Abstract
During embryonic development, maintenance of cell identity and lineage commitment requires the Polycomb-group PRC2 complex, which catalyzes histone H3 lysine 27 trimethylation (H3K27me3). However, the developmental origins of this regulation are unknown. Here we show that H3K27me3 enrichment increases from blastula stages onward in embryos of the Western clawed frog (Xenopus tropicalis) within constrained domains strictly defined by sequence. Strikingly, although PRC2 also binds widely to active enhancers, H3K27me3 is only deposited at a small subset of these sites. Using a Support Vector Machine algorithm, these sequences can be predicted accurately on the basis of DNA sequence alone, with a sequence signature conserved between humans, frogs, and fish. These regions correspond to the subset of blastula-stage DNA methylation-free domains that are depleted for activating promoter motifs, and enriched for motifs of developmental factors. These results imply a genetic-default model in which a preexisting absence of DNA methylation is the major determinant of H3K27 methylation when not opposed by transcriptional activation. The sequence and motif signatures reveal the hierarchical and genetically inheritable features of epigenetic cross-talk that impose constraints on Polycomb regulation and guide H3K27 methylation during the exit of pluripotency.
The spectacular development of a complex multicellular organism is precisely regulated by epigenetic mechanisms. The chromatin landscape has been extensively studied in mammalian cell lines (Bhaumik et al. 2007; Mikkelsen et al. 2007), but little is known about how this landscape emerges during development. During embryogenesis, the Polycomb group (PcG) proteins are essential for patterning and stable lineage commitment (Simon and Kingston 2009; Margueron and Reinberg 2011; Bogdanović et al. 2012). These proteins are associated with two complexes, Polycomb repressive complex 1 and 2 (PRC1, PRC2).
The PRC2 complex is conserved from plants to vertebrates and catalyzes trimethylation of lysine 27 of histone 3 (H3K27me3), which is associated with repression of developmental genes (Schuettengruber et al. 2009). One of the core components, E(Z) homolog 2 (EZH2), contains a SET domain and is responsible for the catalytic action of the PRC2 complex (Cao and Zhang 2004). A number of other proteins interact with PRC2 in a substoichiometric manner (Smits et al. 2013). One of these is JARID2, a JmjC domain-containing protein (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). Localization of PRC2 to the DNA depends on JARID2; however, there is evidence that JARID2 recruitment is in turn dependent on PRC2 (Peng et al. 2009; Shen et al. 2009; Li et al. 2010; Pasini et al. 2010). Although JARID2 can influence the catalytic action of PRC2, the exact mechanism remains unclear.
The H3K27me3 modification can be bound by the PRC1 complex, which causes compaction of the chromatin (Margueron and Reinberg 2011). Although historically, PRC1 has been thought to act downstream from PRC2, there is mounting evidence that PRC1 functions independently of PRC2 (Schoeftner et al. 2006; Tavares et al. 2012).
Much is yet unclear about how the PcG complexes are directed to the chromatin. There is evidence supporting both sequence-directed and sequence-independent mechanisms. In Drosophila, Polycomb response elements (PREs)—with a combination of motifs for a number of transcription factors (TFs) such as PHO, GAGA factor (GAF), and zeste—have been implicated in PRC2 recruitment (Ringrose et al. 2003; Schuettengruber et al. 2009). Although binding of PHO is indeed observed in nearly all Polycomb domains (Schuettengruber et al. 2009), methods to predict PREs based on these motifs perform very poorly (Zeng et al. 2012). In vertebrates, PRE sequences remain largely elusive. The closest vertebrate homolog of PHO, YY1, has been associated with PcG function (Atchison et al. 2003; Woo et al. 2010), but its binding motif is depleted in Polycomb domains (Ku et al. 2008; Liu et al. 2010). Two vertebrate PREs have been identified (Sing et al. 2009; Woo et al. 2010), however, these are relatively large fragments (1.8 and 3 kb), and the exact sequences required for binding and repression are unknown.
Rather than specific TF binding motifs, CpG islands do seem to play a major role in PcG protein recruitment in mammals. GC-rich sequences can recruit PRC2, and CpG islands devoid of activating motifs are sufficient for initial localization of PRC2 and subsequent methylation of H3K27 (Ku et al. 2008; Mendenhall et al. 2010). Similarly, a high density of CpG dinucleotides is sufficient for Polycomb recruitment in mouse (Lynch et al. 2012). One possible mechanism for PRC1 recruitment to CpG islands is through KDM2B, which binds to unmethylated CpGs via its CxxC zinc finger domain (Farcas et al. 2012). Other sequence-based recruitment mechanisms include interactions with sequence-specific TFs such as REST (Dietrich et al. 2012; Arnold et al. 2013) and RUNX1 (Yu et al. 2012).
Finally, several long noncoding RNAs (lncRNAs) have been identified that influence Polycomb-dependent H3K27 methylation (Rinn et al. 2007; Zhao et al. 2008; Tsai et al. 2010). These lncRNAs may serve as scaffolds by providing binding surfaces for assembly of specific histone modification complexes (Tsai et al. 2010).
To study the earliest dynamics of regulation by Polycomb in the context of vertebrate development, we studied the dynamics and activity of PRC2 in Xenopus tropicalis embryos. H3K27 is mostly newly methylated at promoter-distal positions, with a dramatic increase in enrichment from blastula to gastrula. H3K27 methylation is constrained to preexisting DNA methylation-free domains, which can be accurately predicted from the primary DNA sequence using a Support Vector Machine (SVM) algorithm. Specific regulatory sequence motifs can further distinguish between active and repressed methylation-free regions. These conserved, cell-type–independent signals dynamically guide Polycomb repression during the exit of pluripotency at the beginning of gastrulation.
Results
The repressive H3K27me3 modification is newly deposited from blastula stages onward
To characterize the dynamics of H3K27me3 nucleation and spreading in X. tropicalis, we generated epigenetic profiles using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) at four developmental stages. We assayed H3K4me3, H3K27me3, the enhancer mark H3K4me1, the Polr2a subunit of RNA Polymerase II (RNAPII), Ezh2, Jarid2, and a ChIP input track as control (see Methods) (Table 1; Supplemental Table 1).
Table 1.
