Abstract
When ubiquitin (Ub) is attached to membrane proteins on the plasma membrane, it directs them through a series of sorting steps that culminate in their delivery to the lumen of the lysosome where they undergo complete proteolysis. Ubiquitin is recognized by a series of complexes that operate at a number of vesicle transport steps. Ubiquitin serves as a sorting signal for internalization at the plasma membrane and is the major signal for incorporation into intraluminal vesicles of multivesicular late endosomes. The sorting machineries that catalyze these steps can bind Ub via a variety of Ub-binding domains. At the same time, many of these complexes are themselves ubiquitinated, thus providing a plethora of potential mechanisms to regulate their activity. Here we provide an overview of how membrane proteins are selected for ubiquitination and deubiquitination within the endocytic pathway and how that ubiquitin signal is interpreted by endocytic sorting machineries.
The ubiquitination of cell-surface proteins (e.g., receptor tyrosine kinases) leads to their internalization and delivery to lysosomes for degradation. How these signals are read by endocytic sorting machinery is becoming clearer.
HOW PROTEINS ARE SELECTED FOR UBIQUITINATION
Ubiquitin (Ub) is covalently attached to substrate proteins by the concerted action of E2-conjugating enzymes and E3 ligases (Varshavsky 2012). Most of the specificity and regulation of ubiquitination is at the level of the E3 ligases, which vastly outnumber the E2-conjugating enzymes. Ubiquitination results from the transfer of Ub, held either by the E2 or in some cases by the E3 as a high-energy thioester bond, onto a substrate protein, typically on the terminal amide of a substrate acceptor lysine side chain. Owing to the intense interest in the ubiquitination system, many of the simple rules and distinctions concerning different types of ligases, their specificity for forming particular poly-Ub chains, and even the kinds of residues in substrate proteins that can be ubiquitin modified, have been blurred with the discovery of mixed poly-Ub chains, new enzymatic mechanisms for Ub transfer, and ubiquitination of nonlysine residues (Kulathu and Komander 2012; Wenzel and Klevit 2012; McDowell and Philpott 2013). Nonetheless, in basic terms, Ub ligases function as platforms that coordinate substrate recognition with Ub transfer as well as configuring poly-Ub chain topology by the E2-conjugating enzyme. Substrates can be bound directly by the E3 ligase or by adaptor proteins that in turn bind to ligases, and selection of substrate residues that undergo ubiquitination is accomplished by the particular architecture of the substrate/ligase complex that determines the orientation to the thioester-linked Ub. The ubiquitin-peptidases (deubiquitinating enzymes [DUbs]) that remove Ub from substrates also play critical roles in controlling the level of substrate protein ubiquitination. Of the roughly 90 DUbs in humans and 20 DUbs in yeast, many are found associated with Ub ligases or with the endocytic machinery that recognizes ubiquitinated proteins, thus positioning DUbs to regulate the ubiquitination process at a variety of junctures (Table 1) (Clague et al. 2012).
Table 1.
Established and emerging proteins involved in ubiquitin-dependent control of the endocytic system
| Function and characteristics | References | |
|---|---|---|
| Ub ligases | ||
| Nedd4-2 | Member of the larger family of HECT-type ligases that includes NEDD4-1 (ITCH/AIP4), WWP1, WWP2, SMURF1, SMURF2, NEDL1/HECW1, and NEDL2; ubiquitinates cargo such as ENaC and voltge-gated sodium channels. | Fotia et al. 2004; Zhou et al. 2007 |
| Itch | Ubiquitinates CXCR-4 and ErbB-4; also ubiquitinates nonmembrane proteins such as endophillin and PI4-kinase type Iiα that regulate endocytosis. | Angers et al. 2004; Bhandari et al. 2007; Sundvall et al. 2008; Mossinger et al. 2012 |
| Rsp5 | The sole member of the Nedd4 family in yeast; responsible for ubiquitinating the vast majority of membrane proteins that are degraded within vacuole/lysosome. | Belgareh-Touze et al. 2008 |
| Cbl | Major ligase that targets a broad range of receptor tyrosine kinases. | Lipkowitz and Weissman 2011 |
| SCF (β-TRCP) | Ubiquitinates the growth hormone receptor, which has a motif that mimics the phosphorylated recognition motif that is found within other SCF (β-TRCP) substrates. | van Kerkhof et al. 2011; da Silva Almeida et al. 2013 |
| CHIP | Modifies damaged conformationally defective proteins in the plasma membrane to stimulate their lysosomal degradation and also misfolded proteins in the ER to stimulate their degradation in the proteasome. | Apaja et al. 2010; Okiyoneda et al. 2010 |
| MARCH1 | Member of the family of membrane-associated RING-CH ligases; ubiquitinates MHC-II complex. | Ishido et al. 2009 |
| MARCH2 | Can target CFTR and β2-adrenergic receptors for lysosomal degradation. | Han et al. 2012; Cheng and Guggino 2013 |
| MARCH4 | Targets MHC-I for degradation. | Ishido et al. 2009 |
| MARCH8 | Targets TRAIL-R1, transferrin receptor, type I IL-1 receptor, CD44, and CD96. | Eyster et al. 2011; Fujita et al. 2013; van de Kooij et al. 2013 |
| IDOL | Inducible degrader of the LDLR targets members of the LDL receptor family. | Zelcer et al. 2009 |
| MAGE-L2-TRIM27 | Targets the Wiskott-Aldrich syndrome protein member WASH to regulate activity of retromer complex. | Hao et al. 2013 |
| RNF167 | Localized to endosomes and modifies the SNARE Vamp3, thus altering its sorting. | Yamazaki et al. 2013 |
| EEA1 | Early endosomal tethering factor that mediates fusion and binds to E2-conjugating enzymes, which allows it to ubiquitinate itself and alter its function. | Ramanathan et al. 2013 |
| HOPS | Late endosomal tethering factor that mediates fusion. In vivo targets of its ligase activity are unknown. | Yogosawa et al. 2006 |
| Rabex-5 | Exchange factor for Rab5; binds Ub and has activity as a Ub ligase, which can be used to catalyze autoubiquitination. GEF activity may be regulated by ubiquitination and Rabex-5 may recognize ubiquitinated proteins to alter their trafficking. | Mattera et al. 2006; Raiborg et al. 2006; Mattera and Bonifacino 2008 |
| Ub peptidases | ||
| Doa4 | Localized to yeast endosomes via direct and indirect interactions with ESCRT-III; required for releasing Ub from cargo before Ub is sorted and degraded in lysososome/vacuoles. | Richter et al. 2007, 2013 |
| USP8/UBPY | Similar in structure to yeast Doa4; associates with both ESCRT-0 and ESCRT-III and plays complementary roles in sorting of ubiquitinated cargoes into MVBs. | Wright et al. 2011 |
| AMSH | Specific for K63 polyubiquitin chains. AMSH also associates with both ESCRT-0 and ESCRT-III to provide multiple functions during MVB sorting. | Clague et al. 2012 |
| USP20 | Together with USP33, these DUbs associate with β2-adrenergic receptors to promote their deubiquitination and recycling. | Berthouze et al. 2009 |
| USP2 | Can be alternatively spliced to generate proteins with different functions; can inhibit down-regulation of ENaC and EGFR. | Oberfeld et al. 2011; Liu et al. 2013 |
| Ub-binding proteins | ||
| ESCRT-0 | Comprised of Hrs and STAM in animal cells or Vps27 and Hse1 in yeast cells; contains multiple Ub-binding domains and localizes with clathrin and PtdIns-3P on endosomes to function as a Ub-sorting receptor. | Komada and Kitamura 2005 |
| Bro1 | Yeast homolog of HD-PTP and ALIX; contributes to the recruitment of the DUb Doa4 to endosomes via its association with ESCRT-III; also, may work as an alternate ESCRT-0-like Ub-sorting receptor. | Richter et al. 2007, 2013; Pashkova et al. 2013 |
| HD-PTP | Functions in concert with ESCRT-0 and ESCRT-I for MVB sorting. Its Ub binding suggests it functions as a Ub-sorting receptor. | Doyotte et al. 2008; Ali et al. 2013; Pashkova et al. 2013 |
| ALIX | Plays multiple roles in MVB biogenesis, viral budding, cytokinesis, and exosome formation; recognizes ubiquitinated viral gag proteins to promote viral budding; may use its Ub-binding properties for alternate functions as well. | Dowlatshahi et al. 2012; Sette et al. 2013 |
| ESCRT-I | Heterotetramer that bridges associations with ESCRT-0 and ESCRT-II; has multiple cellular functions, many shared with Alix. Several alternative subunits can be combined to make specialized ESCRT-I complexes that allow it to provide specific functions. | Morita 2012 |
| ESCRT-II | Tetrameric complex that binds PtdIns-3P and Ub via the amino terminus of Vps36/Eap45; bridges association of ESCRT-I with ESCRT-III during MVB biogenesis. | Gill et al. 2007; Henne et al. 2012 |
| GGA | Shares domain organization and similar biochemical interactions with ESCRT-0 components; binds Ub via VHS and GAT domains; involved as a Ub-sorting receptor at endosomes and the TGN. | Puertollano and Bonifacino 2004; Scott et al. 2004; Ren and Hurley 2010 |
| Tom1/Tollip | Endosomal complex with multiple Ub-binding domains housed in VHS, C2, and UIMs. | Brissoni et al. 2006; Blanc et al. 2009; Zhu et al. 2012 |
| VPS9 | Exchange factor for yeast Rab5/Vps21; binds Ub via CUE domain, which is required for efficient endocytosis to the vacuole. | Davies et al. 2003; Donaldson et al. 2003 |
| Epsin | ENTH domain family protein involved in internalization from the cell surface in animal and yeast cells; associates with ubiquitinated cargo and may be a major Ub-sorting receptor for clathrin-mediated endocytosis. | Sen et al. 2012 |
| Eps15 | Adaptor for internalization from the plasma membrane; may serve as a Ub-sorting receptor for endocytosis; also undergoes ubiquitination itself, which might regulate its function. | de Melker et al. 2004; Stang et al. 2004; Sigismund et al. 2005 |
EGFR, epidermal growth factor receptor; ER, endoplasmic reticulum: MVBs, multivesicular bodies; TGN, trans-Golgi network.
Ubiquitination of cell-surface membrane proteins initiates their journey to lysosomes for degradation (Fig. 1). Ub does this in part by serving as a signal for internalization and subsequent sorting into the interior of lysosomes. In general, the selection of membrane proteins for ubiquitination can either be due to a programmed biological response (such as ligand-mediated receptor down-regulation) or as a way to remove aberrantly folded or damaged proteins from the cell surface as a quality-control mechanism.
