Abstract
Surveillance by RNA interference is central to controlling the mobilization of transposable elements (TEs). In stem cells, Piwi argonaute (Ago) proteins and associated proteins repress mobilization of TEs to maintain genome integrity. This defense mechanism targeting TEs is termed the Piwi-interacting RNA (Piwi-piRNA) pathway. In this Opinion, we draw attention to the situation that the genomes of cestodes and trematodes have lost the piwi and vasa genes that are hallmark characters of the germline multipotency program. This absence of Piwi-like Agos and Vasa helicases prompts the question: how does the germline of these flatworms withstand mobilization of TEs? Here we present an interpretation of mechanisms likely to defend the germline integrity of parasitic flatworms.
Keywords: Platyhelminthes, Cestoda, Trematoda, piwi, vasa, argonaute, transposable elements, germline, piRNA pathway
During the past decade draft genomes of several species of the phylum Platyhelminthes (see Glossary) have been reported and/or made available in public databases: (i) the freshwater planarian Schmidtea mediterranea (turbellarian); (ii) the blood flukes Schistosoma mansoni, Schistosoma haematobium, and Schistosoma japonicum (trematodes); and (iii) the cyclophyllidean tapeworms Echinococcus multilocularis, Echinococcus granulosus, Taenia solium, and Hymenolepis microstoma (cestodes) [1,2,3,4,5,6]. Closer scrutiny of these genomes revealed that piwi, vasa, and genes encoding group 4 Tudor homologues were absent from the cestode and trematode classes of the Platyhelminthes [7,8]. These otherwise conserved, post-transcriptional regulators are associated with stem cell maintenance and germ cell development in diverse taxa - pre-bilaterians, the platyhelminth order Tricladida, the free-living platyhelminth relatives of the parasitic flatworms, and many others.
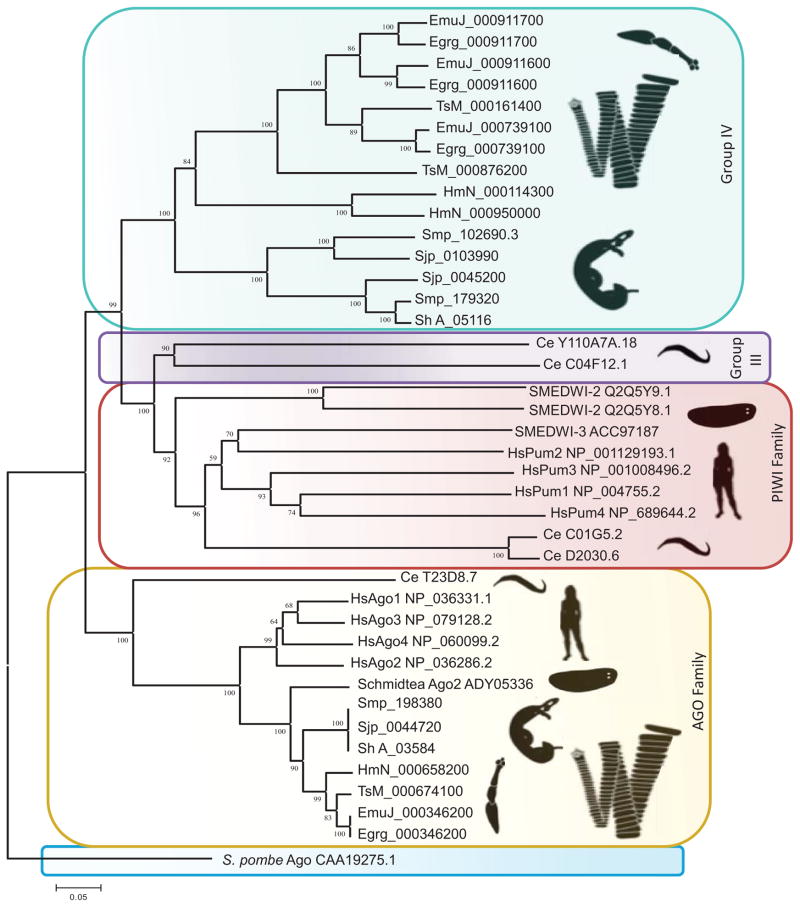
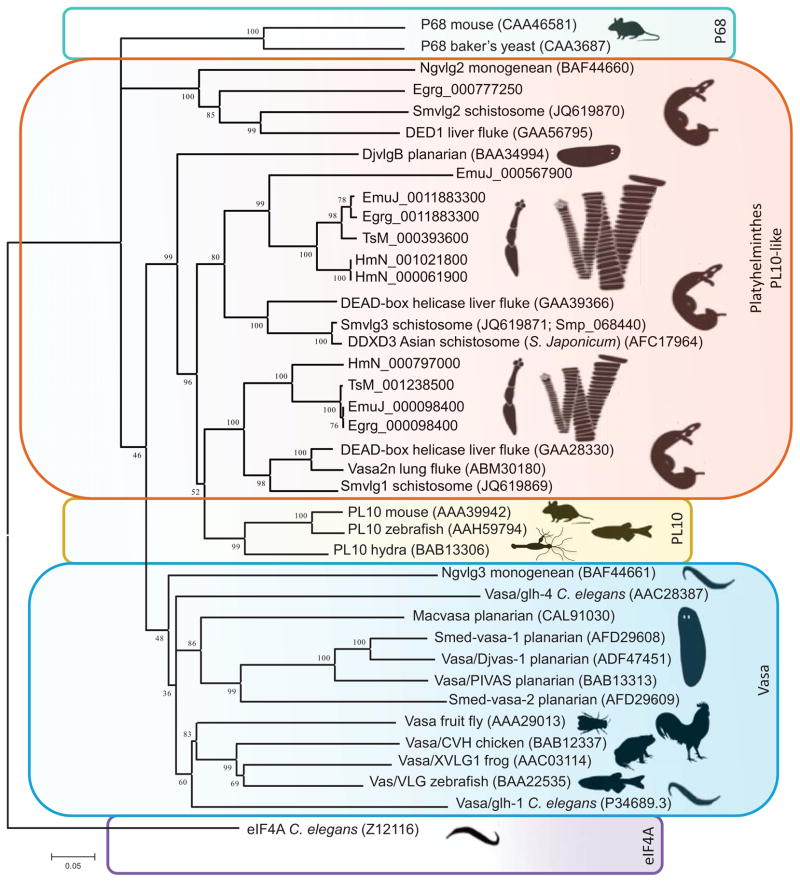
Phylogenetic analysis of the Argonaute (Ago) protein family with six platyhelminth genomes and other informative species revealed the loss of the Piwi subfamily of Argonautes in cestodes and trematodes [3,8]. This contrasts strikingly with planarians, which express Piwi-like Agos in the somatic and germ stem cells [3,8]. Additionally, it has become apparent that cestodes and trematodes evolved a clade of Argonaute proteins exclusive to the parasitic flatworms. This clade is termed the group 4 Argonautes; it is discrete from the Ago-like clade, the Piwi-like clade, and the