Overview of the ChIP-sequencing data in this study

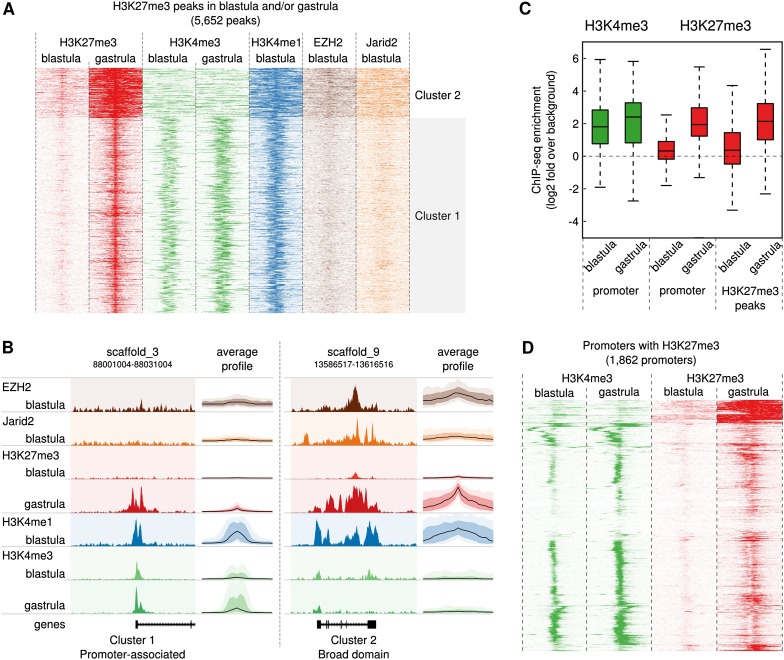
We identified three clusters of temporal H3K27 methylation patterns (Fig. 1A,B; Supplemental Figs. 1, 2). Cluster 1 is promoter-associated, with H3K27me3 constrained to a ∼10-kb region centered at the gene promoter (Fig. 1B, left panel). Cluster 2 shows large domains (up to ∼100 kb) completely covering one or more genes (Fig. 1B, right panel). Cluster 3 consists of a set of small sequences with no clear association with genes (Supplemental Fig. 1). Genes regulated by H3K27me3 are enriched for developmental transcription factors and functions in development, pattern formation, and morphogenesis (Supplemental Table 2). Promoters show very low levels of H3K27me3 at the blastula stage, with a strong increase at gastrulation (Fig. 1C), whereas H3K4me3 is already enriched at the blastula stage. Indeed, a genome-wide analysis of all 1862 promoters that gain H3K27me3 confirms the developmental hierarchy of permissive and repressive modifications (Fig. 1D; Akkers et al. 2009). However, we do observe some sites of early H3K27me3 nucleation. In many broad H3K27me3 domains, methylation of H3K27 is initiated at a local site. The majority of these sites do not correspond to gene promoters and show no H3K4me3 enrichment (Supplemental Figs. 3, 4). We did not observe a significant overlap with noncoding RNA genes (Supplemental Table 4), and TF motif analysis yielded no enriched motifs. The promoter-distal locations of early H3K27 methylation sites, however, raised the question as to how H3K27 methylation relates to both PRC2 and enhancer sites.
Figure 1.
H3K27me3 is newly deposited from blastula stages onward. (A) Heatmap of a k-means clustering analysis (k = 3, Euclidian distance) of H3K27me3 (blastula and gastrula), H3K4me3 (blastula and gastrula), H3K4me1, Ezh2, and Jarid2 (blastula) in 10-kb regions around H3K27me3 peak summits. The rows correspond to the peaks, the x-axis shows the position relative to the peak center. The intensity of the color represents the number of reads in 100-bp windows. The first two clusters are shown: for cluster 3, see Supplemental Figure 1. (B) Average profile and representative example of the two clusters shown in A. The average profiles show the median enrichment (black line) and the 50th and 90th percentiles in a darker and lighter color, respectively. (C) Boxplot showing enrichment of H3K4me3 (green, left) and H3K27me3 (red, right) in 2-kb regions around the TSS for genes marked by H3K27me3 in the blastula or gastrula stage (“promoter”), and all peaks for H3K27me3 in any stage (“H3K27me3 peaks”). The y-axis represents log2 of the fold enrichment compared with randomly selected genomic sequences. The background level (no enrichment; log2 of 0) is marked by a dotted line. (D) H3K4me3 (green) and H3K27me3 (red) enrichment in blastula- and gastrula-stage embryos around transcription start site (TSS; ±5 kb) marked by H3K27me3 in the gastrula. Regions were clustered using hierarchical clustering with the Euclidian distance metric.