Figure 1.
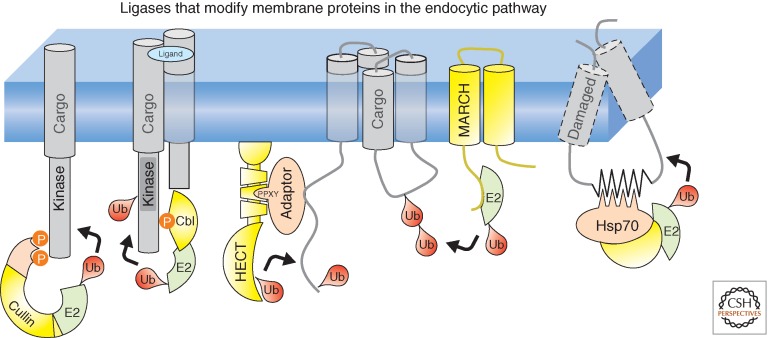
The prominent Ub-ligation complexes that work on cell-surface proteins are pictured here. Cullin-Ring ligases, such as the SCF β-TRCP complex, can modify a subset of receptors (such as the growth hormone receptor) at the cell surface. The Cbl family of ligases has a prominent role in binding to tyrosine phosphorylated receptor tyrosine kinases and ubiquitinating them to induce their ligand-stimulated down-regulation. A wide variety of cell-surface proteins are ubiquitinated by members of the Nedd4 family of ligases that possess a HECT Ub-ligase domain. These ligases can bind directly to substrate proteins or work through an intermediary adaptor protein. MARCH ligases are found in multiple domains of the secretory/endocytic system and can modify a wide variety of membrane proteins. Unfolded and damaged membrane proteins can be recognized by some of the same machinery that recognizes damaged proteins in the endoplasmic reticulum (ER). Pictured here is the CHIP ligase, which functions in a complex with Hsp70 and Hsc70 chaperones.
Ubiquitination of receptor tyrosine kinases (RTKs) is one of the most studied examples for how Ub traffics proteins through the endocytic system. RTKs consist of >50 members in humans and have an essential role in controlling cell proliferation and migration (Lemmon and Schlessinger 2010). Ligand-induced activation of RTKs leads to rapid ubiquitination of several RTKs at the PM by one of the Cbl (for Cas-Br-M ectopic retroviral transforming sequence) RING (really interesting new gene)-type E3 ubiquitin ligase family members (Levkowitz et al. 1999). A canonical example is the epidermal growth factor receptor (EGFR), which upon activation and autophosphorylation of pY1045, can now bind the amino-terminal pY-binding domain (PTB) of Cbl. In addition, receptor ubiquitination is augmented by the adaptor protein, growth factor receptor-bound protein 2 (GRB2), a constitutive partner of Cbl that binds to other pY residues of the EGFR. This facilitates multiple-mono- and Lys63-linked polyubiquitination in a manner dependent on the Ubc4- and Ubc5-conjugating enzymes (Haglund et al. 2003; Huang et al. 2006, 2007). Other RTKs, such as those for hepatocyte growth factor (HGF), vascular endothelial growth factor receptor (VEGFR-1), and platelet-derived growth factor (PDGF) receptors, also rapidly undergo ligand-stimulated ubiquitination by Cbl (Haglund and Dikic 2012). Cbl has other targets as well, such as integrins, which connect cells to the extracellular matrix to control cell adhesion, migration, and proliferation. Ubiquitination of integrins promotes their lysosomal degradation, which in turn prevents the accumulation of ligand-bound integrins within endosomes that might go on to form nonproductive adhesion sites upon exocytosis (Lobert et al. 2010). Ubiquitination of the α5 subunit of the α5β1 fibronectin integrin receptor is triggered by the activation of the FGFR2 (fibroblast growth factor receptor 2) that induces the recruitment of c-Cbl in osteoblasts (Kaabeche et al. 2005).
A group of Ub ligases with emerging importance are the membrane-associated RING-CH (MARCH) ligases, encompassing a family of 11 RING-CH E3 Ub ligases (Nathan and Lehner 2009). MARCH ligases are orthologs of the Kaposi sarcoma-associated herpesvirus (KSVH) immunoevasion ligases K3 and K5 and contain two transmembrane segments and a cytosolic RING-CH domain (Boname and Lehner 2011). Endogenous or overexpressed MARCH ligases play a key role in the ubiquitination-dependent down-regulation and lysosomal degradation of a variety of cell-surface receptors (e.g., MHC-I; K3, K5, MARCH-9 and -4, MHC-II; MARCH-2 and -8, CD4, CD44, and CD98; MARCH-4 and -8) (Nathan and Lehner 2009). The mechanism used by these integral membrane ligases to recognize other membrane protein substrates is intriguing and appears to involve recognition of hydrophobic transmembrane as well as the proximal regions juxtaposed to these transmembrane-spanning segments. A transmembrane hydrophobic patch in concert with a membrane proximal Lys residue at the exofacial interface of HLA-DR constitutes key elements of the recognition surface for MARCH-8, whereas the Ub-acceptor Lys residue has to be located at the proximity of the membrane–cytosol interface (Jahnke et al. 2012; Kajikawa et al. 2012).
Another major class of E3 ligases is the neural precursor-cell-expressed, developmentally down-regulated 4 (Nedd4) family, which contains a catalytic HECT (homologous E6-AP carboxy-terminal) domain (Rotin and Kumar 2009). The Nedd4 family includes yeast Rsp5 and nine human members (e.g., WWP1/2, Smurf1/2, Nedd4, Nedd4-2, and AIP4/Itch) each containing an amino-terminal C2 domain, 2-4 WW domains, and a carboxy-terminal catalytic HECT domain. The WW domains bind PPXY consensus sequences found in substrate proteins. PPXY-containing proteins that undergo this direct recognition mechanism include the heterotrimeric epithelial sodium channel (ENaC), which is down-regulated by Nedd4-2 and mutations within the PPXY motifs stabilize ENaC at the cell surface to cause hypertensive Liddle’s syndrome (Snyder 2009). A similar regulation was recently documented for the PPXY-containing spliced isoform of the ErbB-4 (Carraway 2010). Some WW-binding motifs of substrate proteins are activated by phosphorylation; for instance, phosphorylation of the CXCR-4 carboxy-terminal tail recruits the AIP4/Itch ligase and phosphorylation of the Ste2 GPCR in yeast is required for Rsp5-mediated ubiquitination (Hicke et al. 1998; Marchese et al. 2003; Vina-Vilaseca and Sorkin 2010).
Nedd4 family ligases can also use an indirect mechanism to recognize substrates through the use of adaptor proteins that bind substrates and have their own PPXY motifs allowing them to attach to the ligase WW domains. So far, the biggest collection of adaptors identified is in yeast, which uses an indirect recognition method for targeting Rsp5 to most of its cell-surface membrane proteins. These adaptors include Bsd2 and Sna3, which can mediate Rsp5-catalyzed ubiquitination of manganese and methionine transporters, respectively (Hettema et al. 2004; MacDonald et al. 2012b). A similar set of integral membrane protein Nedd4-family adaptors is found in mammalian cells. Ndfip1/N4WBP5 and Ndfip2/N4WBP5A also contain PPXY motifs and bind to the WW domains of the WWP2 E3 ligase, illustrating the evolutionary conservation of aspects of substrate recognition. This interaction is required for ubiquitination/degradation of the PM metal transporter DMT1/NRAMP2, which plays a critical role in iron homeostasis (Foot et al. 2011). As integral membrane proteins, these adaptors conceivably recognize intermembrane domains analogous to what is proposed for MARCH ligases, although the exact mechanism for how they do this and whether that is sensitive to transporter activity remains to be clarified.
Adaptors also include a larger family of soluble proteins that include Bul1, Bul2, and 11 so-called ART proteins (arrestin-related trafficking) in yeast (O’Donnell 2012). These soluble adaptors, including Bul1/2, belong to the family of α-arrestins. These are related to the better-studied β-arrestin family that has well recognized multiple roles in receptor down-regulation, except that they are equipped with PPXY motifs allowing them to interact with Nedd4-family ligases. Mammalian α-arrestins consist of six arrestin domain-containing adaptors and include ARRDC1-5 and TXNIP. Each Art Rsp5 adaptor is thought to have a repertoire of plasma membrane protein cargo that overlaps to various degrees with that of other Art adaptors. For instance, Bul1/2 has a critical role in ubiquitinating and sorting the Gap1 transporter to the vacuole, whereas Art1 plays a critical role in trafficking the Mup1 transporter (Helliwell et al. 2001; Soetens et al. 2001; Lin et al. 2008). In mammalian cells, ARRDC3 controls ubiquitination and down-regulation of β2 and β3 adrenergic receptors as well as the β4 integrin (Draheim et al. 2010; Nabhan et al. 2010; Patwari et al. 2011; Shea et al. 2012). Recent studies have shown that some α-arrestin proteins are regulated by phosphorylation, in particular kinases downstream from Tor1, thus allowing cell global regulation of plasma membrane protein levels in response to nutrient status (MacGurn et al. 2011; Merhi and Andre 2012). As a consequence of directly binding Nedd4 Ub ligases, all of the α-arrestin-family proteins also undergo ubiquitination. Interestingly, mutants of Art1 or Art4 lacking their major ubiquitinatable lysine target residues function poorly as adaptors for their Can1 and Jen1 transporter substrates suggesting that ubiquitination activates the α-arrestins to bind better to substrate proteins or to the endocytic sorting machinery (Lin et al. 2008; Becuwe et al. 2012).
SELECTION OF DAMAGED PM PROTEINS FOR UBIQUITINATION
Cells also use Ub-dependent endosomal sorting to rid themselves of damaged plasma membrane proteins. To do this, cells must use ligases that can recognize “degrons” indicative of partially unfolded proteins. In yeast cells, damaged membrane proteins—at least those that can escape the endoplasmic reticulum quality-control degradative process (ERAD), are largely targeted by Rsp5, the Nedd4-family ligase that is also responsible for the down-regulation of properly folded transporters and receptors in response to specific biological stimuli. Particular examples include mutant alleles of the Pma1 ATPase, the GPCR Ste2, the cell wall sensor Wsc1, and the arginine permease Can1 (Li et al. 1999; Pizzirusso and Chang 2004; Wang et al. 2011; Zhao et al. 2013). Exactly how Rsp5 recognizes damaged proteins is not yet known, but likely involves the same broad set of adaptor proteins that Rsp5 uses to recognize other substrates. One interesting possibility is that properly folded proteins are induced to form a similar unfolded “degron”-like motif as a trigger for their ubiquitination and down-regulation in response to particular stimuli. For instance, transporter activity itself induces conformational changes that expose arrestin/Rsp5-binding sites and these might mimic small disordered protein segments thought to be exposed upon damaging (Cain and Kaiser 2011; Merhi and Andre 2012; Keener and Babst 2013). Once ubiquitinated, these proteins follow an endosomal sorting complex required for transport (ESCRT)-dependent trafficking route into multivesicular bodies and onto lysosomes for degradation.
