Abstract
Structural studies of proteins normally require large quantities of pure material that can only be obtained through heterologous expression systems and recombinant technique. In these procedures, large amounts of expressed protein are often found in the insoluble fraction, making protein purification from the soluble fraction inefficient, laborious, and costly. Usually, protein refolding is avoided due to a lack of experimental assays that can validate correct folding and that can compare the conformational population to that of the soluble fraction. Herein, we propose a validation method using simple and rapid 1D 1H nuclear magnetic resonance (NMR) spectra that can efficiently compare protein samples, including individual information of the environment of each proton in the structure.
Heterologous expression of proteins in Escherichia coli is the most widespread technique used to produce large amounts of protein for structural studies. However, a sufficient quantity of protein is not always obtained using common, native, non-denaturant protocols. Major problems associated with rapid and high-level expression in bacteria include protein aggregation and formation of inclusion bodies1. In some structural genomics projects2,3, approximately 40% of the expressed targets are found in the insoluble fraction. Although these proteins are usually discarded as viable targets, the use of a simple refolding protocol could resolve the problem. Often, the fraction of soluble protein is not sufficient to make the production of large amounts of protein viable, and, in these cases, use of a refolding protocol can be the best solution to increase protein yield. Several techniques have been applied to refold proteins4,5,6, but the problem of how to solubilize and correctly fold proteins remains a major issue. One of the main difficulties in using refolded proteins is that there are very few experiments that discriminate between different conformational states and guarantee the correct refolding of the protein. Enzymatic7,8,9,10 and biological activity11,12,13 have been widely used to confirm correct protein refolding. However, in general, significant statistical error is associated with these assays, and the structural integrity of the refolded sample cannot be ensured. With respect to direct analysis of protein structure, circular dichroism (CD) is an effective technique only when large changes in secondary structure are present.
High resolution structural studies can be hindered by low yields of expressed protein or lack of an optimized production/purification protocol. For example, obtaining a 3D nuclear magnetic resonance (NMR) spectrum of a 20 kDa protein typically requires 5–10 mg of very pure and high cost isotopically-labeled protein. In x-ray crystallography, large amounts of very concentrated protein are required to test crystallization conditions. In addition, protein purity alone is not always sufficient to ensure successful crystal growth. Even small changes in a protein's three-dimensional structure can disturb crystal formation14,15,16. If an alternative conformation or oligomer attaches to the crystal, it may form a crystal surface that prevents attachment of other protein molecules, poisoning its growth17,18. Proteins can exist as multiple conformations in equilibrium due to many factors, such as structural flexibility, enzymatic activity, cooperativity, and oligomerization. For some experiments, conformational purity may not be important. For example, a biological activity assay of a protein with 20% in an inactive conformation is equivalent to another experiment with 20% less protein, but with 100% in an active conformation. However, in crystallography, NMR, and many other types of studies, this information may be relevant and necessary. A simple technique that can compare and ensure that different protein samples, obtained from different purification protocols, have the same conformational distribution is helpful in such cases.
In this paper, we suggest the use of a simple and fast 1D 1H NMR spectrum to validate refolding protocols before increasing protein production. This technique is able to detect very small conformational changes in protein structure, as 1H NMR is sensitive to changes in the environment of each hydrogen atom of a molecule (see e.g.19). It represents a powerful method for the verification of the spatial structure of (macro)molecules20 and gives us the expected information, and these properties are unique and essential for subsequent structural studies of the analyzed sample.