Enhancers recruit PRC2
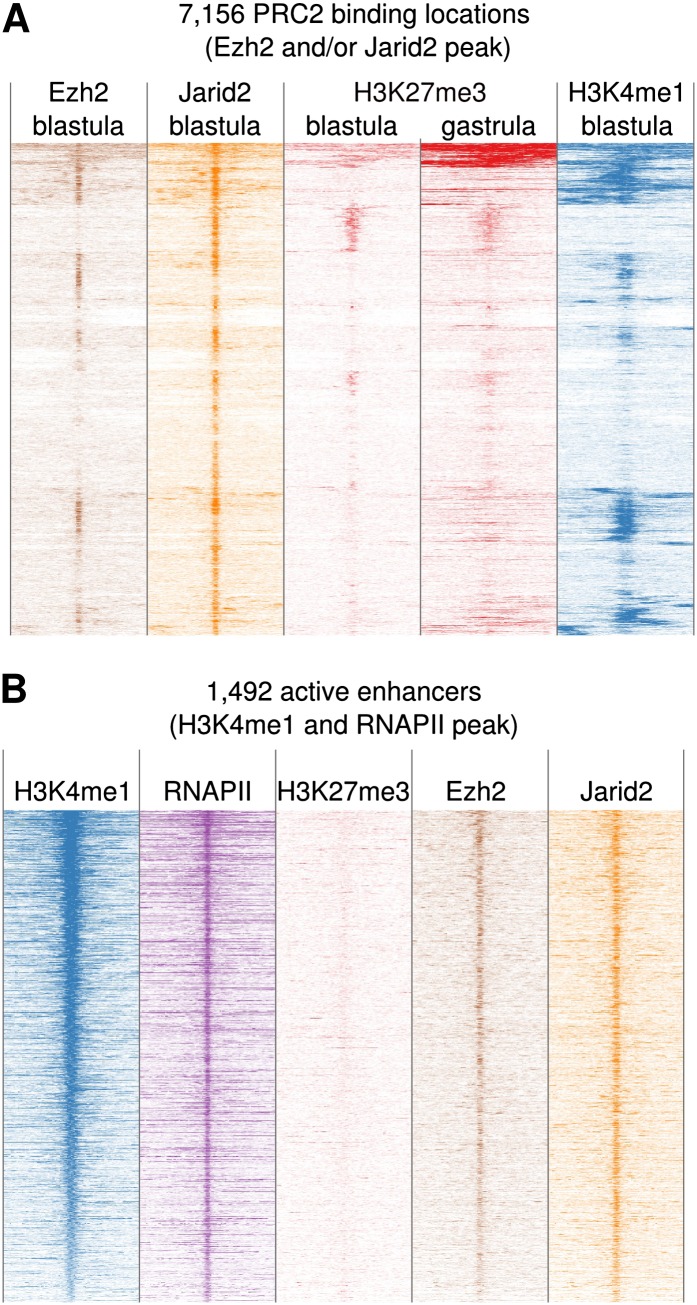
We therefore assessed H3K27me3 dynamics at PRC2 subunits Ezh2 and Jarid2 binding sites. Surprisingly, only a small subset of the PRC2 binding sites gains H3K27me3 during subsequent development to the tailbud stage (Fig. 2A; Supplemental Fig. 5). This suggests that binding of the methyltransferase Ezh2 is either very unstable or not sufficient for H3K27 methylation at many loci. The lack of chromatin-associated H3K27me3 is not explained by depletion of histones, as most regions bound by both Ezh2 and Jarid2 binding have the enhancer mark H3K4me1 (Fig. 2A).
Figure 2.
PRC2 is recruited to the majority of active developmental enhancers. (A) The majority of PRC2 binding sites do not gain H3K27me3. Heatmap of a hierarchical clustering of Ezh2, Jarid2, H3K27me3, and H3K4me1 enrichment around the summits of Ezh2 and Jarid2 peaks. Data are visualized in 100-bp bins in 10-kb regions. (B) PRC2 is recruited to active enhancers. Enrichment of H3K4me1, RNAPII, H3K27me3, Ezh2, and Jarid2 at active enhancers. Data are visualized in 100-bp bins in 10-kb regions and are sorted by H3K4me1. Enhancers are defined as RNAPII peaks in regions with H3K4me1 but not H3K4me3.
To assess the relationship with enhancers, H3K4me1 peaks with RNAPII (Kim et al. 2010), but not H3K4me3, were selected, and the enrichment for H3K27me3, Ezh2, and Jarid2 was determined (Fig. 2B). The results show that PRC2 is recruited to the majority of active enhancers at the pluripotent blastula stage. Consistently, these sites contain motifs of important developmental regulators (Supplemental Fig. 6). The POU, Forkhead, Homeobox, HMG-box, and T-box motif families are all significantly enriched, indicating that enhancers from both pluripotency and lineage specification networks are bound by PRC2.
Comparative analysis of sequence features of vertebrate H3K27me3 domains
To determine the involvement of DNA sequence in establishing H3K27 methylation, we evaluated TF motifs and repeat content in broad H3K27me3 domains that are established by the gastrula stage and, moreover, compared nucleosome positioning signals, GC content, and CpG islands in Xenopus, zebrafish, and human in a comparative analysis.
First, from known TF motifs, only the REST motif was significantly enriched (Supplemental Fig. 7). Although REST is known to associate with Polycomb complexes (Dietrich et al. 2012; Arnold et al. 2013), this motif is present in only ∼1% of H3K27me3 domains. No other TF binding motifs were significantly associated with H3K27me3 domains.
Second, identification of overrepresented repeats shows that large H3K27me3 domains (Cluster 2) are highly enriched for specific simple sequence repeat types (more than 50 times) such as TAGA/TCTA and CA/TG (Supplemental Material; Supplemental Table 3). This may reflect a tendency of increased DNA polymerase slippage in Polycomb-repressed chromatin.
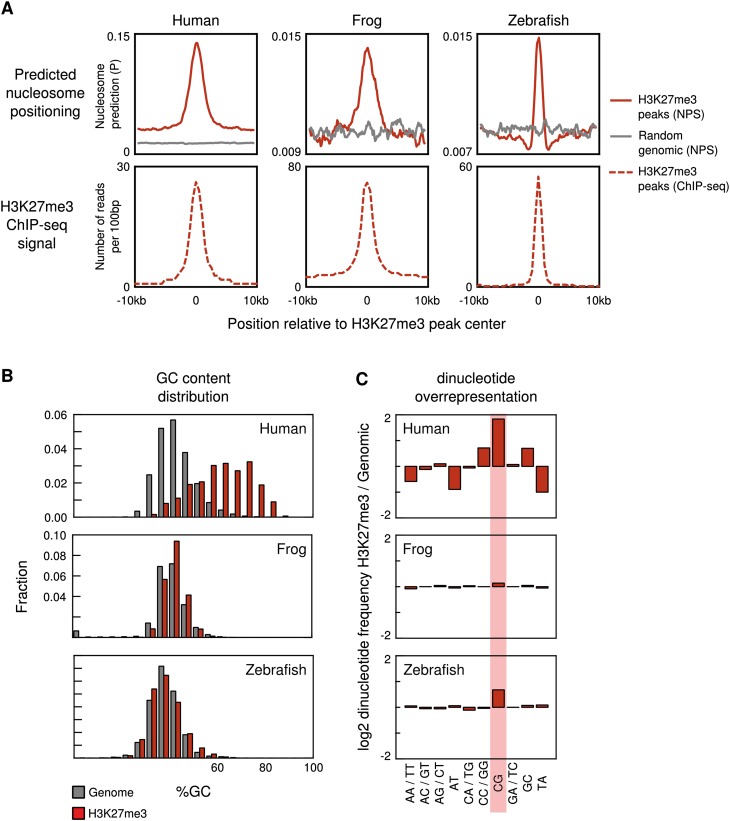
Third, to test the involvement of nucleosome positioning signals, we used a recent model for prediction of nucleosome positions from DNA sequence (van der Heijden et al. 2012). We observed a significantly higher predicted positioning signal in H3K27me3 domains compared with random genomic sequence (Fig. 3A, upper panels) in a pattern that closely follows the H3K27me3 enrichment profile (Fig. 3A, lower panels).