Damaged plasma membrane proteins in animal cells also are trafficked to lysosomes for degradation, but how they are recognized for ubiquitination is somewhat different. Here, many of the components that recognize unfolded membrane proteins at the endoplasmic reticulum also operate at the cell surface. A proteomic analysis isolated components of the ubiquitination machinery in association with the temperature-sensitive form of CD4-λ as a reporter protein. When the reporter was triggered to unfold, it recruited heat shock protein 70 (Hsc70), Hsp90, and CHIP (carboxy-terminal Hsp70 interacting protein) (Apaja et al. 2010). CHIP is an E3 ligase that functions in ER and cytosolic protein quality control (Cyr et al. 2002). CHIP consists of an amino-terminal tetratricopeptide (TPR) domain that binds to Hsc70, Hsp70, and Hsp90 molecular chaperones, a central helical domain mediating CHIP dimerization and a carboxy-terminal U-box domain responsible for binding E2-Ub-conjugating enzyme (Zhang et al. 2005). A similar mechanism appears to be involved in the ubiquitin-dependent degradation of the unfolded mutant (ΔF508) CFTR with exposed misfolded cytosolic domains. Both Hsc70 and Hsp90, together with a subset of cochaperones (DNAJA1 [Hdj2], DNAJB2 [HSJ1], Aha1, and HOP) contribute to efficient CHIP recruitment, ubiquitination, and ESCRT-dependent lysosomal targeting of the mutant CFTR (Okiyoneda et al. 2010). Besides CHIP, other Ub ligases can recognize non-native polypeptides via chaperone interactions. These include Ubr1/2, UBE3A, and Cul5; however, their contribution to plasma membrane quality control has not been addressed (Okiyoneda et al. 2011). There are also precedents for chaperone-independent recognition of non-native client protein as exemplified by CHIP, Hrd1, and San1 activity in the cytoplasm, ER, and nucleus, respectively (Rosser et al. 2007; Kanehara et al. 2010; Rosenbaum et al. 2011). Substrate recognition by San1 is catalyzed by intrinsically disordered amino- and carboxy-terminal domains with embedded conserved recognition motifs (Rosenbaum et al. 2011). It is conceivable that multiple ligases are involved in plasma membrane quality control, each with an overlapping set of possible substrates. This possibility is supported by the finding that loss of CHIP partially inhibits lysosomal trafficking of unfolded CFTR, DRD4, and V2R, whereas depletion of other Ub ligases (e.g., Hrd1 and Gp78) attenuate the ΔF508-CFTR removal from the cell surface (Apaja et al. 2010; Okiyoneda et al. 2010).
BASICS OF Ub-BINDING DOMAINS
Distinct Ub signals are recognized and decoded by Ub-binding domains (UBDs) present in a wide variety of cellular proteins (Ub receptors) that are able to link these signals to specific cellular responses (e.g., endocytosis, degradation, or translocation of the modified protein). Ub operates in the endocytic pathway in two main ways: as a sorting signal on cargo being recognized by UBD-containing sorting receptors, or by modifying the endocytic machinery. To date, around 20 families of UBDs present in around 300 proteins have been described, although the repertoire of proteins with Ub-binding activity is still expanding (Rahighi et al. 2009; Husnjak and Dikic 2012). Most of these UBDs bind to an exposed hydrophobic patch of the Ub molecule surrounding I44 with fairly weak micromolar affinity (Fig. 2). In addition, Ub exists in different conformations that become apparent when comparing the structures of various UBD:Ub complexes (Lange et al. 2008). Thus Ub can dynamically adopt several conformations that can subsequently be captured by UBDs by a variable combination of conformational selection and induced fit (Wlodarski and Zagrovic 2009; Long and Bruschweiler 2011; Peters and de Groot 2012). The weak binding of isolated UBDs forces the cell to apply a number of supportive mechanisms that increase the strength of binding and ensure specific signaling. These include increasing avidity by forming oligomeric complexes that contain multiple UBDs, forming chains of poly-Ub exposing several UBD-binding surfaces, and crowding together binding partners where movement is restricted. Importantly, recognition of Ub in the endocytic pathway mostly occurs on membranes where proteins diffuse in only two dimensions rather than in three. Moreover, Ub receptors can have additional motifs for weakly binding their ubiquitinated partner proteins, which not only increases avidity but also adds to the specificity of target selection. This mechanism can also provide the capacity for “coincidence” detection whereby these binding motifs are only active when Ub binding is instigated.
Figure 2.
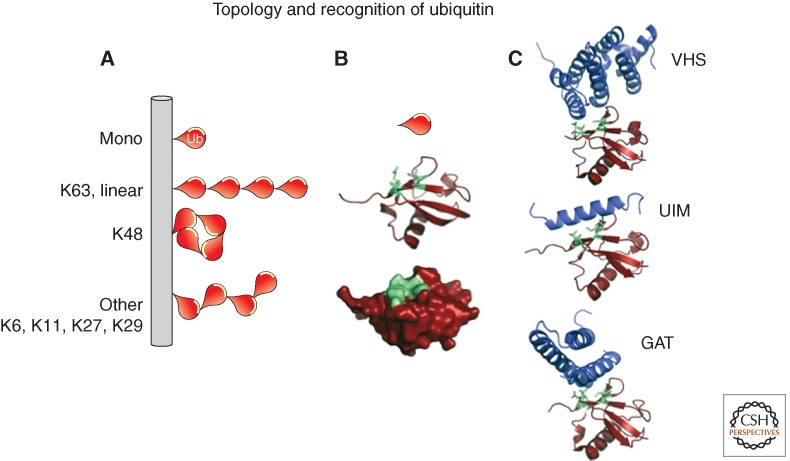
Ub presents a hydrophobic interaction surface, pictured in green, which is responsible for binding to the vast majority of Ub-binding proteins. (A) Scheme of the types of Ub topologies that can be added to substrate proteins. Mono-Ub is a single Ub attached directly to the substrate. Multiple attached Ubs in a chain can be linked via K63 or by the amino terminus to form a linear chain. These chains have an “open” configuration in which the hydrophobic interface of each Ub moiety is highly exposed. Poly-Ub chains linked through K48 adopt a more closed conformation that makes the hydrophobic interface less accessible. Other linkages position this interaction interface in more varied orientations making it more suitable for binding particular subsets of UBD-containing proteins. (B) Structure of Ub (PDBID:1UBQ) with the hydrophobic interaction surface highlighted in green. (C) Examples of three Ub-interaction modules in a complex with Ub. VHS domain: PDBID:3LDZ; UIM:PDBID:1Q0W; GAT:PDBID:1WR6.
There are a wide variety of Ub modifications (mono-, multi-, polyubiquitination of various linkage types), and an important aspect of Ub signaling concerns how the cell distinguishes different topologies. These distinctions are not only relevant for the function of UBD-containing endocytic proteins but also particular DUbs that hydrolyze specific chain types. In isolation and in solution, most known UBDs interact with mono-Ub and do not show a significant preference for a certain chain type (Husnjak and Dikic 2012). Although this preference may be owing to the bias that most of the currently known UBDs were identified and characterized by their ability to bind mono-Ub. A subset of UBDs is able to specifically recognize a particular poly-Ub topology and this is typically achieved by simultaneously binding to two neighboring Ub within the chain that shows a linkage-specific conformation. For example, the UBAN domain of NEMO and the NZF domain of HOIL-1L specifically recognize linear (Met1-linked) Ub chains by making contacts to a surface on one Ub moiety and the canonical hydrophobic surface on the other Ub (Rahighi et al. 2009; Sato et al. 2011). The NZF domains of TAB2 and TAB3 also simultaneously interact with two neighboring Ub moieties, but can do so only when linked via a K63 linkage (Kulathu et al. 2009). Likewise, the K63-specific DUb AMSH, which operates on ubiquitinated endosomal cargo, simultaneously binds two linked Ub moieties to properly position the isopeptide bond for cleavage on its active site (Sato et al. 2008). These examples aside, many UBDs still show some chain preference with many disfavoring binding to K48-linked poly-Ub even though they bind a single molecule of Ub (Rahighi et al. 2009). Structural studies reveal that K48-linked chains adopt a compact conformation that occludes the I44 hydrophobic region. In contrast, K63-linked chains are more extended and the hydrophobic patch is fully exposed. Poly-Ub chains also show a high level of “quaternary” dynamics allowing a variety of dramatically different conformations in solution. Constraining these conformations can directly alter the ability of particular UBDs and DUbs to recognize these chains thus providing additional physical parameters that might regulate Ub recognition in vivo (Ye et al. 2012).
For the most part, mono-Ub and K63-linked chains predominate in the endocytic pathway as sorting signals when attached to cargo proteins, although other linkages such as K11, K29, and K48 may be operational as well (Shields and Piper 2011). In experimental settings, namely, where Ub is permanently attached cargo, a single Ub is sufficient for delivery to lysosomes (Shih et al. 2000; Raiborg et al. 2002; Stringer and Piper 2011). However, as an internalization signal, a single Ub is exceptionally weak and Ub is likely operational only in chains (Barriere et al. 2006; Hawryluk et al. 2006; Bertelsen et al. 2011). For multivesicular body (MVB) sorting, multiple Ubs are the favored signal over mono-Ub (Huang et al. 2006, 2013). In addition, chain formation and disassembly by DUbs are continuously occurring during transport of Ub cargo along the endocytic pathway. Making chains might play a dual role of not only offering better binding to UBD-containing sorting receptors, but serving as a buffer against DUbs encountered along the way that might erode Ub signals before they can engage endosomal sorting machinery (Clague et al. 2012). Similarly, the endocytic machinery, particularly that which contains UBDs, can be ubiquitinated and are predominantly modified by mono-Ub or short K63-linked chains. Mono-Ub is sufficient to induce conformational changes within UBD-containing proteins rendering them unable to recognize and sort ubiquitinated cargo (Hoeller et al. 2006).