C. elegans group 3 Argonautes clade that traditionally constitute the Ago protein family [3,8] (Figure 1). It is also noteworthy that group 4 Tudor proteins, the interactive partners of Piwi, appear to be absent from these parasitic flatworms (flukes, tapeworms) [3]. In like fashion, phylograms of Vasa and related DEAD-box helicases further revealed the absence of orthologues of vasa in trematodes and cestodes [3,9]. Instead, these flatworms have evolved at least two PL10-like genes that cluster together and branch from the PL10 clade of DEAD-box helicases (Figure 2). Moreover, the flukes S. mansoni and Clonorchis sinensis, the cestodes noted above, and the monogenean, Neobenedenia girellae, encode a DEAD-box helicase that might be assignable to the PL10 family or the closely related p68 DEAD-box family [3,8,9,10]. Expression analysis of these latter DEAD-box helicases termed S. mansoni vasa-like gene 2 (Smvlg2) and N. girellae vasa-like gene 2 (Ngvlg2) revealed that they exhibit germline specific expression [9,10]. Silencing by RNA interference (RNAi) of Ngvlg2 led to loss of germ cells, which indicated that the genes perform vasa-like roles during germline development [10]. N. girellae also has a third N. girellae vasa-like gene (Ngvlg3) that groups with vasa orthologues. This is noteworthy from a phylogenetic point of view since it has been proposed that monogeneans are basal to a clade composed of Classes Trematoda and Cestoda [11] and suggests that vasa orthologues had been lost in the common ancestor of flukes and tapeworms. However, transcripts encoding Ngvlg3 mRNA were not detectable while silencing Ngvlg3 did not affect gonad anatomy in the same fashion as silencing Ngvlg1 (PL10-like cluster) and Ngvlg2 [10].
Figure 1.
Evolutionary relationships of platyhelminth Argonautes (AGO). The Neighbor-Joining method was employed to infer relationships using entire amino acid sequences from informative species, including trematodes, and cestodes. Phylogenetic analysis was conducted in MEGA5. Colored boxes highlight families of argonaute proteins, for example, the red region includes Piwi orthologues, and the green colored boxes include group 4 argonaute proteins of cestodes and trematodes. Adapted from [3]. Abbreviations: Egrg, Echinococcus granulosus; EmW, E. multilocularis; HmN, Hymenolepis microstoma; TsM, Taenia solium; Sjp, Schistosoma japonicum; Smp, S. mansoni; Sh, Schistosoma haematobium; Ce, C. elegans; SMED, Schmidtea mediterranea; Hs, Homo sapiens; S. pombe, Schizosaccharomyces pombe.