Figure 3.
Conserved and diverged sequence features of vertebrate H3K27me3 domains. (A) The top panel shows the predicted nucleosome occupancy of H3K27me3 domains (red line) and random genomic regions (gray line). The maximum nucleosome occupancy (van der Heijden et al. 2012) was calculated per 1 kb using a moving window (window size, 1 kb; step size, 100 bp). Shown is the mean of predicted maximum occupancy for all H27me3 domains or random genomic regions. The bottom panel shows the median ChIP-seq enrichment of H3K27me3 domains. (B) %GC of H3K27me3 domains and genomic background in human, Xenopus tropicalis, and zebrafish. (C) Overrepresentation of dinucleotides in human, X. tropicalis, and zebrafish H3K27me3 domains compared with the genomic background (log2 of the fold difference in dinucleotide frequency).
Finally, we analyzed the GC% and CpG enrichment of H3K27me3 domains. In human, as reported (Ku et al. 2008; Mendenhall et al. 2010), the GC% distribution of H3K27me3 domains is highly skewed compared with the genomic background (Fig. 3B, upper panel). In contrast, there is no significant GC% skewing in both frog and zebrafish (Fig. 3B, middle and lower panel). Similarly, the overrepresentation of the CG dinucleotide, which is very conspicuous in human (and mouse; data not shown), is only minor in zebrafish and absent in frog (Fig. 3C). Analysis of H3K27me3 enrichment at CpG islands shows similar results, with H3K27me3 at the majority of human CpG islands in contrast to frog and zebrafish (Supplemental Fig. 8). Therefore, CpG island characteristics do not constitute the pan-vertebrate sequence features of H3K27me3 domains, even though nucleosome positioning signals are conserved in these regions.
Pan-vertebrate DNA sequence signals of H3K27me3 domains
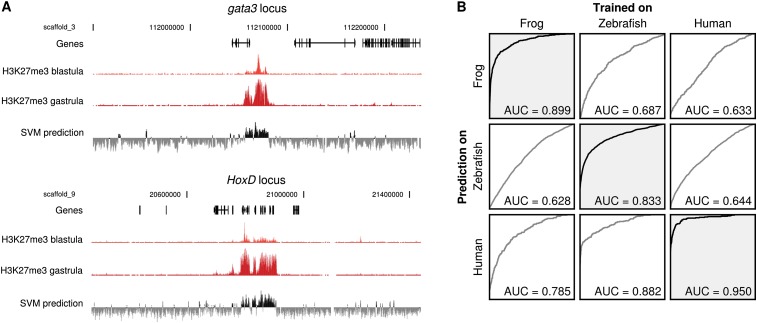
To use an alternative approach to analyze the DNA sequence properties of H3K27 methylation, we applied an SVM algorithm (Lee et al. 2011). This is a supervised machine learning approach that, in this particular case, uses all sequences of length k (k-mers) to distinguish a specific set of sequences from genomic background. It is trained with a subset of the genomic data (positive and negative sequences) to produce a classifier that can be used to predict the status of new sequences. The rest of the data that was not used for training can be used to test the performance of the SVM. We first trained an SVM on Xenopus H3K27me3 domains. Even though we found no enriched TF motifs in these regions except for the REST motif, and even though the GC% is similar to genomic background, an SVM trained on part of the genome is able to accurately classify H3K27me3-marked regions in the remaining part (Fig. 4A; Supplemental Fig. 9). Next, we analyzed the performance of the SVM in a cross-species analysis (Fig. 4B). Shown is the prediction performance as a receiver operating curve (ROC) that plots the fraction of true positives versus the fraction of false positives; a high area under the curve (AUC ∼ 1) corresponds to high accuracy and sensitivity. An SVM trained in any of the three species can predict H3K27me3 domains to a certain degree in a different species, uncovering pan-vertebrate sequence conservation in H3K27 methylation (Fig. 4B). The performance of the SVM is not based purely on GC% and CpG content as it also can distinguish natural H3K27me3 sequences from random regions with an identical dinucleotide background or from regions with a similar GC% sampled from the genome (Supplemental Fig. 10). By use of the SVM output from the three species, we identified motifs that are consistently enriched or depleted in H3K27me3 domains (Supplemental Material; Supplemental Figs. 11, 12). Remarkably, the unique sequence properties captured by the SVM trained on human H1 ES cells also identify cell-type–specific H3K27me3 domains that are not present in H1 ES cells (Supplemental Fig. 13), showing that the SVM detects a sequence-based, cell-type–independent propensity for H3K27 methylation.
Figure 4.
A k-mer Support Vector Machine (SVM) can accurately predict H3K27me3 domains from the primary sequence in vertebrates. (A) Examples of H3K27me3 domain prediction by a k-mer SVM using k = 8 (lower track; black is a positive score, gray a negative score). H3K27me3 enrichment in the blastula and gastrula is visualized in the top two tracks. (B) Interspecies performance of k-mer SVMs (k = 8) trained on different vertebrates. Performance is visualized in a receiver operator curve (ROC) and the ROC area under curve (ROC AUC) is shown as a performance measure. The y-axis shows the sensitivity; the x-axis, 1 − specificity. The columns indicate the species used for training; the rows indicate the species used for evaluation. For instance, the lower left ROC curve indicates the performance of human sequence classification of an SVM trained on frog.