In addition to Ub, some UBDs also bind Nedd8, which is 57% identical to Ub; this offers the possibility that this Ub-like protein might partially substitute for Ub in the endocytic pathway (den Besten et al. 2012). Indeed, there is increasing evidence that Nedd8 plays a role in the endocytosis of EGF and TGF-β receptors (Oved et al. 2006; Zuo et al. 2013). In both cases, neddylation is mediated by the Ub E3-ligase c-Cbl. Neddylation of the EGFR seems to serve the same purpose as its ubiquitination: lysosomal sorting and degradation of EGFR. In contrast, neddylation of TβRII inhibits ubiquitination and degradation by promoting its internalization via EEA1-positive early endosomes rather than the caveolar pathway. It still needs to be clarified to which extent neddylation of transmembrane receptors takes place under endogenous conditions; however, these observations hint at a larger cross talk between Ub and other Ub-like systems.
HOW UBIQUITIN MEDIATES INTERNALIZATION FROM THE PLASMA MEMBRANE
Ub can serve as an internalization signal from the plasma membrane. For most proteins, Ub serves as a sorting signal into clathrin-dependent internalization pathways. When compared with other internalization signals, such as YxxL or DxxxLL motifs that bind the AP-2 clathrin adaptor complex, a single Ub molecule is a paltry internalization signal. Engineered Ub-fusion reporter proteins show that multiple Ubs are required to act as an efficient internalization signal on their own (Barriere et al. 2006, 2007; Hawryluk et al. 2006; Bertelsen et al. 2011). Multiple monoubiqitinations was well as polyubiquitin chains are likely to constitute an efficient internalization signal. The major poly-Ub chain that operates at the internalization step appears to be K63 linked, although other linkages have been implicated as well (Boname and Lehner 2011; Zhang et al. 2013). Ub can also function in the context of other weaker internalization signals such that the aggregate effect of multiple Ub and non-Ub signals become sufficient for incorporation into clathrin-coated vesicles (Kumar et al. 2007). Still for other proteins that undergo ubiquitination and internalization from the plasma membrane, Ub itself may not be the operative internalization signal, but instead plays a role later in the endocytic pathway for other trafficking steps (Shenoy et al. 2001; Huang et al. 2006, 2007).
The two best candidates for endocytic adaptor proteins that can link Ub cargoes to the clathrin-mediated endocytic machinery are Epsin and Eps15, which interact with each other and contain UIM domains (Table 1). Epsin favors binding to K63-polyubiquitin, which correlates well with the requirement of multiple Ubs as an internalization signal (Hawryluk et al. 2006; Sato et al. 2009). Other UBDs are present at sites of CME including ones in CIN85 and Hrs, and others are likely to be uncovered (Soubeyran et al. 2002; Stamenova et al. 2007; Mayers et al. 2013). Because relatively few proteins have been assessed for Ub-dependent internalization, it is possible that these additional UBD-containing proteins operate as adaptors for different subsets of ubiquitinated proteins. Depletion of Epsin or Eps15 inhibits internalization of Ub-fusion chimeric reporters showing their overall importance in the process of Ub-dependent internalization (Hawryluk et al. 2006; Sato et al. 2009). Altering Epsin levels can perturb the dynamics of clathrin-coated vesicle assembly and this activity is curtailed when Epsin lacks its UIM domains (Barriere et al. 2006). The biochemical properties of Epsin and its spatiotemporal characteristics during clathrin-coated vesicle assembly qualify it as the main Ub-receptor candidate for CME internalization. Yet, much of the analysis specifically focused on the Ub-binding property of Epsin has been in the context of overexpression experiments that find a loss in the ability of Epsin lacking its UIMs to affect aspects of CME (Sugiyama et al. 2005). A more complicated picture emerges from experiments in yeast, in which several endocytic adaptors can be substituted with versions lacking their UBDs. Here, internalization of ubiquitinated proteins is blocked as well as internalization of proteins bearing alternate internalization signals suggesting the ability to bind Ub serves purposes beyond capturing ubiquitinated cargo (Dores et al. 2010). Interestingly, experiments in Caenorhabditis elegans have shown the Epsin UIMs to be important for Ub-independent endocytosis of LDLR (Kang et al. 2013). One possibility is that Ub cargoes are not simply passive passengers but that their Ub moiety helps properly assemble and orchestrate the clathrin-coated vesicle formation through interaction with UBDs of the associated machinery. This possibility is supported by experiments showing that the presence of Ub on cargo affects the dynamics of clathrin-coated vesicle (CCV) assembly (Henry et al. 2012). This view is also consistent with experiments on the endocytic Ub-sorting ESCRT apparatus that relies on the presence of ubiquitinated cargo to productively assemble on the surface of endosomes (MacDonald et al. 2012a). Yet another view is that ubiquitination of the internalization machinery might drive a powerful regulatory mechanism that involves formation of multiple intra- and intermolecular Ub-UBD interactions. The outcome would be that their ubiquitination inhibits their capacity to bind to and control the functions of other ubiquitinated targets. Both Epsin and Eps15 undergo ubiquitination in cells via mechanisms that rely on their ability to bind to Ub-associated with Ub-ligation machinery (Polo et al. 2002; Woelk et al. 2006). Experimental demonstration that ubiquitination could have consequences on the function of clathrin vesicle machinery has been obtained via overexpression experiments and chimeric Ub-fusion proteins (Fallon et al. 2006; Hoeller et al. 2006). These experiments provide an important conceptual possibility, yet the precise and likely intricate mechanisms that may regulate the physiological functions of clathrin vesicle machinery via its ubiquitination remain unresolved.
SORTING OF UBIQUITINATED MEMBRANE PROTEINS INTO MULTIVESICULAR ENDOSOMES
Once Ub cargo is delivered to early endosomes it can undergo sorting into intraluminal vesicles (ILVs) or remain on the limiting membrane of endosomes to later be recycled to the plasma membrane or trans-Golgi network (TGN) (Huotari and Helenius 2011). For most proteins, incorporation into intraluminal vesicles seals their degradative fate because this sorting step is generally irreversible. ILV formation continues until the late endosome stage, whereupon fusion to lysosomes allows the ILVs and their protein cargo to be delivered to the proteolytic lumen of the lysosome thus affording complete degradation of integral membrane protein cargo (Fig. 3). The intraluminal vesicles that populate late endosomes make them synonymous with multivesicular endosomes or multivesicular bodies (MVBs); however, not all ILVs contain proteins that are sorted from endosomal membranes and destined for lysosomal degradation. For instance, at least some of these intraluminal membranes serve as storage compartments as exosomes, lamellar bodies, or MHC-II complexes (Babst 2011; Raposo and Stoorvogel 2013). How Ub might be used to target proteins to this alternative ILV compartments is not yet clear and may be independent of Ub as has been shown for the sorting of MHC-II in activated antigen-presenting cells. In addition, the main sorting apparatus for sorting Ub cargo, the ESCRT apparatus, is not critically required for exosome formation or the generation of other classes of ILVs. The machinery responsible for the generation of these alternate ILVs will be interesting to elucidate, especially given the recent recognition of their profound regulatory activity given their capacity to deliver proteins and miRNA to distant cells. Such alternative Ub-independent MVB pathways appear absent from yeast, and the combination of structure/function studies in both yeast and animal cells provide a solid yet still evolving biochemical understanding of the conserved MVB sorting pathway responsible for lysosomal degradation of ubiquitinated membrane proteins.
Figure 3.
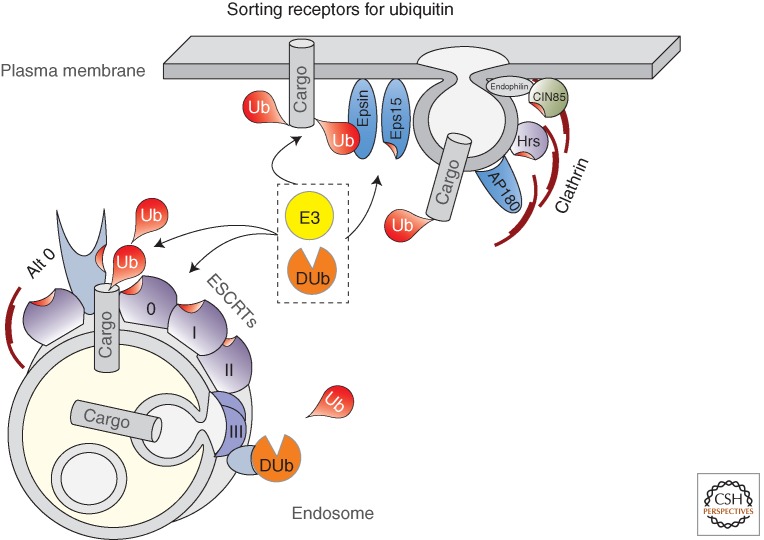
Cohort of proteins involved in recognition and processing of ubiquitinated membrane proteins at the cell surface and at endosomes. At the cell surface, proteins such as Epsin and Eps15 recognize ubiquitinated proteins and recruit them into clathrin-coated vesicles. Other Ub-binding proteins such as CIN85 and Hrs may have a similar role in helping usher ubiquitinated cargo into vesicles. Once ubiquitinated cargo reaches the endosomes, it is recognized by the ESCRT apparatus, composed of ESCRT-0, -I, -II, and -III. The Hrs subunit of ESCRT-0 has multiple Ub-binding domains making it a critical Ub-sorting receptor, which might pass Ub cargo to other ESCRTs that also bear Ub-binding domains. Other endosomal proteins may serve as alternative ESCRT-0-like receptors that deliver ubiquitinated cargo into the larger ESCRT apparatus. These “Alt 0” proteins include GGAs, TOM1, and the Bro1-family of proteins that includes Bro1, Alix, and HD-PTP.