Figure 2.
Evolutionary relationships of Vasa, PL10, p68, and eIF4A. The Neighbor-Joining method was employed to infer relationships using entire amino acid sequences from informative species, including trematodes, and cestodes. Phylogenetic analysis was conducted in MEGA5. Among the DEAD-box helicases, p68 is closely related to Vasa and PL10. The eIF4A served as the out-group. Branch names indicate the common name of the species displayed; GenBank and GeneDB accessions are provided. Colored boxes highlight families of DEAD-box helicases, for example, the blue region includes vasa orthologues, and the peach colored boxes include platyhelminth PL10-like enzymes, etc. Adapted from [9] (with permission) to include PL10-like orthologues of cestodes.
Considered at large, the absence of Piwi, Vasa, and group 4 Tudor proteins indicates that parasitic flatworms (sub-phylum Neodermata, the cestodes, monogeneans and trematodes) have lost the ancestral components, classified traditionally as germline stem cell associated markers that are conserved in metazoans (Box 1). These enzymes and other factors establish and maintain inter alia the multipotency of progenitor germ cells (PGCs), germline stem cells (GSCs), and multipotent progenitor cells including the neoblasts (totipotent adult stem cells) of planarians, a hypothetical regulatory network collectively referred to as the germline multipotency program (GMP) [12,13]. Furthermore, it has been proposed that these factors are ancestrally linked in the GMP because together they participate in protection of the genome from the mobilization of endogenous and exogenous transposable elements (TEs), an ostensibly essential role in germline stem cells and somatic stem cells devoted to the production of progenitors for tissue renewal [14,15,16]. It is relevant to note here that approximately 30 to 50% of the genomes of the three major species of human schistosomes and other trematodes consists of TEs and other repetitive elements, although substantially less of the genome of (at least cyclophyllidean) cestodes is composed of these elements (~2 – 11%) [3,4,5,6,17] (Box 1).
Box 1. Outstanding questions.
Might the loss of Piwi/Vasa have been less crucial in evolution of the Cestoda and Trematoda if the canonical pathways in which proteins participate had already changed in their common ancestor?
Given the marked dissimilarity of the TE content of the genomes of flukes (35%) and tapeworms (2%), might tapeworms and flukes have different mechanisms of combating TEs, or highly different efficiencies?
Did flukes and tapeworms co-opt the stem cell specific siRNA pathway?
This opinion highlights the absence of piwi, the piRNA pathway, and vasa from flukes and tapeworms, and speculates on alternative mechanisms that might defend the integrity of the germline of the parasitic flatworms against TEs.
piRNA pathway - guardian of the germline
Piwi and associated proteins, including Vasa, are involved in the functions and synthesis of a novel class of small non-coding RNAs of 24–31 nucleotides in length termed Piwi-interacting RNAs (piRNAs). With model species, it became clear that the piRNA pathway was restricted to germ cells and, in some cases, gonadal somatic cells [13]. Deciphering the function of Piwi in the context of the silencing of TEs has mainly relied on findings in Drosophila and mammalian cell lines and, to a lesser extent, zebrafish and C. elegans [12,18]. The piRNAs associate with Piwi to establish an active piRNA-induced silencing complex (piRISC) that recognizes and silences TEs [15,19,20]. This process includes a Ping-Pong amplification cycle for piRNA biogenesis that amplifies piRNAs, increasing the quantity of piRISC available to silence TEs. The system functions to protect the integrity of the germline genome [15,16,19,21,22,23,24,25]. Piwi proteins and piRNAs may also play roles in epigenetic and post-transcriptional regulation of genes in fruit flies and mice [26,27,28,29,30,31,32,33]. Whereas most loci encoding piRNAs are located in introns and exons, similar to miRNA genes, ~20% of piRNAs map to genomic loci enriched with repetitive sequences and TEs [19,34,35].
A specific function for Vasa in the piRNA pathway remains to be determined but it has been linked to the biogenesis of piRNAs [36,37,38,39,40,41,42]. In Drosophila, Vasa is required for nuage ( = chromatoid body) assembly, the site of piRNA biosynthesis, and in Vasa-deficient fruit flies, the Piwi proteins Agp3 and Aub fail to localize to the nuage particles, and synthesis of piRNAs by the Ping-Pong cycle is markedly diminished [43]. Moreover, TE activities increase in germ cells due to defective de novo methylation [41]. Immunoprecipitation studies demonstrated that primary transcripts of piRNAs clusters associate with Vasa, suggesting that Vasa is involved in transportation of precursor piRNAs from the nucleus to the cytoplasm [44,45,46]. C. elegans and mice exhibit similar phenomena [20,36,47,48,49]. In pull-down studies, the mouse orthologue of Vasa, Mouse Vasa-homologue (Mvh), binds with the Piwi proteins Mili, Miwi, and Miwi2. In addition, Mvh-deficient testes fail to silence TEs in germ cells, as do Vasa-deficient fruit flies, [20,36]. It is also notable that orthologues of vasa of C. elegans, Drosophila, and mice have been linked to Dicer. Although the specific mechanism(s) by which Vasa contributes to short interfering RNA (siRNA) rather than microRNA (miRNA) pathways remains unclear, it may facilitate export of precursor RNAs, in like fashion to its anticipated role with piRNAs [30,50,51,52,53,54].