We applied the proposed method to P21 from Trypanosoma cruzi, an 18 KDa protein that contains six cysteine residues that are all expected to form disulfide bonds. P21 is a ubiquitous secreted protein of T. cruzi, the causative parasite of Chagas disease. P21 was found to serve as part of host cell invasion mechanism21, triggering actin polymerization on the host cell via interactions with the CXCR4 chemokine receptor22, which is present on the cellular membrane. Previous studies21 showed that recombinant P21 (P21-His6, with an N-terminal histidine tag) adheres to HeLa cells in a dose-dependent manner. Additionally, invasion of HeLa cells by T. cruzi metacyclic trypomastigotes (tissue cultured trypomastigotes, TCT) and extracellular amastigotes (EA) is greatly increased in the presence of P21-His6. Furthermore, P21-His6 upregulates the phagocytosis of zymosan particles in mice peritoneal macrophages22, indicating that P21 is involved in the phagocytic mechanism. More than 95% of expressed P21-His6 is insoluble in Escherichia coli, making the yield of the soluble protein insufficient for biological and structural studies. To overcome this limitation, we extracted P21-His6 from inclusion bodies using urea and refolded it. Since recombinant P21 is fused to a histidine tag, we used nickel affinity chromatography to refold the protein by applying a slow urea gradient. This technique has been used successfully to refold proteins in many cases23,24,25,26,27 because protein absorption on the solid matrix prevents aggregation during the process through separation of individual protein molecules5. To assess the available refolding validation techniques, we compared parasite invasion and phagocytosis of particles induced by refolded (rP21-His6) and soluble (sP21-His6) protein. As a second approach, refolded and soluble protein structures were compared by circular dichroism (CD) to find information regarding the state of the proteins' secondary structures. Finally, 1D 1H NMR spectra of the soluble and refolded proteins were compared in order to confirm the correct refolding of P21-His6 at atomic resolution. Comparison of conformational identity between the soluble and refolded preparations is an essential analysis needed to perform structural studies using the refolded material, and NMR spectroscopy is the only one of these methods that addresses that question.
Results
The yield of P21-His6 was greatly improved when the protein was recovered from inclusion bodies and refolded. In order to compare the yield of soluble to refolded protein, we expressed recombinant P21-His6 in 8 L 2xYT medium. Expression produced 24.9 g of dry weight bacteria and 8.1 g of insoluble pellet after lysis and centrifugation. Most of the protein was present in the insoluble fraction. sP21-His6 was purified from the soluble fraction in a single step of Nickel affinity chromatography; however, only 0.68 mg of purified protein was obtained. The refolding procedure recovered 57.76 mg of rP21-His6 from inclusion bodies, a yield 85 times greater than purification from the soluble fraction.
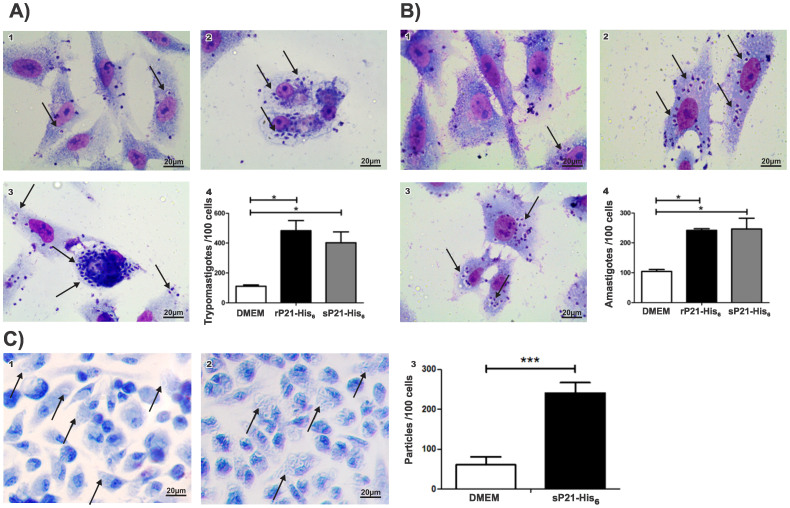
Results from invasion and phagocytosis experiments demonstrated that soluble and refolded P21-His6 have similar biological properties. In the presence of sP21-His6, invasion of Vero cells by TCT (Figure 1A) was increased approximately 3.6-fold, and invasion by EA (Figure 1B) was increased approximately 2.4-fold. Similar results were obtained for rP21-His6 (4.4-fold increase for TCT and 2.3-fold increase for EA). The difference between sP21-His6 and rP21-His6 was not significant. Phagocytosis of zymosan particles by macrophages was increased 3.5-fold with the addition of 40 μg/mL sP21-His6 (Figure 1C) and 3.5-fold with the addition of 40 μg/mL rP21-His622. The similarity of these increases suggests that refolded and soluble protein exhibit the same biological properties.
Figure 1.