Many of the proteins required for the biogenesis of ILVs and Ub-dependent sorting were first identified in yeast genetic screens (Coonrod and Stevens 2010). Loss of these proteins, often referred to as class E Vps proteins, cause defects in MVB formation, sorting of Ub cargo into the lysosome (vacuole) lumen, and cause a cohort of other phenotypes including enlarged late endosomes that accumulated recycling Golgi proteins and vacuolar membrane proteins. Collectively, these proteins define the ESCRT apparatus (endosomal sorting complex required for transport) comprised of discrete complexes (ESCRTs -0, -I, -II, and -III) as well as accessory factors that regulate ESCRT function. The ESCRT apparatus couples cargo recognition and sorting with the formation and scission of ILVs (Hurley and Hanson 2010). ILV formation is distinct from other vesicle formation events such as those formed by COP-I or clathrin because vesicle formation proceeds by pushing membranes out of the cytosol rather than pulling vesicle buds into it. In addition, ESCRT proteins are not stoichiometric components of the finished vesicle and have a robust mechanism(s) to ensure that they are not consumed in the vesicle-making process, although how this is achieved remains unclear currently. Finally, ILVs must have lipid composition that is functionally distinct from that of the limiting membrane, which allows the lysosomal hydrolases to selectively destroy ILV but not the limiting membrane. A wealth of structural studies, combined with liposome-based reconstitution of key events in vitro has suggested a concerted mechanism whereby lipids undergo sorting and phase separation into a microdomain constrained by an interconnected ESCRT apparatus clustered at the neck of forming ILVs (Hurley and Hanson 2010). A spiral polymer of ESCRT-III subunits forms at the bud neck to allow ILVs to pinch off into the lumen. This same membrane constriction activity of ESCRT-III that is harnessed for ILV formation on endosomes is also used for budding of retroviruses from the plasma membrane and membrane scission during cytokinesis in animal cells and some Archea (Hanson et al. 2008; Lata et al. 2008; Lindas et al. 2008; Samson et al. 2008; Wollert et al. 2009; Henne et al. 2012, 2013). Before ILV scission, Ub can be removed from cargo to help replenish the cytosolic pool of Ub. This process has been best observed in yeast, whereby the Doa4 deubiquitinating enzyme is recruited to ESCRT-III via the accessory factor Bro1 (Luhtala and Odorizzi 2004). Loss of Doa4 causes Ub to accumulate in the vacuole lumen and be depleted from the cytosol, which in turn impacts a number of Ub-dependent processes (Amerik et al. 2000). Interestingly, the activity of Doa4 appears to be regulated in response to Ub levels indicating that disposal of Ub via the MVB pathway may be used as a regulatory device to alter the activity of the entire ubiquitin/proteasome system (Kimura et al. 2009). The best candidate mammalian Doa4 ortholog is Usp8/UBPY, which can also bind ESCRT-III; however, its role in rescuing Ub from lysosomal degradation has not been specifically determined (Wright et al. 2011). Indeed, USP8/UBPY as well as a similarly positioned DUb, AMSH, can bind both ESCRT-0 and ESCRT-III and a host of experiments looking at different cargoes suggest they can play a role in deubiquitinating cargo before MVB sorting to promote cargo recycling (McCullough et al. 2004; Mizuno et al. 2005; Bowers et al. 2006; Berlin et al. 2010; Zhou et al. 2013), cargo deubiquitination after sorting as a way to release them from the ESCRT apparatus and promote sorting into MVBs (Alwan and van Leeuwen 2007; Balut et al. 2011), or as a way of deubiquitinating ESCRT components to rescue them from degradation or inactive conformations (Row et al. 2007; Sierra et al. 2010). One of the interesting aspects of cargo deubiquitination in the final stages of sorting is that it predicts that cargo is sorted into a domain capable of trapping Ub cargo such that it cannot escape upon deubiquitination. It is unknown what constitutes such a cargo trap; however, a tight meshwork of polymerized ESCRT-III in the initial stages of vesicle constriction has been proposed (Henne et al. 2012).
Ub cargo at endosomes is first recognized by ESCRT-0, composed of Vps27 and Hse1 in yeast and of Hrs and STAM1/2 in humans. ESCRT-0 has a number of biochemical features that allow it to function as the initial receptor for Ub cargo (Shields and Piper 2011). First, it has several UBDs: one within each of its two amino-terminal VHS domains, three UIMs found in the middle of both proteins, and an additional UBD within the SH3 domain of STAM2 (Lange et al. 2012a). The UBDs within ESCRT-0 are required for its association with Ub cargo and its ability to sort that cargo into MVB ILVs. Hrs also binds an isoform of Eps15, another UBD-containing protein, which localizes to endosomes and may help convey a subset of internalized proteins to the ESCRT-0 complex. ESCRT-0 also binds PtdIns3-P via an FYVE domain and clathrin with its carboxyl terminus, allowing it to localize to specialized subregions within early endosomes where proteins are segregated for recycling versus degradation.
ESCRT-0 also binds directly to ESCRT-I and can initiate recruitment of ESCRT-I to membranes. ESCRT-I binds to and recruits ESCRT-II, which then directs the assembly and activity of the ESCRT-III vesicle scission machinery. ESCRT-I and -II also have UBDs. ESCRT-I is a heterotetramer. Only four subunit exist for composing yeast ESCRT-1, whereas humans have additional isoforms that may help specialize ESCRT-I for a variety of cellular functions. ESCRT-I binds Ub via a UEV domain within TSG101 and a triplet of UBA domains within UBAP1, two subunits that are functionally orthologous to yeast Vps23 and Mvb12, respectively, which also bind Ub. UBAP1 is one of three alternative Mvb12 orthologs that can be incorporated into the mammalian ESCRT-I heterotetramer, but is the one with the highest Ub-binding activity and appears to be the one most suited for sorting Ub cargo into MVBs (Stefani et al. 2011; Agromayor et al. 2012). Mutations that inactivate Ub binding by yeast Vps23 and Mvb12 or by mammalian UBAP1 block sorting of Ub cargo suggesting that ESCRT-I also functions as a Ub-sorting receptor (Shields et al. 2009; Stefani et al. 2011). ESCRT-II binds Ub via its Vps36/Eap45 subunit, which also binds endosomal PtdIns-3P. The yeast and mammalian Vps36 bind Ub via different mechanisms with an NZF domain mediating Ub binding by yeast Vps36 and mammalian Vps36/Eap45 binding via a pleckstrin homology domain variant termed a GLUE domain (Shields and Piper 2011). It is not yet clear whether Ub binding by ESCRT-II is critical for sorting Ub cargo. Depletion of mammalian ESCRT-II does not cause dramatic sorting defects comparable to ESCRT-I depletion and the contribution of its Ub-binding activity in the sorting process has not been determined (Bowers et al. 2006). Yeast ESCRT-II is essential for MVB sorting; however, loss of its Ub-binding activity produces only minor to moderate sorting defects.
Sequential Orchestration of ESCRT Function
The fact that the many UBDs within the ESCRT apparatus bind to similar overlapping surfaces on Ub and that cargo can be sorted into ILVs when it is modified only by a single Ub or short polyubiquitin chain supports the idea that individual ESCRTs work in sequence to bind Ub cargo and pass it onto successive acting ESCRTs. This sequential sorting process is orchestrated by the ability of ESCRTs to directly interact with ESCRT-0 binding, activating, and transferring cargo to ESCRT-I, and similarly ESCRT-I to ESCRT-II. The ability of ESCRT-I and -II to form a supercomplex and the lack of a clear necessary role for Ub binding by ESCRT-II suggests ESCRT-I and -II function together rather than individually in sequence, as a secondary Ub-sorting receptor that receives cargo from ESCRT-0 (Amerik et al. 2006; Shields et al. 2009; Boura et al. 2012). The idea that these work in sequence is supported by immunolocalization experiments showing a differential distribution of ESCRT-0 and ESCRT-I, live cell imaging that shows sequential assembly of ESCRTs at the sites of viral budding and cytokinesis, and biochemical studies in yeast mutants where assembly intermediates can be accumulated (Babst et al. 2002; Bache et al. 2003; Elia et al. 2011; Guizetti et al. 2011; Jouvenet et al. 2011). In addition, several other proteins that are structurally related to ESCRT-0 have been proposed to work in parallel to ESCRT-0 as alternative early Ub-cargo receptors. These include Tom1, Tom1L1, and GGA3, which bind clathrin, Ub, and ESCRT-I supporting the idea that several ESCRT-0-like complexes gather Ub cargo and pass it to a central ESCRT-I/-II supercomplex (Shields and Piper 2011; Haglund and Dikic 2012). Recently, other contenders for Ub-sorting receptors have been identified including Bro1, Alix, and HD-PTP, which bind Ub, and interact with ESCRT-I and ESCRT-III (Ali et al. 2013; Pashkova et al. 2013). These proteins could potentially offer an additional sorting pathway for Ub cargo in parallel with ESCRT-0.
Exactly how ESCRTs are spatially coordinated to move Ub cargo has yet to be determined. This aspect has not been reconstituted in vitro and our understanding remains largely speculative. Any model must account for two features: getting Ub cargo into the forming ILV and getting the sorting receptors off of it. Current models favor a variation of a concentric model for ILV formation, guided by the circular structures generated by the ESCRT-III polymer that is ultimately responsible for vesicle scission (Nickerson et al. 2007; Henne et al. 2012; Rozycki et al. 2012). Here, ESCRT-I and -II are either on the outside of a forming vesicle neck, pushing cargo into the center, or are already in the center being surrounded by a polymerizing ESCRT-III.
Roles of Multiple UBDs
The ESCRT apparatus has multiple UBDs that could potentially provide a number of functions. One would be to recognize a wide variety of Ub cargoes, each presenting Ub in a variety of contexts (Mayers et al. 2011). Multiple UBDs would also boost the avidity of the ESCRTs for Ub cargo. Indeed, the affinity of individual ESCRT UBDs for mono-Ub is quite low and crowding them together would help increase their ability to gather Ub cargo; likewise, disassembly of the ESCRT apparatus would provide a mechanism to disengage from Ub cargo (Haglund and Dikic 2012). Another possibility is that multiple UBDs may specify recognition of polyubiquitin chains. Endocytic cargoes are typically marked with short K63 polyubiquitin chains, and yeast mutants incapable of making K63 chains have major defects in MVB sorting (Lauwers et al. 2010). Specificity for K63-linked chains, as opposed to multiple-mono-Ub, has been suggested by the cooperative binding K63 poly-Ub shows for the multiple UBDs within ESCRT-0 or a single UBD within yeast Vps36 (Kulathu et al. 2009; Lange et al. 2012b). However, these effects are moderate and a single Ub on cargo is sufficient to confer sorting into ILVs, indicating that the role of K63 chains may be to simply increase the number of mono-Ub sorting signals attached to cargo (Ren and Hurley 2010; Stringer and Piper 2011).