The piRNA pathway likely arose early in the evolution of the Metazoa [55]. In pre-bilaterians such as poriferans and ctenophorans, and in the marine annelid Platynereis and some other lophotrochozoans, piwi is also expressed in multipotent somatic stem cells [12]. Functional data from species of lophotrochozoans are largely absent, except for some planarians. However, whereas it is likely that metazoans at large express piRNAs, the piRNA pathway is predominantly expressed in germ cells. Hence it is noteworthy that the planarian piRNA pathway occurs primarily in neoblasts, the adult totipotent stem cells, and is not restricted to germ cells [18]. Three Piwi orthologues, SMEDWI-1, SMEDWI-2, and SMEDWI-3 occur in S. mediterranea [18,56], and two, SMEDWI-2 and SMEDWI-3, are essential to neoblast function, regeneration, homeostasis, and piRNA expression [18,57,58]. Deep sequencing identified a diverse population of small RNAs of ~32 nt (range 25–35) in length in S. mediterraneath at resemble piRNAs of insects and vertebrates [18,59]. These small RNAs map to thousands of loci in the planarian genome; 20–30% mapped to TEs, with the remainder mapping to introns and exons of genes of unknown functions [18,58]. Deeper investigation is needed to clarify the piRNA pathway in planarians, such as pull-down experiments with SMEDWI protein specific antibodies to confirm physical interactions among the small RNAs and the SMEDWI proteins characteristic of bona fide piRNAs of the Ping-Pong piRNA biogenesis pathway. Similar observations have been made in Dugesia japonica that, like S. mediterranea, has inherited three piwi genes, DjPiwi-1, -2, and -3 [60,61].
Have other Argonautes assumed Piwi roles in flukes and tapeworms?
Although Piwi is absent from the parasitic flatworms, schistosomes express several Argonautes including a protein termed Argonaute 2 (Ago2), which is a member of the group 4 Argonautes exclusive to flukes and tapeworms. Argonaute 2 of S. mansoni (SmAgo2) is expressed in the ovary, vitelline gland, testes, the neoblast-like stem cells of the adult schistosomes, and germinal cells of the sporocyst [7,62,63]. The neoblast-like stem cells may be totipotent and exhibit a similar molecular signature to planarian neoblasts [7,62,63 ]. Gene silencing approaches indicate that SmAgo2 is required for maintenance of germinal cells and the proliferation in sporocysts [63]. There are small non-coding RNAs (sncRNAs) of 20–21 nt that associate with Argonaute 2 (SjAgo2) in S. japonicum, an sncRNA size reminiscent of endogenous short interfering RNAs (endo-siRNAs) rather than miRNAs or piRNAs [64]. Follow-up analysis of the sncRNA population in S. japonicum eggs ruled out piRNAs with the dominant species of small RNAs between 18 – 23 nt [64]. These endo-siRNAs arise from TEs, predominately non-long terminal repeat (LTR) retrotransposons and LTR retrotransposons [64]. These observations including the failure to identify a piwi gene indicate the absence of a canonical piRNA pathway in schistosomes. However, might there exist in its place, a non-canonical piRNA pathway? This would seem to be necessary given the need for a mechanism to regulate the mobility of TEs that comprise sizeable fractions of the genome [4] to guard the integrity of the genome of the germ cells of these parasitic flatworms. SmAgo2/SjAgo2 may perform as a guardian that suppresses the TE activities in germinal cells (Figure 3).
Figure 3.