(A) sP21-His6 and rP21-His6 affect invasion of Vero cells by T. cruzi TCT and EA. Light microscopy of (a) TCT invasion and (B) EA invasion after treatment with (1) DMEM (control, not treated), (2) 40 μg/mL of rP21-His6 and (3) 40 μg/mL of sP21-His6. (4) Quantification. Arrows in (1), (2), and (3) indicate internalized parasites. (C) Phagocytosis of zymosan particles by mice peritoneal macrophages in the presence of sP21-His6. Light microscopy of (1) control (untreated) cells and (2) cells treated with 40 μg/mL sP21-His6. (3) Quantification of (1) and (2). Arrows in (1) and (2) indicate internalized particles.
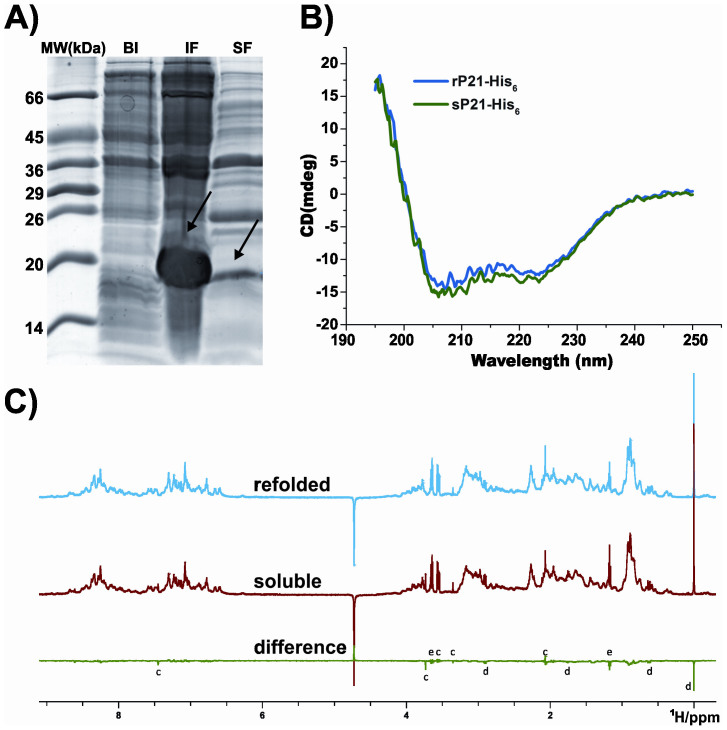
Finally, CD and NMR experiments validated the correct refolding of P21-His6. The CD spectra showed equivalent secondary structure composition for both preparations (Figure 2B). Comparison between 1H NMR spectra of sP21-His6 and rP21-His6 showed exactly the same chemical shifts and intensities for every proton in the structure (Figure 2C). In this way, both samples could be compared directly and were found to be identical to one another, with respect to both correct folding and conformational ensemble. To better evaluate the similarities (or differences) between the spectra of both samples, a difference spectrum was calculated and is shown in Figure 2C (green line). The larger differences can be assigned to non-protein components of the sample (residual ethanol from concentrator rinsing, small differences in DSS concentration, low molecular weight contaminants). These differences underscore the sensitivity of the method and the identity of both protein conformations and ensembles. In addition, up field shifts around 0 ppm in both spectra indicate a hydrophobic nucleus in P21-His6, showing that the structure is folded.
Figure 2.
(A) SDS-PAGE of bacterial lysate, showing the predominate expression of P21-His6 in the insoluble fraction. Lanes: (MW) molecular weight markers, (BI) lysate before IPTG induction, (IF) insoluble fraction after induction, and (SF) soluble fraction after induction. Arrow indicates the position of P21-His6. (B) Circular dichroism (CD) spectra of refolded and soluble P21-His6. Superposition of rP21-His6 (blue) and sP21-His6 (green) CD spectra at 20°C. Refolded and soluble protein samples have similar secondary structure composition. (C) Comparison between 1D1H NMR spectra of sP21-His6 and rP21-His6, showing exactly the same structure for both proteins. Spectrum of refolded (blue) and soluble (red) protein; spectra difference after normalization (green). The marked peaks indicate (c) low weight contaminants, (d) DSS, and (e) residual ethanol.
