Rev. 1998;18:259–96. doi: 10.1002/(sici)1098-1128(199807)18:4<259::aid-med3>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 5.Shelanski ML, Gaskin F, Cantor CR. Microtubule assembly in the absence of added nucleotides. Proc Natl Acad Sci USA. 1973;70:765–8. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaskin F. Analysis of microtubule assembly kinetics using turbidimetry. Methods Mol Biol. 2011;777:99–105. doi: 10.1007/978-1-61779-252-6_7. [DOI] [PubMed] [Google Scholar]
- 7.Bonne D, Heusele C, Simon C, Pantaloni D. 4',6-Diamidino-2-phenylindole, a fluorescent probe for tubulin and microtubules. J Biol Chem. 1985;260:2819–25. [PubMed] [Google Scholar]
- 8.Khalil MW, Sasse F, Lunsdorf H, Elnakady YA, Reichenbach H. Mechanism of action of tubulysin, an antimitotic peptide from myxobacteria. Chembiochem. 2006;7:678–83. doi: 10.1002/cbic.200500421. [DOI] [PubMed] [Google Scholar]
- 9.Ma YM, Zhou YB, Xie CM, Chen DM, Li J. Novel microtubule-targeted agent 6-chloro-4-(methoxyphenyl) coumarin induces G(2)-M arrest and apoptosis in HeLa cells. Acta Pharmacol Sin. 2012;33:407–17. doi: 10.1038/aps.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhalla KN. Microtubule-targeted anticancer agents and apoptosis. Oncogene. 2003;22:9075–86. doi: 10.1038/sj.onc.1207233. [DOI] [PubMed] [Google Scholar]
- 11.Ross-Macdonald P, de Silva H, Guo Q, et al. Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol Cancer Ther. 2008;7:3490–8. doi: 10.1158/1535-7163.MCT-08-0826. [DOI] [PubMed] [Google Scholar]
- 12.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–8. [PubMed] [Google Scholar]
- 13.Li CM, Lu Y, Ahn S, Narayanan R, Miller DD, Dalton JT. Competitive mass spectrometry binding assay for characterization of three binding sites of tubulin. J Mass Spectrom. 2010;45:1160–6. doi: 10.1002/jms.1804. [DOI] [PubMed] [Google Scholar]