H3K27 methylation in preexisting DNA methylation-free regions
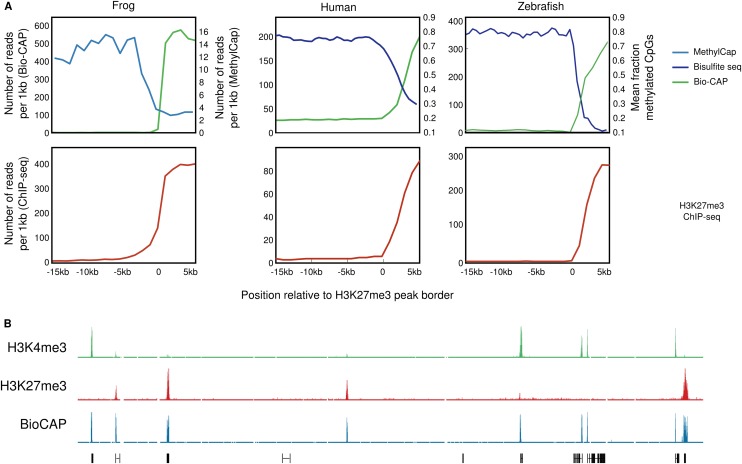
We previously showed that H3K27me3 and DNA methylation tend to be mutually exclusive using capture of methylated DNA by a MBD domain (MethylCap) (Bogdanović et al. 2011). Indeed, when the average MethylCap signal is visualized over H3K27me3 peak borders, we observe a paucity in DNA methylation in H3K27me3-enriched regions (Fig. 5A, left panel). Recently, it was shown by CXXC affinity capture (Bio-CAP) that nonmethylated islands (NMIs), rather than CpG islands, are a conserved feature of vertebrate genomes (Long et al. 2013). Indeed, the Bio-CAP signal confirms that H3K27me3 domains are unmethylated (Fig. 5A, left panel). In human and zebrafish, H3K27me3 is similarly enriched in regions depleted of DNA methylation (Fig. 5A). The NMIs in Xenopus do not correspond to the classic definition of CpG islands (cf. Fig. 3B; Long et al. 2013). Strikingly, we find that H3K27me3 is deposited during gastrulation in preexisting (blastula-stage) DNA methylation-free regions (Supplemental Fig. 14). However, this only concerns a subset of NMIs, as many NMIs have already gained H3K4me3 by the blastula stage without gaining H3K27me3. In Xenopus, all NMIs gain one or both of these histone modifications (Fig. 5B). Taken together, a paucity in DNA methylation is a conserved feature of H3K27me3 domains that is shared with many H3K4me3-enriched promoters, raising the question of which features could distinguish between unmethylated regions that gain and those that do not gain H3K27me3.
Figure 5.
H3K27 methylation in DNA methylation-free regions. (A) The methylation state (top panel) and H3K27me3 enrichment (bottom panel) visualized over H3K27me3 domain borders. The methylation state assessed by MethylCap (X. tropicalis; light blue) or bisulfite sequencing (human and zebrafish; dark blue) is shown, as well as the Bio-CAP signal (green). For H3K27me3 ChIP-seq, MethylCap, and Bio-CAP, the number of reads per 1 kb is shown from −15 kb to +5 kb relative to the H3K27me3 domain border. The bisulfite seq signal indicates the mean fraction of methylated CpGs. For data sets, see Supplemental Table 6. (B) Screenshot of a representative profile of H3K4me3 (green), H3K27me3 (red), and Bio-CAP (blue) ChIP-seq enrichment in X. tropicalis blastula-stage embryos.
TF motifs distinguish unmethylated regions that do or do not gain H3K27me3
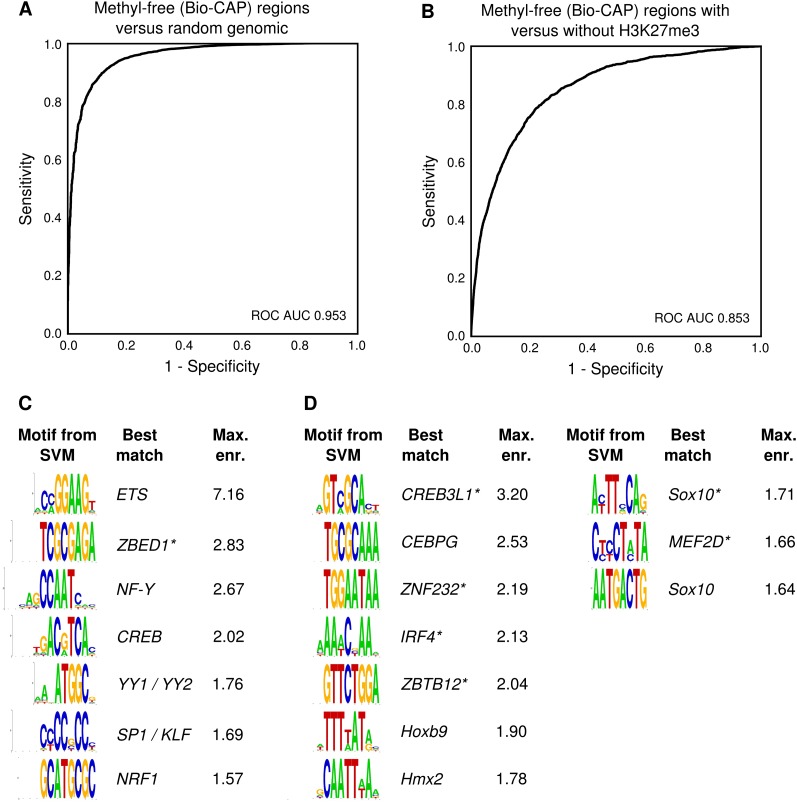
The H3K27me3-marked sequences form a subset of the NMIs in all three species. More precisely, H3K27me3 decorates DNA methylation-free regions at many developmentally regulated genes, whereas unmethylated islands near other genes, including housekeeping genes, gain H3K4me3 but not H3K27me3 (Fig. 5B). We therefore trained an SVM on DNA methylation-free regions and find it performs very well (ROC AUC 0.953) (Fig. 6A). Only 19% of the NMIs in Xenopus gain H3K27me3. However, an SVM specifically trained to distinguish between NMIs with and without H3K27me3 is able to separate them with high performance (ROC AUC 0.853) (Fig. 6B). Interestingly, the k-mers with the highest and lowest weights of the SVM correspond to important regulatory motifs (Fig. 6C,D). NMIs without H3K27me3 are characterized by motifs of housekeeping TFs such as ETS, NFY, CREB, and YY1 (Fig. 6C). These correspond to the motifs that were previously identified in mouse CpG islands without H3K27me3 (Ku et al. 2008). However, we find that NMIs that gain H3K27me3 show enrichment of motifs for developmental regulators such as Sox and Homeobox TFs (Fig. 6D). Therefore, TF motifs distinguish unmethylated DNA elements that do or do not acquire H3K27me3.