Discussion
In this study, we applied a one-step refolding procedure to P21-His6, which increased the yield of the protein 85-fold. This refolding method is based on immobilization of the P21-His6 on Nickel-affinity resin, preventing protein aggregation during the process. Refolding requires approximately 14 hours (which can be performed overnight with no need for human interference) and uses less than 0.5 L of buffer. It is less laborious and quicker than other refolding techniques, such as dialysis.
We used pre-existing biological information about P21 to develop protein-specific assays that could be applied to both refolded and soluble P21-His6 fractions. Both samples increased parasite invasion and phagocytosis of zymosan particles at similar rates, suggesting that they behaved identically with respect to biological activity. This information would be sufficient in a majority of biological studies, but is not sufficient for structural studies, in which correct folding and conformational homogeneity of samples are essential.
As more structure-focused approaches, CD and NMR spectra provided structural comparisons between sP21-His6 and rP21-His6. Although CD measurements provide information about protein secondary structure, no information at an atomic resolution can be provided. This method is more useful for identifying incorrectly folded proteins by detecting differences in the spectra, because even if two spectra are identical, we cannot confirm that structures are equal. Simple one-dimensional 1H NMR overcomes this limitation by providing information about the environment of each 1H atom of the protein, allowing a comparison of samples obtained by different purification methods at atomic resolution. With this simple 1D 1H NMR measurement, we confirmed that sP21-His6 and rP21-His6 have exactly the same structures and identical conformational equilibria. In addition, more important information can be obtained from the spectrum: the good dispersion of the peaks over the ppm scale indicates the presence of secondary structure, and the existence of high-field shifted peaks around the reference (DSS at 0 ppm) indicates the presence of a hydrophobic core in the protein. Therefore, the structural and the biological data were in agreement, as expected.
We propose the use of simple and fast 1D 1H NMR spectra to validate refolding protocols before increasing protein production. This technique is able to detect very small conformational changes in protein structure, as 1H NMR is sensitive to changes in the environment of each hydrogen atom of a molecule. By searching the REFOLD database28, we found that 34 of 1001 refolded proteins used this NMR spectroscopy in some way. In the majority of cases, the studies were driven by NMR, and the refolded protein was produced and isotopically labeled for analysis. Only three proteins from the database used simple 1D 1H NMR spectra, two of them for visual inspection29,30, and one for comparison with the natural modified protein31. In a recent study32, the authors compared the 1D 1H NMR spectra from the refolded protein and the protein purified from soluble fraction. They concluded that the structures were exactly the same because the spectra were identical (no spectral difference was presented). In a recent review33 focused on tools used to evaluate protein conformations, NMR is briefly mentioned as one of the leading techniques of structural biology. The authors said that NMR “still requires very sophisticated equipment, appreciable amounts of sample, and specialist human resources that keep them away from the pipeline in protein production and control. Newer techniques (or new applications of well-established techniques) are being developed to complement these traditional biophysical assays and to provide information on the success of protein folding.” We want to stress that the proposed method uses the detailed information present in NMR spectra in a simple way, to answer a simple question. If NMR (as x-ray crystallography) is analyzed as a tool to determine three-dimensional protein structure, then it presents the difficulties pointed out in the aforementioned review; however, this is not the case for the 1D 1H spectra needed for conformational comparisons of two samples.
Conclusion
One-dimensional homonuclear 1H NMR spectroscopy requires no special sample production and little time for measurement. Typically, a high quality spectrum of a 500 μL protein sample (~100 μM of a small protein) on a 600 MHz spectrometer can be obtained in less than 30 minutes. Most of the time is spent on adjustment of the equipment and definition of experimental parameters. Using spectrometers with a cryoprobe is desirable, but not required. Spectra processing and comparison is very quick and can be performed immediately after acquisition using spectrometer software.