The other possibility is that multiple UBDs may serve as a means to regulate ESCRT activity. Indeed, elimination of multiple UBDs within ESCRT-0 or the ESCRT-I/II supercomplex cause defects beyond merely loss of Ub-cargo sorting. Recent studies indicate that multiple UBDs may provide a coincidence mechanism for the ESCRT apparatus to properly assemble wherein the availability of proper lipids, membrane-associated ESCRTs, and Ub cargo are simultaneously required before sorting and ILV formation can ensue (MacDonald et al. 2012a). Other functions for UBDs within the ESCRT apparatus have been proposed based on the observation that many ESCRT components become ubiquitinated themselves (Erpapazoglou et al. 2012). Often, UBDs confer ubiquitination owing to the fact that proteins with UBDs can bind Ub chains that bring them into the proximity of Ub ligases forming those chains. A potential role for ubiquitination of the machinery is that it could mediate interaction with UBDs in either intra- or intermolecular interactions (Haglund and Dikic 2012). Artificial ubiquitination of ESCRTs can inactivate them. Moreover, ESCRTs-0 and -I associate with a variety of Ub ligases and DUbs and their depletion and overexpression hint at a variety of regulatory roles apart from merely modifying Ub cargo (Wright et al. 2011). Nevertheless, preventing ubiquitination of ESCRT-0 and ESCRT-I does not block their ability to execute MVB sorting indicating that the role of ESCRT ubiquitination may be for an ancillary function (such as controlling their levels via targeting to the proteasome) or providing an additional layer of regulation (Shields and Piper 2011).
CONCLUDING REMARKS AND FUTURE DIRECTIONS
The last decade has established a compelling paradigm for how Ub mediates trafficking events within the endocytic pathway by acting as a sorting signal. This paradigm is replete with cellular machinery that recognizes Ub cargo and sorts into a variety of transport intermediates. In broad terms, this process works by aggregation of multiple low-affinity UBDs within a particular sorting apparatus, which generally recognizes Ub cargo modified by several Ub moieties, often presented as short K63-linked polyubiquitin chains. This basic framework is helpful; however, many aspects are significantly underdetermined and more work is required for us to transition from a basic and generic process we can describe now to a more accurate, sophisticated, and nuanced understanding that accounts for the complexity of the endocytic system. Such an understanding is becoming more approachable both experimentally and conceptually with the development of new tools to manipulate and measure ubiquitination and with an increasing appreciation for the expanding effects ubiquitination can exert and the specialized functions different polyubiquitin chains can fulfill.
In terms of understanding Ub as a lysosomal sorting signal, the vast majority of studies have focused on well-folded and well-behaved cell-surface proteins that undergo “down-regulation” and delivery to the lysosome in response to a particular trigger. Far less is known about how damaged proteins are selected and sorted toward lysosomes and whether different machinery is required to process these proteins. In addition, if Ub works as a sorting signal for internalization from the cell surface, as a sorting signal from the TGN, or possibly at other locales distinct from the MVB, it makes sense that a Ub-sorting signal might be used only transiently, as a stepwise sorting signal to discrete locales within the endocytic system without committing cargo entirely to lysosomal degradation. Yet, few experimental strategies are configured to test for this possibility.
The endocytic machinery that recognizes Ub cargo is largely coming into focus. Our most complete understanding is of the ESCRT apparatus, which functions on endosomes to mediate sorting into ILVs of multivesicular late endosomes. We have far less understanding of how Ub cargoes are recognized and sorted at the plasma membrane, TGN, and elsewhere within the endocytic system. Despite our detailed structural understanding of the ESCRT apparatus, a better understanding remains to be ascertained in terms of knowing all the proteins that interact with Ub cargo, whether some are specialized for different types of cargo, what they do to move that cargo into ILVs, and how their activity can be stimulated or inhibited. Moreover, several players within the Ub/endosomal system have multiple functions within the cell (such as the ESCRTs that function in MVB sorting, cytokinesis, and transcription) or multiple roles in the same process (such as the AMSH, USP8, and Bro1 during MVB sorting) making it difficult for overexpression or underexpression experiments to deconvolute their contributions. Exploiting the emerging structural details for how these components interact to make specific mutants defective in particular functional aspects should help parse these roles more precisely.
Another challenge is posed by the nature of how Ub is recognized, which is typically accomplished via an expanding variety of domains that often bind with poor affinity sometimes in the millimolar range. Indeed the plethora of low-affinity UBDs that are emerging not only presents the daunting prospect that Ub may control a large swath of biology we have yet to fathom but also poses the experimental obligation to show that such UBDs indeed contribute to a Ub-dependent biological process. The low affinity of UBDs typically obliges them to work in a larger context, in concert with other UBDs or perhaps within protein complexes that undergo a conformational change in response to its own ubiquitination. A seductive observation is that most UBDs make the proteins they reside in susceptible to ubiquitination, affording them a vast potential for different modes of regulation. For instance, the GTP exchange factor for Rab5, Rabex5, has proposed roles as a Ub ligase against itself and other targets in the endocytic pathway and as a Ub-binding protein that engages Ub cargo as well as a ubiquitinated form of itself—all of which may impinge on the ability of Rabex-5 to activate Rab5 (Mattera et al. 2006; Mattera and Bonifacino 2008; Xu et al. 2010; Yan et al. 2010; Aikawa et al. 2012). Such discoveries promise a rich and interesting future. Nonetheless, they pose a significant challenge because the physiological relevance of these models can be difficult to dissect clearly.
Footnotes
Editors: Sandra L. Schmid, Alexander Sorkin, and Marino Zerial
Additional Perspectives on Endocytosis available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Agromayor M, Soler N, Caballe A, Kueck T, Freund SM, Allen MD, Bycroft M, Perisic O, Ye Y, McDonald B, et al. 2012. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure 20: 414–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa Y, Hirakawa H, Lee S 2012. Spatiotemporal regulation of the ubiquitinated cargo-binding activity of Rabex-5 in the endocytic pathway. J Biol Chem 287: 40586–40597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N, Zhang L, Taylor S, Mironov A, Urbe S, Woodman P 2013. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol 23: 453–461 [DOI] [PubMed] [Google Scholar]
- Alwan HA, van Leeuwen JE 2007. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J Biol Chem 282: 1658–1669 [DOI] [PubMed] [Google Scholar]
- Amerik AY, Nowak J, Swaminathan S, Hochstrasser M 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell 11: 3365–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik A, Sindhi N, Hochstrasser M 2006. A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J Cell Biol 175: 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers A, Ramjaun AR, McPherson PS 2004. The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J Biol Chem 279: 11471–11479 [DOI] [PubMed] [Google Scholar]
- Apaja PM, Xu H, Lukacs GL 2010. Quality control for unfolded proteins at the plasma membrane. J Cell Biol 191: 553–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M 2011. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol 23: 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3: 283–289 [DOI] [PubMed] [Google Scholar]
- Bache KG, Brech A, Mehlum A, Stenmark H 2003. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol 162: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balut CM, Loch CM, Devor DC 2011. Role of ubiquitylation and USP8-dependent deubiquitylation in the endocytosis and lysosomal targeting of plasma membrane KCa3.1. FASEB J 25: 3938–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere H, Nemes C, Lechardeur D, Khan-Mohammad M, Fruh K, Lukacs GL 2006. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in mammalian cells. Traffic 7: 282–297 [DOI] [PubMed] [Google Scholar]
- Barriere H, Nemes C, Du K, Lukacs GL 2007. Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol Biol Cell 18: 3952–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Leon S 2012. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol 196: 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh-Touze N, Leon S, Erpapazoglou Z, Stawiecka-Mirota M, Urban-Grimal D, Haguenauer-Tsapis R 2008. Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem Soc Trans 36: 791–796 [DOI] [PubMed] [Google Scholar]
- Berlin I, Schwartz H, Nash PD 2010. Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8.STAM complex. J Biol Chem 285: 34909–34921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen V, Sak MM, Breen K, Rodland MS, Johannessen LE, Traub LM, Stang E, Madshus IH 2011. A chimeric pre-ubiquitinated EGF receptor is constitutively endocytosed in a clathrin-dependent, but kinase-independent manner. Traffic 12: 507–520 [DOI] [PubMed] [Google Scholar]
- Berthouze M, Venkataramanan V, Li Y, Shenoy SK 2009. The deubiquitinases USP33 and USP20 coordinate β2 adrenergic receptor recycling and resensitization. EMBO J 28: 1684–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Trejo J, Benovic JL, Marchese A 2007. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J Biol Chem 282: 36971–36979 [DOI] [PubMed] [Google Scholar]
- Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F 2009. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic 10: 161–171 [DOI] [PubMed] [Google Scholar]
- Boname JM, Lehner PJ 2011. What has the study of the K3 and K5 viral ubiquitin E3 ligases taught us about ubiquitin-mediated receptor regulation? Viruses 3: 118–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Chung HS, Herrick DZ, Canagarajah B, Cafiso DS, Eaton WA, Hummer G, Hurley JH 2012. Solution structure of the ESCRT-I and -II supercomplex: Implications for membrane budding and scission. Structure 20: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, Lehner PJ, Luzio JP 2006. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem 281: 5094–5105 [DOI] [PubMed] [Google Scholar]
- Brissoni B, Agostini L, Kropf M, Martinon F, Swoboda V, Lippens S, Everett H, Aebi N, Janssens S, Meylan E, et al. 2006. Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr Biol 16: 2265–2270 [DOI] [PubMed] [Google Scholar]
- Cain NE, Kaiser CA 2011. Transport activity-dependent intracellular sorting of the yeast general amino acid permease. Mol Biol Cell 22: 1919–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway KL III 2010. E3 ubiquitin ligases in ErbB receptor quantity control. Semin Cell Dev Biol 21: 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Guggino W 2013. Ubiquitination and degradation of CFTR by the E3 ubiquitin ligase MARCH2 through its association with adaptor proteins CAL and STX6. PLoS ONE 8: e68001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Liu H, Urbe S 2012. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell 23: 457–467 [DOI] [PubMed] [Google Scholar]
- Coonrod EM, Stevens TH 2010. The yeast vps class E mutants: The beginning of the molecular genetic analysis of multivesicular body biogenesis. Mol Biol Cell 21: 4057–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Hohfeld J, Patterson C 2002. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci 27: 368–375 [DOI] [PubMed] [Google Scholar]
- da Silva Almeida AC, Hocking HG, Boelens R, Strous GJ, van Rossum AG 2013. βTrCP interacts with the ubiquitin-dependent endocytosis motif of the GH receptor in an unconventional manner. Biochem J 453: 291–301 [DOI] [PubMed] [Google Scholar]