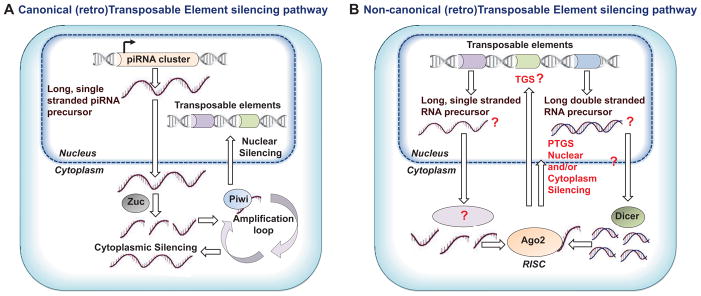
(Retro)Transposable Element (TE) silencing pathways. (A) Canonical TE silencing pathway; piRNAs are transcribed from genomic loci termed piRNA clusters. Cleavage of these single strand transcripts by Zucchini (Zuc) generates an initial pool of primary piRNAs, which are loaded into Piwi. Secondary piRNAs are generated in a slicer-dependent amplification loop. Transposon transcripts are cleaved through this amplification cycle, accomplishing the silencing of cytoplasmic TEs. Piwi associated with piRNAs translocates to the nucleus and drives nuclear silencing. Adapted from [69] and [70]). (B) Hypothetical non-canonical TE silencing pathway in trematodes and cestodes. Long, single stranded RNA precursors [65] and/or long double stranded precursors are cleaved by an unknown enzyme and/or Dicer, respectively. Small non-coding RNA, that is, endo-siRNA-like molecules interacting with Argonaute 2 (Ago 2), which belongs to group 4 of Argonaute proteins in parasitic flatworms, constitute the RNA induced silencing complex (RISC) [64] that would drive transcriptional gene silencing (TGS) and/or post-transcriptional gene silencing (PTGS) of TEs.
Scrutiny of the draft genomes of E. multilocularis, E. granulosus, T. solium, and H. microstoma supports the prediction that piwi does not occur in tapeworms; the piwi gene has not been identified in these genomes [3,6]. In the ancestor of Echinococcus and Taenia, the group 4 argonaute gene was subjected to two recent rounds of duplication, resulting in four copies with > 90% identity at the nucleotide level, and organized as two couples of tandemly arranged copies, but without identity in promoter regions. One copy is clearly a pseudogene with a highly fragmented coding region, and it is not clear yet how many of these genes are functional [3]. Nevertheless, a long non-coding RNA family, which is massively transcribed from repetitive elements in the genomes of E. multilocularis and E. granulosus, has been identified as potential precursors of piRNA-like small RNAs [65]. These sncRNAs need to be investigated more fully as do a population of sncRNAs between 25 and 30 nt in length that has been identified, but not analyzed, in studies on miRNAs in E. granulosus [66]. Taken together, these findings indicate it is unlikely that a traditional piRNA pathway including Piwi is conserved in cestodes or trematodes.
Might other PL10-like helicases perform the role of Vasa?
Whereas vasa is absent from the genomes of cestodes and trematodes [3,7,9], three PL10-like genes, Smvlg1, Smvlg2, and Smvlg3 have been characterized in S. mansoni. Expression profiles of these DEAD-box helicases suggest they perform roles similar to Vasa including functions related to the GMP and to stem cell maintenance [7,9,63]. Silencing of Smvlg3 indicated that this PL10-like enzyme is necessary for proliferation and maintenance of germinal cells in the sporocyst, a population of cells that shares a molecular signature with neoblasts (adult totipotent stem cells) of planarians [63]. Smvlg1, Smvlg2, and Smvlg3 may have assumed the role of vasa and display signatures similar to the GMP of other metazoans together with the neoblasts of planarians. In light of these observations, it is plausible that these PL10-like RNA helicases in schistosomes could perform the role of Vasa in the piRNA pathway (Box 1). Notably, unambiguous orthologues of Smvlg1, Smvlg2, and Smvlg3 also evolved in the genomes of tapeworms [3] (Figure 1).
Concluding remarks: how do these pathogens flourish without Piwi and Vasa?
Accordingly, how might flukes and tapeworms maintain genome integrity without piwi and vasa? Perhaps these parasitic platyhelminths evolved a germline, stem cell-specific endogenous siRNA pathway to perform the roles of the piRNA pathway that is active in other metazoans, such as planarians (Figure 3A). We hypothesize the operation of a non-canonical TE silencing pathway in trematodes and cestodes. Here, long, single stranded RNA precursors [65] and/or long double stranded precursors might be cleaved by an unknown enzyme and/or Dicer. Small non-coding RNA, that is, endo-siRNA-like molecules interacting with Argonaute 2 (Ago 2), which belongs to group 4 of Argonaute proteins in parasitic flatworms, constitute the RNA induced silencing complex (RISC) [64] that would drive transcriptional gene silencing (TGS) and/or post-transcriptional gene silencing (PTGS) of TEs (Figure 3B). A summary of the admittedly scant information described so far concerning alternative piRNAs includes the 20–21 nt family in S. japonicum and the abundant piRNA-like RNA population of E. granulosus [65]; and both proteins and RNAs must be involved in such a pathway. Functional genomics analyses utilizing RNAi and transgenesis [67,68] to define pathways, enzymes and ncRNAs that participate in silencing of TEs represent potentially fruitful routes of investigation to address this enigma as do RNA-Seq and genome methylation status analyses of the endogenous TEs of these worms, and immunoprecipitations to characterize binding partners [64].
Highlights.
Flukes and cestodes do not have a piRNA pathway
How might these flatworm pathogens defend the germline genome?