Finally, simple precautions should be taken for the preparation of NMR samples. The NMR buffer should be of neutral to acidic pH and low ionic strength. High pH results in less protonated amides, causing loss of signal. Low salt concentration (up to 300 mM) is required to minimize radio wave reflections during NMR pulses that can cause heating of the sample (tuning and matching of the spectrometer probe head). 1H NMR is very sensitive to non-water exchangeable hydrogens of small molecules, and, as a consequence, the use of hydrogen-containing buffer agents (like Tris, Acetate, MES, and HEPES) and some other molecules (glycerin, DTT, etc.) should be avoided, or these reagents can be replaced by their deuterated equivalents. Sodium or potassium phosphate buffer is recommended, since the hydrogen atoms of these compounds are exchangeable with water, resulting in a signal that is coincident with the water resonance that should be suppressed. Moreover, extensive care with contaminants should be taken, as 1H NMR can detect contaminant signals, even in small concentrations.
Methods
Protein expression and cell lysis
P21-His6 was expressed in Escherichia coli BL21 cells transformed with a pET-28a(+) (Novagen) expression vector, which contains an N-terminal His6-tag followed by a thrombin recognition site. Expression was carried out in 8 L of 2xYT medium (16 g tryptone, 10 g yeast extract, 5 g NaCl per 1 L medium) containing 35 μg/mL kanamycin. Bacteria were incubated at 37°C until the optical density at 600 nm (OD600) reached ~0.7, at which point expression was induced by addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cultures were then incubated for an additional 3 hours at 37°C. Cells were harvested by centrifugation at 6,000 × g for 40 minutes at 4°C and resuspended in lysis buffer (50 mM Tris, 300 mM NaCl, 20 mM imidazole, pH 8.0) containing 1 mg/mL lysozyme. The solution was maintained on ice for 30 min before cells were lysed by ultrasound (15 cycles of 40 s at 40 W, 40 s interval). The lysate solution was subjected to centrifugation (14,000 × g for 40 min at 4°C), and both supernatant and pellet were used for native and denaturing purification of the protein, respectively.
Native P21-His6 purification from the soluble fraction (sP21-His6)
The soluble fraction was added to 2 mL of Ni-NTA resin (Qiagen) and extensively washed with lysis buffer. To remove low-affinity bound proteins, a second wash with wash buffer (50 mM Tris, 300 mM NaCl, 50 mM imidazole, pH 8.0) was performed. sP21-His6 was eluted with elution buffer I (50 mM Tris, 300 mM NaCl, 160 mM imidazole, pH 8.0), concentrated, and dialyzed against buffer II (50 mM Tris, 50 mM NaCl, pH 8.0).
Denaturing P21-His6 purification and refolding from the insoluble fraction (rP21-His6)
In order to extract the expressed protein from inclusion bodies, the lysate pellet was resuspended in urea buffer (50 mM Tris, 300 mM NaCl, 5 mM imidazole, 6 M urea, 5% (v/v) glycerol, pH 8.0) and stirred overnight at room temperature. After centrifugation (14,000 × g for 40 min at 4°C), the supernatant, containing P21-His6 extracted from the inclusion bodies, was immobilized in a column containing 5 mL Ni-NTA resin fixed on an Äkta Purifier system. After extensive washing with urea buffer, the urea concentration was slowly decreased with a linear buffer gradient (−7 mM/min of urea, 0.5 mL/min)23. To remove glycerol and additional impurities, the column was washed with wash buffer until no protein signal was detected. rP21-His6 was eluted with elution buffer II (50 mM Tris, 300 mM NaCl, 220 mM imidazole, pH 8.0), concentrated, and dialyzed against buffer II (see above).
Cell lines and cultures
Vero cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Cultilab), 10 mg/mL streptomycin (Sigma), 100 U/mL penicillin (Sigma), and 40 mg/mL gentamycin (Sigma) at 37°C in a humidified atmosphere containing 5% CO2. Peritoneal macrophages from BALB/C mice were harvested from the peritoneal cavity in 5 mL DMEM. Mice were stimulated with 1 mL 3% thioglycollate medium 3 days prior to harvest.
Parasites
To generate tissue-cultured trypomastigotes (TCT) from the CL strain, confluent monolayers of Vero cells were infected with metacyclic trypomastigotes and maintained for at least two weeks to establish the intracellular cycle. Extracellular amastigotes (EA) were derived from trypomastigotes by axenic differentiation. Infected Vero cell cultures were subjected to centrifugation (2500 × g, 5 minutes), and the pellet was resuspended in liver infusion tryptose (LIT) medium. Finally, the resuspension was incubated for 18 hours at 37°C to generate at least 95% pure extracellular amastigotes.