- Davies BA, Topp JD, Sfeir AJ, Katzmann DJ, Carney DS, Tall GG, Friedberg AS, Deng L, Chen Z, Horazdovsky BF 2003. Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. J Biol Chem 278: 19826–19833 [DOI] [PubMed] [Google Scholar]
- de Melker AA, van der Horst G, Borst J 2004. Ubiquitin ligase activity of c-Cbl guides the epidermal growth factor receptor into clathrin-coated pits by two distinct modes of Eps15 recruitment. J Biol Chem 279: 55465–55473 [DOI] [PubMed] [Google Scholar]
- den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ 2012. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol 19: 511–516, S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Yin H, Gekakis N, Supek F, Joazeiro CA 2003. Ubiquitin signals protein trafficking via interaction with a novel ubiquitin binding domain in the membrane fusion regulator, Vps9p. Curr Biol 13: 258–262 [DOI] [PubMed] [Google Scholar]
- Dores MR, Schnell JD, Maldonado-Baez L, Wendland B, Hicke L 2010. The function of yeast epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic 11: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, Sundquist WI, Kopito RR 2012. ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev Cell 23: 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P 2008. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci 105: 6308–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draheim KM, Chen HB, Tao Q, Moore N, Roche M, Lyle S 2010. ARRDC3 suppresses breast cancer progression by negatively regulating integrin β4. Oncogene 29: 5032–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci 108: 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erpapazoglou Z, Dhaoui M, Pantazopoulou M, Giordano F, Mari M, Leon S, Raposo G, Reggiori F, Haguenauer-Tsapis R 2012. A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol Biol Cell 23: 2170–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyster CA, Cole NB, Petersen S, Viswanathan K, Fruh K, Donaldson JG 2011. MARCH ubiquitin ligases alter the itinerary of clathrin-independent cargo from recycling to degradation. Mol Biol Cell 22: 3218–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L, Belanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, et al. 2006. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI3K-Akt signalling. Nat Cell Biol 8: 834–842 [DOI] [PubMed] [Google Scholar]
- Foot NJ, Leong YA, Dorstyn LE, Dalton HE, Ho K, Zhao L, Garrick MD, Yang B, Hiwase D, Kumar S 2011. Ndfip1-deficient mice have impaired DMT1 regulation and iron homeostasis. Blood 117: 638–646 [DOI] [PubMed] [Google Scholar]
- Fotia AB, Ekberg J, Adams DJ, Cook DI, Poronnik P, Kumar S 2004. Regulation of neuronal voltage-gated sodium channels by the ubiquitin-protein ligases Nedd4 and Nedd4-2. J Biol Chem 279: 28930–28935 [DOI] [PubMed] [Google Scholar]
- Fujita H, Iwabu Y, Tokunaga K, Tanaka Y 2013. Membrane-associated RING-CH (MARCH) 8 mediates the ubiquitination and lysosomal degradation of the transferrin receptor. J Cell Sci 126: 2798–2809 [DOI] [PubMed] [Google Scholar]
- Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL 2007. Structural studies of phosphoinositide 3-kinase-dependent traffic to multivesicular bodies. Biochem Soc Symp 74: 47–57 [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331: 1616–1620 [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I 2012. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci 125: 265–275 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Han SO, Xiao K, Kim J, Wu JH, Wisler JW, Nakamura N, Freedman NJ, Shenoy SK 2012. MARCH2 promotes endocytosis and lysosomal sorting of carvedilol-bound β2-adrenergic receptors. J Cell Biol 199: 817–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR 2013. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 152: 1051–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk MJ, Keyel PA, Mishra SK, Watkins SC, Heuser JE, Traub LM 2006. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic 7: 262–281 [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Losko S, Kaiser CA 2001. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J Cell Biol 153: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Zhao Y, Emr SD 2012. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151: 356–371 [DOI] [PubMed] [Google Scholar]
- *.Henne WM, Stenmark H, Emr SD 2013. Molecular mechanisms of the membrane scultping ESCRT pathway. Cold Spring Harb Perspect Biol 5: a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry AG, Hislop JN, Grove J, Thorn K, Marsh M, von Zastrow M 2012. Regulation of endocytic clathrin dynamics by cargo ubiquitination. Dev Cell 23: 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Valdez-Taubas J, Pelham HR 2004. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J 23: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Zanolari B, Riezman H 1998. Cytoplasmic tail phosphorylation of the α-factor receptor is required for its ubiquitination and internalization. J Cell Biol 141: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, et al. 2006. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol 8: 163–169 [DOI] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A 2006. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell 21: 737–748 [DOI] [PubMed] [Google Scholar]
- Huang F, Goh LK, Sorkin A 2007. EGF receptor ubiquitination is not necessary for its internalization. Proc Natl Acad Sci 104: 16904–16909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Zeng X, Kim W, Balasubramani M, Fortian A, Gygi SP, Yates NA, Sorkin A 2013. Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proc Natl Acad Sci 110: 15722–15727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A 2011. Endosome maturation. EMBO J 30: 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI 2010. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat Rev Mol Cell Biol 11: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Dikic I 2012. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem 81: 291–322 [DOI] [PubMed] [Google Scholar]
- Ishido S, Goto E, Matsuki Y, Ohmura-Hoshino M 2009. E3 ubiquitin ligases for MHC molecules. Curr Opin Immunol 21: 78–83 [DOI] [PubMed] [Google Scholar]
- Jahnke M, Trowsdale J, Kelly AP 2012. Structural requirements for recognition of major histocompatibility complex class II by membrane-associated RING-CH (MARCH) protein E3 ligases. J Biol Chem 287: 28779–28789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol 13: 394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaabeche K, Guenou H, Bouvard D, Didelot N, Listrat A, Marie PJ 2005. Cbl-mediated ubiquitination of α5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J Cell Sci 118: 1223–1232 [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Li PC, Goto E, Miyashita N, Aoki-Kawasumi M, Mito-Yoshida M, Ikegaya M, Sugita Y, Ishido S 2012. The intertransmembrane region of Kaposi’s sarcoma-associated herpesvirus modulator of immune recognition 2 contributes to B7-2 downregulation. J Virol 86: 5288–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Xie W, Ng DT 2010. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol 188: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YL, Yochem J, Bell L, Sorensen EB, Chen L, Conner SD 2013. Caenorhabditis elegans reveals a FxNPxY-independent low-density lipoprotein receptor internalization mechanism mediated by epsin1. Mol Biol Cell 24: 308–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener JM, Babst M 2013. Quality control and substrate-dependent downregulation of the nutrient transporter Fur4. Traffic 14: 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yashiroda H, Kudo T, Koitabashi S, Murata S, Kakizuka A, Tanaka K 2009. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell 137: 549–559 [DOI] [PubMed] [Google Scholar]
- Komada M, Kitamura N 2005. The Hrs/STAM complex in the downregulation of receptor tyrosine kinases. J Biochem 137: 1–8 [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Komander D 2012. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 13: 508–523 [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D 2009. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol 16: 1328–1330 [DOI] [PubMed] [Google Scholar]
- Kumar KG, Barriere H, Carbone CJ, Liu J, Swaminathan G, Xu P, Li Y, Baker DP, Peng J, Lukacs GL, et al. 2007. Site-specific ubiquitination exposes a linear motif to promote interferon-α receptor endocytosis. J Cell Biol 179: 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL 2008. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 320: 1471–1475 [DOI] [PubMed] [Google Scholar]
- Lange A, Castaneda C, Hoeller D, Lancelin JM, Fushman D, Walker O 2012a. Evidence for cooperative and domain-specific binding of the signal transducing adaptor molecule 2 (STAM2) to Lys63-linked diubiquitin. J Biol Chem 287: 18687–18699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Ismail MB, Riviere G, Hologne M, Lacabanne D, Guilliere F, Lancelin JM, Krimm I, Walker O 2012b. Competitive binding of UBPY and ubiquitin to the STAM2 SH3 domain revealed by NMR. FEBS Lett 586: 3379–3384 [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W 2008. Helical structures of ESCRT-III are disassembled by VPS4. Science 321: 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B 2010. The ubiquitin code of yeast permease trafficking. Trends Cell Biol 20: 196–204 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 4: 1029–1040 [DOI] [PubMed] [Google Scholar]
- Li Y, Kane T, Tipper C, Spatrick P, Jenness DD 1999. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol Cell Biol 19: 3588–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135: 714–725 [DOI] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R 2008. A unique cell division machinery in the Archaea. Proc Natl Acad Sci 105: 18942–18946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkowitz S, Weissman AM 2011. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 11: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zanata SM, Kim J, Peterson MA, Di Vizio D, Chirieac LR, Pyne S, Agostini M, Freeman MR, Loda M 2013. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene 32: 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert VH, Brech A, Pedersen NM, Wesche J, Oppelt A, Malerod L, Stenmark H 2010. Ubiquitination of α5β1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev Cell 19: 148–159 [DOI] [PubMed] [Google Scholar]
- Long D, Bruschweiler R 2011. In silico elucidation of the recognition dynamics of ubiquitin. PLoS Comput Biol 7: e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N, Odorizzi G 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol 166: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Buchkovich NJ, Stringer DK, Emr SD, Piper RC 2012a. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep 13: 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Stringer DK, Piper RC 2012b. Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting. Traffic 13: 586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Smolka MB, Emr SD 2011. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147: 1104–1117 [DOI] [PubMed] [Google Scholar]
- Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell 5: 709–722 [DOI] [PubMed] [Google Scholar]
- Mattera R, Bonifacino JS 2008. Ubiquitin binding and conjugation regulate the recruitment of Rabex-5 to early endosomes. EMBO J 27: 2484–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Tsai YC, Weissman AM, Bonifacino JS 2006. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J Biol Chem 281: 6874–6883 [DOI] [PubMed] [Google Scholar]
- Mayers JR, Fyfe I, Schuh AL, Chapman ER, Edwardson JM, Audhya A 2011. ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J Biol Chem 286: 9636–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Wang L, Pramanik J, Johnson A, Sarkeshik A, Wang Y, Saengsawang W, Yates JR III, Audhya A 2013. Regulation of ubiquitin-dependent cargo sorting by multiple endocytic adaptors at the plasma membrane. Proc Natl Acad Sci 110: 11857–11862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clague MJ, Urbe S 2004. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol 166: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell GS, Philpott A 2013. Non-canonical ubiquitylation: Mechanisms and consequences. Int J Biochem Cell Biol 45: 1833–1842 [DOI] [PubMed] [Google Scholar]