Fluke genomes are replete with mobile genetic elements
Acknowledgments
We thank Dr. Matty Knight for insightful comments on the manuscript. Support for doctoral studies (DES) from the Institute of Biomedical Sciences of The George Washington University is gratefully acknowledged. These studies were supported by NIH-NIAID award R01AI072773 (the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the NIH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
- Ago clade of Argonautes
the Ago subfamily of Argonaute proteins interacts with miRNAs and siRNAs. Ago proteins, which are expressed in all tissues, exhibit three structural features: PAZ (Piwi, Argonaute, and Zwille) domain, the MID (middle) domain, and the C-terminal PIWI domain
- Argonaute (Ago)
catalytic component in RNA-induced silencing complexes (RISC) responsible for coordinating downstream gene-silencing events. Argonaute proteins bind to different classes of small non-coding RNAs, i.e., microRNAs (miRNAs), small interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs)
- Argonaute protein family
includes the Ago and Piwi clades. C. elegans contain specific group 3 argonaute proteins, termed WAGO proteins, as well as the Ago and Piwi clade. Parasitic flatworms evolved a group 4 Ago clade
- DEAD-box RNA helicases
member of Superfamily 2 (SF2) of helicases. Characterized by presence of 12 conserved motifs: Q-motif, Motif I, Motif Ia, motif Ib, motif Ic, motif II, motif III, motif IV, motif IVa, motif V, motif Va, and motif VI. Motif II contains the amino acid sequence DEAD (Asp-Glu-Ala-Asp), from which family name derived. The motifs are involved in ATP binding, hydrolysis, ATP-dependent RNA binding, and helicase activities. DEAD-box helicases occur in all cells; central to metabolism of RNA, from transcription to decay
- Germline stem cells
germline stem cells have the capacity to generate all cells in the body. Under normal fate the germline stem cells are unipotent, producing gametes and self-renewing to maintain the generative population
- Group 4 Tudor proteins
Tudor domains were initially identified as a common structural motif, approximately 60 amino acids, in the Drosophila Tudor protein. The motif has now been identified in a wide variety of organisms. These Tudor domain-containing proteins bind to proteins with methylated arginine and lysine residues and play a role in epigenetics, gene expression, and regulation of small RNAs. There are four different functional groups of Tudor proteins distinguished by the sequence flanking the Tudor domain. The group 4 Tudor proteins have been shown to interact with the methylated RG/RA box of Piwi proteins and are involved in piRNA biogenesis by promoting the formation of Piwi ribonucleoprotein (piRNP) complexes
- microRNAs (miRNAs)
endogenous non-coding RNAs (~22 nt) that regulate gene expression at transcriptional and post-transcriptional level. Produced from genes or introns, processed from long primary transcripts. miRNAs target and bind complementary sequences in the 3′–untranslated region of mRNAs, leading to gene silencing via translation repression and/or mRNA degradation. Mature miRNAs associate with Ago clade of Argonautes
- Neighbor-Joining Method
a method for construction of phylograms(evolutionary trees) that is distanced based and uses bottom-up clustering
- Neoblasts
pluripotent adult stem cells with capacity for indefinite self-renewal, totipotency; drive long-term tissue homeostasis. Endow planarians with ability to regenerate. Neoblast-like stem cells occur in tapeworms and schistosomes. Characteristics include roundish-to-ovoid cells with a high nuclear-to-cytoplasmic ratio, large nucleolus, exhibit chromatoid bodies (RNA-protein bodies near nuclei), and numerous free ribosomes
- Neodermata
the sub-phylum of the Platyhelminthes that includes the Classes Monogenea, Trematoda and Cestoda. All members of the Neodermata are parasites
- Nuage
term used to describe the electron dense, ribonucleoprotein rich, perinuclear granule organelle-like structure in germ cells of Drosophila that plays a role in the piRNA pathway and piRNA-mediated silencing of transposons. The structure has been described in other animals and is termed chromatoid bodies, germinal granules, P granules, and mitochondrial cloud in other species. The nuage/chromatoid body has been described in the neoblasts of planarians
- Ping-Pong amplification cycle
a mechanism for piRNA biogenesis hypothesized from observations in Drosophila and later in other organisms. This mechanism obviates the need for RNA-dependent RNA polymerase. The mechanism is initiated by mature sense primary piRNAs, derived from transposable elements and transcripts from genomic loci termed piRNA clusters, bound to proteins of the Piwi clade which recognize complementary sequences, antisense transcripts from the same piRNA cluster. The antisense piRNAs are cleaved, generating secondary piRNAs with new 5′-ends targeted by another Piwi protein. The 3′ end is then trimmed and then the mature antisense secondary piRNAs are now able to target sense transcripts from the piRNA clusters generating new sense piRNAs
- Piwi clade of Argonautes
subfamily of Argonautes; typically expressed in germline and adult somatic stem cells of invertebrates; responsible for regeneration e.g. neoblasts in planarians. Does not occur in parasitic flatworms. Piwi, like Ago, has three main structural motifs: PAZ (Piwi, Argonaute, and Zwille) domain, the MID (middle) domain, and the C-terminal PIWI domain. Piwis associate with small non-coding RNAs termed piwi-interacting RNAs (piRNAs). Essential for biogenesis and/or stability of piRNAs, the piRNA biogenesis pathway, and the Ping-Pong amplification cycle
- Piwi-interacting RNAs (piRNAs)