Animals and ethics
Male or female BALB/c mice were six to eight weeks old and were maintained under standard conditions on a 12 hour light, 12 hour dark cycle in a temperature controlled room (25 ± 2°C) with food and water ad libitum. All experimental protocols were approved by the Ethics Committee from Universidade Federal de Uberlândia. Moreover, maintenance and care of these animals complied with the guidelines of the Laboratory Animal Ethics Committee from the Institution. Animal euthanasia was performed in accordance with international welfare grounds, according to the American Veterinary Medical Association Guidelines on Euthanasia.
Host cell invasion assay
A volume of 500 μL Vero cell suspension (1.5 × 105 cells) was added to multiwell plates containing sterile glass coverslips and incubated overnight. The invasion assay was performed by adding EA (20 parasites per cell) or TCT (10 parasites per cell) to plates in the presence or absence of 40 μg/mL sP21-His6 or 40 μg/mL rP21-His6. The plates were incubated for 2 hours at 37°C in a humidified incubator containing 5% CO2. After incubation, the cells were gently washed three times with PBS, fixed with Bouin's solution, and stained with Giemsa.
Phagocytosis assay
Zymosan particles (10 mg/mL, Sigma) were resuspended in sterile PBS, sonicated for 15 minutes, and pelleted by centrifugation. To determine particle concentration, a Neubauer chamber was used for counting. Resident peritoneal macrophages were seeded in cell culture well plates (5 × 105 cells/well) containing 13 mm round glass coverslips and were then incubated overnight in a humidified atmosphere containing 5% CO2 at 37°C to allow the cells to adhere. Non-adherent cells were removed by washing with PBS before adding DMEM containing 10% FBS. To perform the phagocytosis assay, zymosan particles were added to wells (20 particles/each macrophage fixed) in the presence or absence of 40 μg/mL sP21-His6. Particles were incubated with macrophages for 2 hours. The cells were then washed with PBS, fixed in Bouin's solution, and stained with Giemsa. Results were expressed as the number of internalized particles/100 cells. These results were compared to those previously determined for rP21-His6 using the same protocol22.
Circular dichroism spectropolarimetry (CD)
Spectra were recorded on a Jasco 815 CD spectrometer using 0.1 cm thick quartz cells. Samples containing 0.15 mg/mL sP21-His6 or rP21-His6 in 10 mM sodium phosphate buffer (pH 7.4) were analyzed at 20°C to assess structural similarity.
Nuclear magnetic resonance spectroscopy (NMR)
Samples of sP21-His6 and rP21-His6 were concentrated to the same concentration (1.3 mg/mL, 70 μM in a final volume of 500 μL) and then simultaneously dialyzed against 2 L NMR buffer (50 mM potassium phosphate, 150 mM KCl, pH 6.0) at 4°C. The NMR buffer was changed twice to ensure that the samples were in exactly the same buffer conditions for NMR measurements, as 1H signals are very sensitive to changes in their environment. DSS (100 μM, for spectrum referencing) and D2O (5% v/v), for tuning and matching the spectrometer probe head) were added to both samples. 1D 1H spectra were recorded at 303 K in a high-resolution, small bore 600 MHz Bruker Avance III spectrometer. Since the spectrometer was equipped with a TCI cryoprobe, a 4× enhancement in the signal intensity was achieved. The intense water signal around 4.8 ppm, typical for an aqueous protein sample, was suppressed using a standard watergate W5 pulse sequence. To ensure spectra with a good signal to noise ratio were obtained for the diluted samples, each 1D experiment was acquired with a total of 256 scans, resulting in an experimental time of approximately 17 minutes per spectrum. Proton chemical shifts were referenced (0 ppm) to the 1H resonance frequency of the sharp and intense signal of the methyl groups in DSS. Time-domain data were acquired and processed with an exponential line broadening of 1 Hz using the Bruker BioSpin spectrometer control software TopSpin 3.1. The same software was used for the spectra analysis and for obtaining the difference spectrum.