- Merhi A, Andre B 2012. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol Cell Biol 32: 4510–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M 2005. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell 16: 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E 2012. Differential requirements of mammalian ESCRTs in multivesicular body formation, virus budding and cell division. FEBS J 279: 1399–1406 [DOI] [PubMed] [Google Scholar]
- Mossinger J, Wieffer M, Krause E, Freund C, Gerth F, Krauss M, Haucke V 2012. Phosphatidylinositol 4-kinase IIα function at endosomes is regulated by the ubiquitin ligase Itch. EMBO Rep 13: 1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Pan H, Lu Q 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the β2-adrenergic receptor. EMBO Rep 11: 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan JA, Lehner PJ 2009. The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp Cell Res 315: 1593–1600 [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Russell MR, Odorizzi G 2007. A concentric circle model of multivesicular body cargo sorting. EMBO Rep 8: 644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberfeld B, Ruffieux-Daidie D, Vitagliano JJ, Pos KM, Verrey F, Staub O 2011. Ubiquitin-specific protease 2-45 (Usp2-45) binds to epithelial Na+ channel (ENaC)-ubiquitylating enzyme Nedd4-2. Am J Physiol Renal Physiol 301: F189–F196 [DOI] [PubMed] [Google Scholar]
- O’Donnell AF 2012. The running of the Buls: Control of permease trafficking by α-arrestins Bul1 and Bul2. Mol Cell Biol 32: 4506–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL 2010. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T, Apaja PM, Lukacs GL 2011. Protein quality control at the plasma membrane. Curr Opin Cell Biol 23: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, et al. 2006. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem 281: 21640–21651 [DOI] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Sunshine AB, Rich M, Dunham MJ, Yu L, Piper RC 2013. The yeast Alix Homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev Cell 25: 520–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, Zhang Y, Dobrin R, Cohen DE, Larsen PR, et al. 2011. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metab 14: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JH, de Groot BL 2012. Ubiquitin dynamics in complexes reveal molecular recognition mechanisms beyond induced fit and conformational selection. PLoS Comput Biol 8: e1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzirusso M, Chang A 2004. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast. Mol Biol Cell 15: 2401–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416: 451–455 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS 2004. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol 6: 244–251 [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394–398 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Slagsvold T, Stenmark H 2006. A new side to ubiquitin. Trends Biochem Sci 31: 541–544 [DOI] [PubMed] [Google Scholar]
- Ramanathan HN, Zhang G, Ye Y 2013. Monoubiquitination of EEA1 regulates endosome fusion and trafficking. Cell Biosci 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W 2013. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol 200: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hurley JH 2010. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J 29: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, West M, Odorizzi G 2007. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J 26: 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CM, West M, Odorizzi G 2013. Doa4 function in ILV budding is restricted through its interaction with the Vps20 subunit of ESCRT-III. J Cell Sci 126: 1881–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, et al. 2011. Disorder targets misorder in nuclear quality control degradation: A disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell 41: 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM 2007. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem 282: 22267–22277 [DOI] [PubMed] [Google Scholar]
- Rotin D, Kumar S 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10: 398–409 [DOI] [PubMed] [Google Scholar]
- Row PE, Liu H, Hayes S, Welchman R, Charalabous P, Hofmann K, Clague MJ, Sanderson CM, Urbe S 2007. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem 282: 30929–30937 [DOI] [PubMed] [Google Scholar]
- Rozycki B, Boura E, Hurley JH, Hummer G 2012. Membrane-elasticity model of Coatless vesicle budding induced by ESCRT complexes. PLoS Comput Biol 8: e1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD 2008. A role for the ESCRT system in cell division in archaea. Science 322: 1710–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S 2008. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455: 358–362 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S 2009. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Fujita H, Yoshikawa A, Yamashita M, Yamagata A, Kaiser SE, Iwai K, Fukai S 2011. Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc Natl Acad Sci 108: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, Kearney WR, Robertson AD, Boman AL, Piper RC 2004. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat Cell Biol 6: 252–259 [DOI] [PubMed] [Google Scholar]
- Sen A, Madhivanan K, Mukherjee D, Aguilar RC 2012. The epsin protein family: Coordinators of endocytosis and signaling. Biomol Concepts 3: 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette P, Nagashima K, Piper RC, Bouamr F 2013. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea FF, Rowell JL, Li Y, Chang TH, Alvarez CE 2012. Mammalian α-arrestins link activated seven transmembrane receptors to Nedd4 family e3 ubiquitin ligases and interact with β-arrestins. PLoS ONE 7: e50557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ 2001. Regulation of receptor fate by ubiquitination of activated β2-adrenergic receptor and β-arrestin. Science 294: 1307–1313 [DOI] [PubMed] [Google Scholar]
- Shields SB, Piper RC 2011. How ubiquitin functions with ESCRTs. Traffic 12: 1306–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R 2009. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol 185: 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Sloper-Mould KE, Hicke L 2000. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J 19: 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra MI, Wright MH, Nash PD 2010. AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J Biol Chem 285: 13990–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S 2005. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci 102: 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder PM 2009. Down-regulating destruction: Phosphorylation regulates the E3 ubiquitin ligase Nedd4-2. Sci Signal 2: pe41. [DOI] [PubMed] [Google Scholar]
- Soetens O, De Craene JO, Andre B 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J Biol Chem 276: 43949–43957 [DOI] [PubMed] [Google Scholar]
- Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I 2002. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature 416: 183–187 [DOI] [PubMed] [Google Scholar]
- Stamenova SD, French ME, He Y, Francis SA, Kramer ZB, Hicke L 2007. Ubiquitin binds to and regulates a subset of SH3 domains. Mol Cell 25: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stang E, Blystad FD, Kazazic M, Bertelsen V, Brodahl T, Raiborg C, Stenmark H, Madshus IH 2004. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol Biol Cell 15: 3591–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani F, Zhang L, Taylor S, Donovan J, Rollinson S, Doyotte A, Brownhill K, Bennion J, Pickering-Brown S, Woodman P 2011. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol 21: 1245–1250 [DOI] [PubMed] [Google Scholar]
- Stringer DK, Piper RC 2011. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol 192: 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S, Kishida S, Chayama K, Koyama S, Kikuchi A 2005. Ubiquitin-interacting motifs of Epsin are involved in the regulation of insulin-dependent endocytosis. J Biochem 137: 355–364 [DOI] [PubMed] [Google Scholar]
- Sundvall M, Korhonen A, Paatero I, Gaudio E, Melino G, Croce CM, Aqeilan RI, Elenius K 2008. Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc Natl Acad Sci 105: 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Kooij B, Verbrugge I, de Vries E, Gijsen M, Montserrat V, Maas C, Neefjes J, Borst J 2013. Ubiquitination by the membrane-associated RING-CH-8 (MARCH-8) ligase controls steady-state cell surface expression of tumor necrosis factor-related apoptosis inducing ligand (TRAIL) receptor 1. J Biol Chem 288: 6617–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kerkhof P, Westgeest M, Hassink G, Strous GJ 2011. SCFTrCP acts in endosomal sorting of the GH receptor. Exp Cell Res 317: 1071–1082 [DOI] [PubMed] [Google Scholar]
- Varshavsky A 2012. The ubiquitin system, an immense realm. Annu Rev Biochem 81: 167–176 [DOI] [PubMed] [Google Scholar]
- Vina-Vilaseca A, Sorkin A 2010. Lysine 63-linked polyubiquitination of the dopamine transporter requires WW3 and WW4 domains of Nedd4-2 and UBE2D ubiquitin-conjugating enzymes. J Biol Chem 285: 7645–7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Thibault G, Ng DT 2011. Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity. J Biol Chem 286: 29376– 29387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Klevit RE 2012. Following Ariadne’s thread: A new perspective on RBR ubiquitin ligases. BMC Biol 10: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarski T, Zagrovic B 2009. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc Natl Acad Sci 106: 19346–19351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, Polo S 2006. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol 8: 1246–1254 [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH 2009. Membrane scission by the ESCRT-III complex. Nature 458: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Berlin I, Nash PD 2011. Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination. Cell Biochem Biophys 60: 39–46 [DOI] [PubMed] [Google Scholar]
- Xu L, Lubkov V, Taylor LJ, Bar-Sagi D 2010. Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr Biol 20: 1372–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Schonherr C, Varshney GK, Dogru M, Hallberg B, Palmer RH 2013. Goliath family E3 ligases regulate the recycling endosome pathway via VAMP3 ubiquitylation. EMBO J 32: 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Jahanshahi M, Horvath EA, Liu HY, Pfleger CM 2010. Rabex-5 ubiquitin ligase activity restricts Ras signaling to establish pathway homeostasis in Drosophila. Curr Biol 20: 1378–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Blaser G, Horrocks MH, Ruedas-Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D 2012. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogosawa S, Kawasaki M, Wakatsuki S, Kominami E, Shiba Y, Nakayama K, Kohsaka S, Akazawa C 2006. Monoubiquitylation of GGA3 by hVPS18 regulates its ubiquitin-binding ability. Biochem Biophys Res Commun 350: 82–90 [DOI] [PubMed] [Google Scholar]
- Zelcer N, Hong C, Boyadjian R, Tontonoz P 2009. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH 2005. Chaperoned ubiquitylation—Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell 20: 525–538 [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu M, Scotti E, Chen ZJ, Tontonoz P 2013. Both K63 and K48 ubiquitin linkages signal lysosomal degradation of the LDL receptor. J Lipid Res 54: 1410–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Macgurn JA, Liu M, Emr S 2013. The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. Elife 2: e00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Patel SV, Snyder PM 2007. Nedd4-2 catalyzes ubiquitination and degradation of cell surface ENaC. J Biol Chem 282: 20207–20212 [DOI] [PubMed] [Google Scholar]
- Zhou R, Tomkovicz VR, Butler PL, Ochoa LA, Peterson ZJ, Snyder PM 2013. Ubiquitin-specific peptidase 8 (USP8) regulates endosomal trafficking of the epithelial Na+ channel. J Biol Chem 288: 5389–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wang L, Luo X, Zhang Y, Ding Q, Jiang X, Wang X, Pan Y, Chen Y 2012. Tollip, an intracellular trafficking protein, is a novel modulator of the transforming growth factor-β signaling pathway. J Biol Chem 287: 39653–39663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Huang F, Chiang YJ, Li M, Du J, Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, et al. 2013. c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-β type II receptor. Mol Cell 49: 499–510 [DOI] [PubMed] [Google Scholar]