Figure 6.
Transcription factor motifs predict the H3K27me3 state of DNA methylation-free regions. (A) ROC curve of the classification of X. tropicalis DNA methylation-free regions (Bio-CAP peaks) compared with random genomic regions using a k-mer SVM (k = 8). (B) ROC curve of the classification of DNA methylation-free regions (Bio-CAP peaks) using a k-mer SVM (k = 8). The regions are classified as either gaining H3K27me3 or remaining devoid of H3K27me3. (C) Motifs from k-mers with a high SVM weight, which are enriched in (active) DNA methylation-free regions without H3K27me3. The best match to known motifs is shown; uncertain matches are marked with an asterisk. (D) Motifs from k-mers with a low SVM weight that are enriched in (repressed) DNA methylation-free regions with H3K27me3.
Blastula-stage H3K27me3 peaks function as repressive elements in Xenopus and mouse
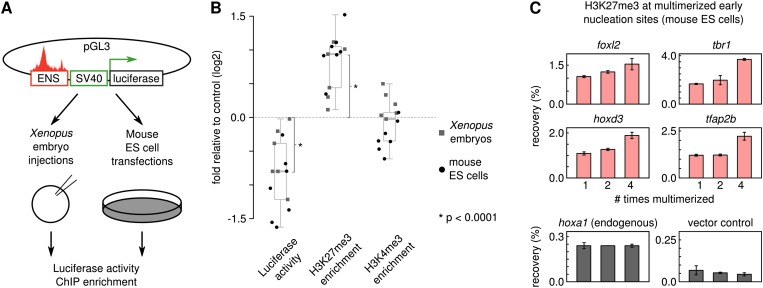
We wondered if regions carrying the SVM sequence signature could function as repressive elements. We selected sites with a positive SVM score that showed H3K27me3 enrichment in blastula-stage embryos. These sites (∼1 kb) were cloned into a pGL3 reporter vector and used for injection in Xenopus embryos and for transfection of mouse ES cells (Fig. 7A). In both experimental systems, these sites significantly repress luciferase expression (P < 0.001) and show H3K27me3 enrichment (P < 0.001), but no difference in H3K4me3 enrichment (Fig. 7B). This confirms that the pan-vertebrate conserved sequence signature we identified indeed directs H3K27 methylation and repression across species. None of the selected sites are CpG islands, and the mean GC% is 38%, lower than the overall GC% of both the Xenopus and the mouse genome (∼40%).
Figure 7.
Early H3K27me3 nucleation sites encode conserved repressive capacity. (A) Repression assay of H3K27me3 early nucleation sites in Xenopus embryos and mouse ES cells. (B) Early nucleation sites function as repressive elements. Shown is luciferase activity normalized to Renilla as fold change relative to control, H3K27me3 ChIP-qPCR enrichment relative to control, and H3K4me3 ChIP-qPCR enrichment relative to control (log2 scale; significance, Wilcoxon signed rank test; Xenopus, light square dots; ES cells, dark round dots). (C) H3K27me3 enrichment increases with length of susceptible sequence. Shown is the H3K27me3 ChIP-qPCR recovery for four SVM-positive H3K27me3 nucleation sites, an endogenous locus (hoxa1), and a vector backbone control.
H3K27 methylation is influenced by nucleosome density and by allosteric activation of Ezh2 (Margueron et al. 2009; Yuan et al. 2012). To assess the influence of sequence length, we tested the effect of multimerization on four H3K27me3 initiation sites in mouse ES cells. Figure 7C shows the H3K27me3 ChIP-qPCR recovery of the same 1-kb fragment multimerized one, two, or four times. The recovery increases almost linearly with the length of the inserted fragment, though the background signal of the vector backbone shows no such increase.
Discussion
Our interspecies analysis of Polycomb regulation uncovered principles of H3K27 methylation during embryonic development involving epigenetic cross-talk, sequence features, and a hierarchy of activation and repression. The developmental timing of H3K27 methylation from the pluripotent blastula stage onward in Xenopus embryos is consistent with a function in stable differentiation and lineage commitment (Bogdanović et al. 2012). The dynamics support reestablishment of repressive chromatin rather than inheritance from sperm or oocytes. This has previously been suggested for zebrafish (Lindeman et al. 2011) and is supported by analysis of histone modifications by mass spectrometry (Schneider et al. 2011).
The PRC2 complex is present at the majority of active developmental enhancers, possibly recruited by binding of PRC2 to enhancer RNAs (eRNAs) that are expressed at enhancers (Kanhere et al. 2010; Kim et al. 2010). However, only a subset of these enhancers gain the H3K27me3 mark. This implies additional regulation, such as activation or inhibition of Ezh2 through posttranslational modifications (Cha et al. 2005; Lan et al. 2007; Wei et al. 2011). Also, PRC2 may be ineffective in H3K27 methylation at these sites because of H3K27 acetylation, a hallmark of active enhancers.
Our results indicate that Polycomb recruitment in vertebrates is not necessarily mediated through a PRE, defined by a combination of repressive binding motifs. We find that overall susceptibility to H3K27 methylation is linked to specific sequence properties, absence of DNA methylation, and binding sites for cell-type–specific transcriptional activators. The conserved sequence signature, however, marks the Polycomb-regulated subgenome in a cell-type–independent fashion and may reflect a propensity for H3K27 methylation. Part of the sequence component reflects the presence of constitutive DNA methylation-free domains. This is very significant as binding of PRC2 to nucleosomes is counteracted by methylated DNA (Bartke et al. 2010). PRC2 binding and H3K27 methylation are also inhibited by gene activation, especially by the presence of the H3K4me3 permissive mark (Schmitges et al. 2011).
In humans, the conserved sequence signature correlates with CpG islands, which previously have been implicated in Polycomb recruitment (Ku et al. 2008; Mendenhall et al. 2010; Lynch et al. 2012). In contrast, Polycomb-regulated sequences in Xenopus and zebrafish do not exhibit a strong CpG or GC% enrichment. However, they are devoid of DNA methylation and can be accurately identified using an SVM. In addition, Xenopus H3K27me3 nucleation sites show repressive capacity in mouse ES cells, even though these sequences do not correspond to CpG islands.