Author Contributions
M.A.S. performed re-folding purification, the biological experiments, acquired, analyzed and interpreted the data and drafted the manuscript; F.B.T. performed in-column re-folding purification and NMR experiments, acquired, analyzed and interpreted the data and drafted the manuscript; H.H.T.M. performed soluble fraction purification, CD and biological experiments, acquired, analyzed and interpreted the data and drafted the manuscript; A.A.R., F.C.M., T.M.C., P.C.B., R.T.S., C.P. & R.G.B.G. performed biological experiments; D.B. & R.A.M. critically revised the manuscript and contributed to its intellectual content; C.E.M. designed the in-column refolding purification and NMR experiments, drafted the manuscript and critically revised the manuscript and contributed to its intellectual content; C.V.S. designed the biological experiments, drafted the manuscript and critically revised the manuscript and contributed to its intellectual content; E.H. designed soluble fraction purification and CD experiments, drafted the manuscript and critically revised the manuscript and contributed to its intellectual content. All the authors approved the final version to be submitted.
Acknowledgments
This study was supported by grants and fellowships from FAPEMIG, CAPES, FAPESP (grants 2010/51867-6, 2012/21153-7), CNPq and INBEQMeDI. CVS was supported by grants from Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG - Processes numbers: APQ-00621-11, APQ-00305-12); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (Process number: 23038.005295/2011-40).
References
- Juergen B. et al. Quality control of inclusion bodies in Escherichia coli. Microb. Cell Fact. 9, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christendat D. et al. Structural proteomics of an archaeon. Nat. Struct. Biol. 7, 903–909 (2000). [DOI] [PubMed] [Google Scholar]
- Service R. F. Structural genomics - Tapping DNA for structures produces a trickle. Science 298, 948–950 (2002). [DOI] [PubMed] [Google Scholar]
- Singh S. M. & Panda A. K. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 99, 303–310 (2005). [DOI] [PubMed] [Google Scholar]
- Eiberle M. K. & Jungbauer A. Technical refolding of proteins: Do we have freedom to operate? Biotech. J. 5, 547–559 (2010). [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Yamamoto E., Mannen T. & Nagamune T. Protein refolding using chemical refolding additives. Biotech. J. 8, (2013). [DOI] [PubMed] [Google Scholar]
- Belorgey D., Crowther D. C., Mahadeva R. & Lomas D. A. Mutant neuroserpin (S49P) that causes familial encephalopathy with neuroserpin inclusion bodies is a poor proteinase inhibitor and readily forms polymers in vitro. J. Biol. Chem. 277, 17367–17373 (2002). [DOI] [PubMed] [Google Scholar]
- Ward R. J. et al. Refolding and purification of bothropstoxin-1, a Lys49 - Phospholipase A(2) homologue, expressed as inclusion bodies in Escherichia coli. Protein Expression Purif. 21, 134–140 (2001). [DOI] [PubMed] [Google Scholar]
- Nishiyama Y. et al. Refolding from denatured inclusion bodies, purification to homogeneity and simplified assay of MGDG synthases from land plants. Protein Expression Purif. 31, 79–87 (2003). [DOI] [PubMed] [Google Scholar]
- Sharma S. K., Goloubinoff P. & Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochem. Biophys. Res. Commun. 372, 341–345 (2008). [DOI] [PubMed] [Google Scholar]
- Li D. W. et al. Refolding and characterization of recombinant human GST-PD-1 fusion protein expressed in Escherichia coli. Acta Biochim. Biophys. Sin. 36, 141–146 (2004). [DOI] [PubMed] [Google Scholar]
- Jang S. H., Kang D. K., Chang S. I., Scheraga H. A. & Shin H. C. High level production of bovine angiogenin in E-coli by an efficient refolding procedure. Biotechnol Lett. 26, 1501–1504 (2004). [DOI] [PubMed] [Google Scholar]
- Gustafsson E. et al. Purification of truncated and mutated Chemotaxis Inhibitory Protein of Staphylococcus aureus-an anti-inflammatory protein. Protein Express. Purif. 63, 95–101 (2009). [DOI] [PubMed] [Google Scholar]
- Rosenberger F. Protein crystallization. J. Cryst. Growth 166, 40–54 (1996). [Google Scholar]
- Rosenberger F., Vekilov P. G., Muschol M. & Thomas B. R. Nucleation and crystallization of globular proteins - What we know and what is missing. J. Cryst. Growth 168, 1–27 (1996). [Google Scholar]
- Lorber B., Skouri M., Munch J. P. & Giege R. The influence of impurities on protein crystallization: the case of lysozyme. J. Cryst. Growth 128, 1203–1211 (1993). [Google Scholar]
- Heijna M. C. R., van Enckevort W. J. P. & Vlieg E. Growth inhibition of protein crystals: A study of lysozyme polymorphs. Cryst. Growth Des. 8, 270–274 (2008). [Google Scholar]
- McPherson A., Malkin A. J., Kuznetsov Y. G. & Koszelak S. Incorporation of impurities into macromolecular crystals. J. Cryst. Growth 168, 74–92 (1996). [Google Scholar]
- Wüthrich K. in NMR of Proteins and Nucleic Acids (John Wiley & Sons, New York, 1986). [Google Scholar]
- Kalbitzer H. R. Method for verifying the correct spatial structure of molecules by means of NMR spectroscopy. US Patent 20060155480 filed 11 jan. 2005, and issued 13 Jul. 2006.