small RNAs of 24–30 nt in length thought to arise from long single stranded RNA precursors derived from transposable elements (TEs), intergenic regions, and protein-coding genes. piRNAs unction in preserving genome integrity and stability, and to play a role in antiviral immunity. piRNAs associated with specific Piwi proteins and together function in silencing TEs. Piwi proteins function in germline development, gametogenesis, germline stem cell maintenance, and meiosis
- PL10
an ATP-dependent DEAD-box RNA helicase, also termed DEAD-box helicase 3 (DDX3) closely related to Vasa. PL10 is not restricted to the germline or somatic stem cells, Conserved in eukaryotes from yeast, plants to animals. Participates in RNA metabolism, tissue differentiation, embryogenesis, asexual reproduction, cell regeneration, tumorigenesis, apoptosis, cooption in viral pathology and innate immune
- Platyhelminthes
Phylum Platyhelminthes includes parasitic clades Cestoda (tapeworms), Trematoda (flukes), and Monogenea (mostly ectoparasites of fish), and the mostly free-living planarian-like worms of order Tricladida
- Small interfering RNAs (siRNAs)
class of small non-coding RNAs (~20–25 nucleotides) notable for their role in the RNA interference pathway. Derived from endogenous loci or foreign genetic material and involved in silencing of invading viruses, transposable element activity, and in some cases protein-coding genes
- Transposable element (TE)
DNA elements that can move from location to another within the genome of a cell, these elements can move either by cut-and-paste mechanism (DNA transposons) or indirectly through an RNA intermediate, i.e. copy-and-paste mechanism (retrotransposons)
- Vasa
ATP-dependent DEAD-box RNA helicase; archetypal germ cell marker; termed DEAD-box helicase 4 (Ddx4).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schistosoma japonicum Genome S, Functional Analysis C. The Schistosomajaponicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496:57–63. doi: 10.1038/nature12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young ND, Jex AR, Li B, Liu S, Yang L, et al. Whole-genome sequence of Schistosoma haematobium. Nature genetics. 2012;44:221–225. doi: 10.1038/ng.1065. [DOI] [PubMed] [Google Scholar]
- 5.Protasio AV, Tsai IJ, Babbage A, Nichol S, Hunt M, et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis. 2012;6:e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng H, Zhang W, Zhang L, Zhang Z, Li J, et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nature genetics. 2013;45:1168–1175. doi: 10.1038/ng.2757. [DOI] [PubMed] [Google Scholar]
- 7.Collins JJ, 3rd, Wang B, Lambrus BG, Tharp ME, Iyer H, et al. Adult somatic stem cells in the human parasite Schistosoma mansoni. Nature. 2013;494:476–479. doi: 10.1038/nature11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y. Phylogenetic analysis of the Argonaute protein family in platyhelminths. Molecular phylogenetics and evolution. 2013;66:1050–1054. doi: 10.1016/j.ympev.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Skinner DE, Rinaldi G, Suttiprapa S, Mann VH, Smircich P, et al. Vasa-Like DEAD-Box RNA Helicases of Schistosoma mansoni. PLoS Negl Trop Dis. 2012;6:e1686. doi: 10.1371/journal.pntd.0001686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohashi H, Umeda N, Hirazawa N, Ozaki Y, Miura C, et al. Expression of vasa (vas)-related genes in germ cells and specific interference with gene functions by double-stranded RNA in the monogenean, Neobenedenia girellae. International journal for parasitology. 2007;37:515–523. doi: 10.1016/j.ijpara.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Park JK, Kim KH, Kang S, Kim W, Eom KS, et al. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes) BMC evolutionary biology. 2007;7:11. doi: 10.1186/1471-2148-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juliano C, Wessel G. Developmental biology. Versatile germline genes. Science. 2010;329:640–641. doi: 10.1126/science.1194037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juliano CE, Swartz SZ, Wessel GM. A conserved germline multipotency program. Development (Cambridge, England) 2010;137:4113–4126. doi: 10.1242/dev.047969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alie A, Leclere L, Jager M, Dayraud C, Chang P, et al. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Developmental biology. 2011;350:183–197. doi: 10.1016/j.ydbio.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature reviews Molecular cell biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 16.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science (New York, NY) 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Chen W, Wang X, Liu H, Chen Y, et al. The carcinogenic liver fluke, Clonorchis sinensis: new assembly, reannotation and analysis of the genome and characterization of tissue transcriptomes. PloS one. 2013;8:e54732. doi: 10.1371/journal.pone.0054732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 20.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & development. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 22.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 24.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 26.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & development. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng W, Lin H. Miwi, a Murine Homolog of Piwi, Encodes a Cytoplasmic Protein Essential for Spermatogenesis. Developmental cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 28.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 29.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes & development. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Current biology: CB. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 31.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 32.Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development (Cambridge, England) 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- 33.