Interestingly, we find that constitutive and cell-type–specific TF motifs distinguish between different DNA methylation-free regions that do or do not acquire H3K27 methylation. Supporting this model, a single point mutation in a binding site of the activating SP1 TF can result in repression by Polycomb (Caputo et al. 2013). The data suggest a genetic default mechanism for this type of facultative heterochromatin, in which default H3K27 methylation in domains of unmethylated DNA can be counteracted by dynamic and cell-type–dependent gene activation. The default nature of this repression is significant in light of the hierarchy between activation and repression, as H3K4me3 is acquired before localized H3K27 methylation represses multilineage gene expression (Akkers et al. 2009).
Our results indicate that it is not GC richness by itself that causes H3K27 methylation but, rather, associated sequence features and absence of DNA methylation in combination with an absence of activating signals. After initial recruitment of PRC2, the H3K27me3 modification spreads on unmethylated susceptible sequences due to the activating effect of nucleosome density (Yuan et al. 2012) and an allosteric positive feedback loop (Margueron et al. 2009). Indeed, the predicted nucleosome occupancy is significantly higher in H3K27me3 domains compared with the genomic average. During subsequent development and differentiation, Polycomb action may be complemented by gene-specific targeting mechanisms involving REST or other repressors.
Binding of PRC2 is reduced by DNA methylation, and H3K27me3 is also inhibited by other histone modifications (Bartke et al. 2010; Pasini et al. 2010; Schmitges et al. 2011), overriding the genetic-default state in different cell types. The underlying genomic sequence signature may function as a genetically inheritable constraint on Polycomb target selection, and the interplay with other epigenetic modifications may elegantly explain the stability of cell lineage commitment. An H3K27 methylation-default state within DNA methylation-free regions that are not transcriptionally activated may establish two major features of epigenetic regulation of development: (1) de novo reconstitution of the epigenetic landscape during early development and (2) locking cell lineage commitment by raising the activation threshold of multilineage gene expression after initial lineage specification at the onset of gastrulation.
Methods
Published data
All previously published, public data sets used in this study are summarized in Supplemental Table 6.
Animal procedures
X. tropicalis and Xenopus laevis embryos were obtained by in vitro fertilization, dejellied in 3% cysteine, and collected at the following Nieuwkoop-Faber stages: nine (blastula), 12 (gastrula), 16 (neurula), and 30 (tailbud).
Chromatin immunoprecipitation and antibodies
Chromatin for chromatin immunoprecipitation was prepared as previously described (Jallow et al. 2004; Akkers et al. 2012). The following antibodies were used: anti-H3K4me3 (Abcam ab8580), anti-H3K27me3 (Upstate/Millipore 07-449), anti-H3K4me1 (Diagenode CS-037-100), Jarid2 (Abcam ab48137), EZH2 (Active Motif 39103), and POLR2A (Diagenode AC-055-100).
For all ChIP-seq samples, three independent biological replicates of different chromatin isolations were pooled.
Sequencing and alignment
Sequencing samples were prepared according to the manufacturer's protocol (Illumina). Shortly, adapter sequences were ligated, the library was size-selected (200–300 bp), and amplified. The sequencing (36 cycles) was carried out on a Genome Analyzer IIx (Illumina) except for the Input track, which was sequenced on a HiSeq 2000 (Illumina). Reads passing the Illumina chastity filter were aligned to the X. tropicalis genome (version JGI 7.1) using BWA (Li and Durbin 2009). For processing and manipulation of SAM/BAM files, SAMtools was used (Li et al. 2009). All duplicate reads and reads mapping to repeat regions were removed.
Detection of enriched regions
We used PeakRanger version 1.15 (Feng et al. 2011) with the following parameters: FDR 1 × 10−6 (H3K27me3, H3K4me3) or 1 × 10−4 (Ezh2, Jarid2, RNAPII, H3K4me1), ext_length 300. All peaks were called relative to an input control track. Only scaffolds >2 Mb were included in the analysis.
Generation of profiles and heatmaps
All heatmaps and bandplot profiles were generated using fluff (http://simonvh.github.com/fluff). This Python package uses pysam (http://code.google.com/p/pysam/), pybedtools (Dale et al. 2011), and HTSeq (http://www-huber.embl.de/users/anders/HTSeq/) for BAM and BED manipulations and Pycluster (de Hoon et al. 2004) for k-means and hierarchical clustering. For all heatmap clustering, the Euclidian distance metric was used. For hierarchical clustering, we used the pairwise complete-linkage function.
Analysis of sequence features
As input for all sequence feature analysis, we used H3K27me3 domains of at least 1 kb from gastrula embryos (stage 12, X. tropicalis; shield, zebrafish) or ES cells (H1-hESC, human). To identify enriched motifs, we used “maxenr” from the GimmeMotifs package (van Heeringen and Veenstra 2011) to scan for all vertebrate motifs from JASPAR. All motifs present in at least 1% of the sequences with an enrichment of at least three times compared with random background sequences were reported. For the repeat analysis, see Supplemental Material. We calculated the average maximum predicted nucleosome occupancy using an empirical statistical mechanics model (Supplemental Material; van der Heijden et al. 2012). An equal number of regions was randomly selected from the same genome and analyzed in the same manner.
Bio-CAP analysis
Peaks were called using MACS 2 (Zhang et al. 2008) relative to the input. The SVM was trained on 1-kb peaks centered around the Bio-CAP peak summit. For motif analysis, the 30 8-mers with either highest or lowest weight were clustered and matched to known motifs using GimmeMotifs (van Heeringen and Veenstra 2011). After clustering, enrichment was determined, and only motifs with an enrichment of at least 1.5 times are shown in Figure 6.
SVM analysis of H3K27me3 domains
For SVM analysis, we employed the method previously described for enhancer prediction (Lee et al. 2011). We used 1-kb H3K27me3 regions as positive sequences and randomly selected genomic sequences with no H3K27me3 ChIP-seq enrichment as negative sequences. For training we used all chromosomes except scaffold_9 (Xenopus) and chr2 (humans, zebrafish), which were used for validation and assessment of (cross-species) performance. In addition, we performed a 10-fold cross-validation procedure using random partitioning of training (90%) and evaluation (10%) data sets to obtain the average performance (mean ROC AUC) of the SVM on X. tropicalis H3K27me3 regions.