- da Silva C. V. et al. Characterization of a 21 kDa protein from Trypanosoma cruzi associated with mammalian cell invasion. Microb. Infect. 11, 563–570 (2009). [DOI] [PubMed] [Google Scholar]
- Rodrigues A. A. et al. A Recombinant Protein Based on Trypanosoma cruzi P21 Enhances Phagocytosis. PLoS ONE 7, e51384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier G., Bakalara N. & Santarelli X. On-column refolding of an insoluble histidine tag recombinant exopolyphosphatase from Trypanosoma brucei overexpressed in Escherichia coli. J. Chromatogr. B 786, 305–309 (2003). [DOI] [PubMed] [Google Scholar]
- Glynou K., Ioannou P. C. & Christopoulos T. K. One-step purification and refolding of recombinant photoprotein aequorin by immobilized metal-ion affinity chromatography. Protein Express. Purif. 27, 384–390 (2003). [DOI] [PubMed] [Google Scholar]
- Barb A. W., Wang X. & Prestegard J. H. Refolded recombinant Siglec5 for NMR investigation of complex carbohydrate binding. Protein Express. Purif 88, 183–189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.-Y., Chen L.-J. & Sun Y. Refolding and purification of histidine-tagged protein by artificial chaperone-assisted metal affinity chromatography. J. Chromatogr. A 1216, 5207–5213 (2009). [DOI] [PubMed] [Google Scholar]
- Hutchinson M. H. & Chase H. A. Adsorptive refolding of histidine-tagged glutathione S-transferase using metal affinity chromatography. J. Chromatogr. A 1128, 125–132 (2006). [DOI] [PubMed] [Google Scholar]
- Chow M. K. M. et al. The REFOLD database: a tool for the optimization of protein expression and refolding. Nucleic Acids Res. 34, D207–D212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. et al. Characteristics of a de-novo designed protein. Protein Sci. 3, 419–427 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrzenjak A., Frank S., Maderegger B., Sterk H. & Kostner G. M. Apo(a)-kringle IV-type 6: expression in Escherichia coli, purification and in vitro refolding. Protein Eng. 13, 661–666 (2000). [DOI] [PubMed] [Google Scholar]
- Assadi-Porter F. M., Aceti D. J., Cheng H. & Markley J. L. Efficient production of recombinant brazzein, a small, heat-stable, sweet-tasting protein of plant origin. Arch. Biochem. Biophys. 376, 252–258 (2000). [DOI] [PubMed] [Google Scholar]
- Ogura K. et al. Practical applications of hydrostatic pressure to refold proteins from inclusion bodies for NMR structural studies. Protein Eng. Des. Sel. 26, 409–416 (2013). [DOI] [PubMed] [Google Scholar]
- Manta B., Obal G., Ricciardi A., Pritsch O. & Denicola A. Tools to evaluate the conformation of protein products. Biotech. J. 6, 731–741 (2011). [DOI] [PubMed] [Google Scholar]