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 34.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 35.O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes & development. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development (Cambridge, England) 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 38.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophilamelanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, et al. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA. 2010;16:2503–2515. doi: 10.1261/rna.2270710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mechanisms of development. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 41.Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, et al. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophilamelanogaster. RNA biology. 2004;1:54–58. [PubMed] [Google Scholar]
- 42.Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Current biology: CB. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klattenhoff C, Xi H, Li C, Lee S, Xu J, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen H. UAP56- a key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB reports. 2009;42:185–188. doi: 10.5483/bmbrep.2009.42.4.185. [DOI] [PubMed] [Google Scholar]
- 46.Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development (Cambridge, England) 2010;137:1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Updike D, Strome S. P granule assembly and function in Caenorhabditiselegans germ cells. Journal of andrology. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voronina E, Seydoux G. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development (Cambridge, England) 2010;137:1441–1450. doi: 10.1242/dev.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beshore EL, McEwen TJ, Jud MC, Marshall JK, Schisa JA, et al. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Developmental biology. 2011;350:370–381. doi: 10.1016/j.ydbio.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, et al. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Developmental cell. 2010;18:102–113. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 52.Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Current biology: CB. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 53.Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. Journal of cell science. 2006;119:2819–2825. doi: 10.1242/jcs.03022. [DOI] [PubMed] [Google Scholar]
- 54.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, et al. Critical roles for Dicer in the female germline. Genes & development. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annual Review of Cell and Developmental Biology. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 57.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 58.Resch AM, Palakodeti D. Small RNA pathways in Schmidtea mediterranea. The International journal of developmental biology. 2012;56:67–74. doi: 10.1387/ijdb.113436ar. [DOI] [PubMed] [Google Scholar]
- 59.Friedlander MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, et al. High-resolution profiling and discovery of planarian small RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rossi L, Salvetti A, Lena A, Batistoni R, Deri P, et al. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Development genes and evolution. 2006;216:335–346. doi: 10.1007/s00427-006-0060-0. [DOI] [PubMed] [Google Scholar]
- 61.Rossi L, Salvetti A, Marincola FM, Lena A, Deri P, et al. Deciphering the molecular machinery of stem cells: a look at the neoblast gene expression profile. Genome biology. 2007;8:R62. doi: 10.1186/gb-2007-8-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cogswell AA, Collins JJ, 3rd, Newmark PA, Williams DL. Whole mount in situ hybridization methodology for Schistosoma mansoni. Molecular and biochemical parasitology. 2011;178:46–50. doi: 10.1016/j.molbiopara.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang B, Collins JJ, 3rd, Newmark PA. Functional genomic characterization of neoblast-like stem cells in larval Schistosoma mansoni. eLife. 2013;2:e00768. doi: 10.7554/eLife.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai P, Piao X, Hou N, Liu S, Wang H, et al. Identification and Characterization of Argonaute Protein, Ago2 and Its Associated Small RNAs in Schistosoma japonicum. PLoS Negl Trop Dis. 2012;6:e1745. doi: 10.1371/journal.pntd.0001745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parkinson J, Wasmuth JD, Salinas G, Bizarro CV, Sanford C, et al. A transcriptomic analysis of Echinococcus granulosus larval stages: implications for parasite biology and host adaptation. PLoS Negl Trop Dis. 2012;6:e1897. doi: 10.1371/journal.pntd.0001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cucher M, Prada L, Mourglia-Ettlin G, Dematteis S, Camicia F, et al. Identification of Echinococcus granulosus microRNAs and their expression in different life cycle stages and parasite genotypes. International journal for parasitology. 2011;41:439–448. doi: 10.1016/j.ijpara.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 67.Rinaldi G, Eckert SE, Tsai IJ, Suttiprapa S, Kines KJ, et al. Germline transgenesis and insertional mutagenesis in Schistosoma mansoni mediated by murine leukemia virus. PLoS pathogens. 2012;8:e1002820. doi: 10.1371/journal.ppat.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beckmann S, Grevelding CG. Paving the way for transgenic schistosomes. Parasitology. 2012;139:651–668. doi: 10.1017/S0031182011001466. [DOI] [PubMed] [Google Scholar]
- 69.Lucas KJ, Myles KM, Raikhel AS. Small RNAs: a new frontier in mosquito biology. Trends in parasitology. 2013;29:295–303. doi: 10.1016/j.pt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & development. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]