ES cell culture
Male mouse embryonic stem cells (E14) were cultured according to standard SIGTR protocols (http://www.sanger.ac.uk/resources/mouse/sigtr/). Briefly, cells were grown on 0.1% gelatin-coated plates and maintained in high-glucose Dulbecco's modified Eagle's medium (Gibco, Invitrogen) supplemented with 15% FBS (PAA Laboratories), 1% penicillin/streptomycin (PS; Gibco), 0.0035% β-mercaptoethanol (Sigma-Aldrich), and 0.2% LIF (Chemicon).
Transient transfection and luciferase assays
Selected regions of ∼1 kb in size were amplified from genomic DNA (Supplemental Table 5) and cloned into the pGL3-promoter vector (Invitrogen). Plasmid DNA was purified using the Plasmid Maxi Kit (Qiagen) or the Wizard Plus SV miniprep DNA purification system (Promega) and transfected in E14 cells using Lipofectamine 2000 (Invitrogen) or microinjected into one-cell-stage X. laevis embryos. Luciferase expression was measured 48 h after transfection (ES cells) or at the gastrula stage (X. laevis). Whole-cell protein extracts were prepared in reporter lysis buffer (Promega), and luciferase assays were performed according to manufacturer's protocol (Promega). Significance was calculated using the one-tailed Wilcoxon ranked-sum test.
Quantitative (RT-) PCR
PCR reactions were performed on a MyiQ single-color real-time PCR detection system (Bio-Rad) using iQ SYBR green supermix (Bio-Rad). Primer sequences are available in the Supplemental Material.
Data access
The sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE41161. Visualization tracks are available at our website (http://veenstra.ncmls.nl).
Supplementary Material
Acknowledgments
We thank H. Stunnenberg for sequencing facilities, E. Gazdag for preparing input DNA, and J. van Noort for access to the nucleosome positioning algorithm. This work was funded by the US National Institutes of Health (grants 1R01HD054356 and 1R01HD069344 to G.J.C.V.) and the Netherlands Organization for Scientific Research (NWO-ALW grant 863.12.002 to S.J.v.H.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.159608.113.
Freely available online through the Genome Research Open Access option.
References
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs K-J, Stunnenberg HG, Veenstra GJC 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell 17: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkers RC, Jacobi UG, Veenstra GJC 2012. Chromatin immunoprecipitation analysis of Xenopus embryos. Methods Mol Biol 917: 279–292 [DOI] [PubMed] [Google Scholar]
- Arnold P, Schöler A, Pachkov M, Balwierz PJ, Jørgensen H, Stadler MB, van Nimwegen E, Schübeler D 2013. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res 23: 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J 22: 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A 2007. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14: 1008–1016 [DOI] [PubMed] [Google Scholar]
- Bogdanović O, Long SW, van Heeringen SJ, Brinkman AB, Gómez-Skarmeta JL, Stunnenberg HG, Jones PL, Veenstra GJ 2011. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res 21: 1313–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanović O, van Heeringen SJ, Veenstra GJC 2012. The epigenome in early vertebrate development. Genesis 50: 192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14: 155–164 [DOI] [PubMed] [Google Scholar]
- Caputo VS, Costa JR, Makarona K, Georgiou E, Layton DM, Roberts I, Karadimitris A 2013. Mechanism of Polycomb recruitment to CpG islands revealed by inherited disease-associated mutation. Hum Mol Genet 22: 3187–3194 [DOI] [PubMed] [Google Scholar]
- Cha T-L, Zhou BP, Xia W, Wu Y, Yang C-C, Chen C-T, Ping B, Otte AP, Hung M-C 2005. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310: 306–310 [DOI] [PubMed] [Google Scholar]
- Dale RK, Pedersen BS, Quinlan AR 2011. Pybedtools: A flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27: 3423–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N, Rappsilber J, Södersten E, Hansen K 2012. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet 8: e1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. 2012. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 1: e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Grossman R, Stein L 2011. PeakRanger: a cloud-enabled peak caller for ChIP-seq data. BMC Bioinformatics 12: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJC 2004. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci 101: 13525–13530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed Polycomb target genes and interact with Polycomb Repressive Complex-2. Mol Cell 38: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Lee D, Karchin R, Beer MA 2011. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res 21: 2167–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata C, Mathavan S, Müller F, Aleström P, et al. 2011. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell 21: 993–1004 [DOI] [PubMed] [Google Scholar]
- Liu Y, Shao Z, Yuan G-C 2010. Prediction of Polycomb target genes in mouse embryonic stem cells. Genomics 96: 17–26 [DOI] [PubMed] [Google Scholar]
- Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grützner F, Odom DT, Patient R, Ponting CP, et al. 2013. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife 2: e00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, Smith AJH, Gobbi MD, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. 2012. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE 2010. GC-rich sequence elements recruit PRC2 in mammalian ES Cells. PLoS Genet 6: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura J-M, Paro R 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell 5: 759–771 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Arteaga-Salas JM, Mentele E, David R, Nicetto D, Imhof A, Rupp RAW 2011. Stage-specific histone modification profiles reveal global transitions in the Xenopus embryonic epigenome. PLoS ONE 6: e22548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan G-C, Lee Y, Orkin SH 2009. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP 2009. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885–897 [DOI] [PubMed] [Google Scholar]
- Smits AH, Jansen PWTC, Poser I, Hyman AA, Vermeulen M 2013. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res 41: e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden T, van Vugt JJFA, Logie C, van Noort J 2012. Sequence-based prediction of single nucleosome positioning and genome-wide nucleosome occupancy. Proc Natl Acad Sci 109: E2514–E2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeringen SJ, Veenstra GJC 2011. GimmeMotifs: A de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics 27: 270–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chen Y-H, Li L-Y, Lang J, Yeh S-P, Shi B, Yang C-C, Yang J-Y, Lin C-Y, Lai C-C, et al. 2011. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 13: 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE 2010. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Mazor T, Huang H, Huang H-T, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, et al. 2012. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell 45: 330–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. 2012. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337: 971–975 [DOI] [PubMed] [Google Scholar]
- Zeng J, Kirk BD, Gou Y, Wang Q, Ma J 2012. Genome-wide polycomb target gene prediction in Drosophila melanogaster. Nucleic Acids Res 40: 5848–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.