Abstract
The term “epigenetics” was originally used to denote the poorly understood processes by which a fertilized zygote developed into a mature, complex organism. With the understanding that all cells of an organism carry the same DNA, and with increased knowledge of mechanisms of gene expression, the definition was changed to focus on ways in which heritable traits can be associated not with changes in nucleotide sequence, but with chemical modifications of DNA, or of the structural and regulatory proteins bound to it. Recent discoveries about the role of these mechanisms in early development may make it desirable to return to the original definition of epigenetics.
“Epigenetics” originally denoted processes that allow a single fertilized egg to develop into numerous highly differentiated tissues. Many now use the term to refer just to chromatin modifications, but the original definition may be most appropriate.
1. INTRODUCTION
The history of epigenetics is linked with the study of evolution and development. But during the past 50 years, the meaning of the term “epigenetics” has itself undergone an evolution that parallels our dramatically increased knowledge of the molecular mechanisms underlying regulation of gene expression in eukaryotes. Our present definitions of epigenetics reflect our understanding that although the complement of DNA is essentially the same in all of an organism’s somatic cells, patterns of gene expression differ greatly among different cell types, and these patterns can be clonally inherited. This has led to a working definition of epigenetics as “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” (Riggs et al. 1996; Riggs and Porter 1996). More recently added to this definition is the constraint that initiation of the new epigenetic state should involve a transient mechanism separate from the one required to maintain it (Berger et al. 2009). Until the 1950s, however, the word epigenetics was used more broadly (and less precisely) to categorize all of the developmental events leading from the fertilized zygote to the mature organism—that is, all of the regulated processes that, beginning with the genetic material, shape the final product (Waddington 1953). This concept had its origins in the much earlier studies in cell biology and embryology, beginning in the late 19th century, which laid the groundwork for our present understanding of the relationship between genes and development. There was a long debate among embryologists about the nature and location of the components responsible for carrying out the developmental plan of the organism. In trying to make sense of a large number of ingenious but ultimately confusing experiments involving the manipulation of cells and embryos, embryologists divided into two schools: Those who thought that each cell contained preformed elements that enlarged during development (“preformationism”), and those who thought the process involved chemical reactions among soluble components that executed a complex developmental plan (“epigenesis”). These views focused on the relative importance of the nucleus and cytoplasm in the developmental process. Although the definition that we choose for epigenetics has changed to accommodate our increasing knowledge, it is important to remember that the original problem was: How can a single fertilized egg give rise to a complex organism with cells of varied phenotypes?
Following Fleming’s discovery of the existence of chromosomes in 1879, experiments by many investigators, including Wilson and Boveri, provided strong evidence that the developmental program resided in the chromosomes. Thomas Hunt Morgan (Morgan 1911) ultimately provided the most persuasive proof of this idea through his demonstration of the genetic linkage of several Drosophila genes to the X chromosome. From that point onward, rapid progress was made in creating linear chromosome maps in which individual genes were assigned to specific sites on the Drosophila chromosomes (Sturtevant 1913). Of course, the questions of classic “epigenesis” remained: What molecules within the chromosomes carried the genetic information, how did they direct the developmental program, and how was the information transmitted during cell division? It was understood that both nucleic acid and proteins were present in chromosomes, but their relative contributions were not obvious; certainly, no one believed that the nucleic acid alone could carry all of the developmental information. Furthermore, earlier questions persisted about the possible contribution of the cytoplasm to developmental events. Evidence from Drosophila genetics (see Section 2) suggested that heritable changes in phenotype could occur without corresponding changes in the “genes.” This debate was dramatically altered by the identification of DNA as the primary carrier of genetic information. Ultimately, it became useful to redefine epigenetics so as to distinguish heritable changes that arise from sequence changes in DNA from those that do not. Further refinements of the definition have accompanied more detailed understanding of underlying mechanisms, but it may not be useful at this point to attempt ultimate precision in describing very complex regulatory processes in which “epigenetic” and “nonepigenetic” components are entwined.
2. CLUES FROM GENETICS AND DEVELOPMENT
Whatever the vagaries of the definition, the ideas and scientific data that underlie the present concept of epigenetics have been accumulating steadily since the early part of the 20th century. In 1930, H.J. Muller (Muller 1930) described a class of Drosophila mutations he called “eversporting displacements” (“eversporting” denoting the high rate of phenotypic change). These mutants involved chromosome translocations (displacements), but “even when all parts of the chromatin appeared to be represented in the right dosage—though abnormally arranged—the phenotypic result was not always normal.” In some of these cases, Muller observed flies that had mottled eyes. He thought that this was probably attributable to a “genetic diversity of the different eye-forming cells,” but further genetic analysis led him to connect the unusual properties with chromosomal rearrangement, and to conclude that “chromosome regions, affecting various characters at once, are somehow concerned, rather than individual genes or suppositious ‘gene elements.’” Over the next 10 to 20 years, strong evidence provided by many laboratories (Hannah 1951) confirmed that this variegation arose when rearrangements juxtaposed the white gene with heterochromatic regions.
During that period, chromosomal rearrangements of all kinds were the object of a great deal of attention. It was apparent that genes were not completely independent entities; their function could be affected by their location within the genome—as amply shown by the many Drosophila mutants that led to variegation, as well as by other mutants involving translocation to euchromatic regions, in which more general (nonvariegating) position effects could be observed. The role of transposable elements in plant genetics also became clear, largely through the work of McClintock (McClintock 1965), although they are probably not involved in normal development.
A second line of reasoning came from the study of developmental processes. It was evident that during development there was a divergence of phenotypes among differentiating cells and tissues, and it appeared that such distinguishing features, once established, could be clonally inherited by the dividing cells. Although it was understood at this point that cell-specific programming existed, and that it could be transmitted to daughter cells, how this was done was less clear.
A number of mechanisms could be imagined, and were considered. For those with a biochemical point of view, a cell was defined by the multiple interdependent biochemical reactions that maintained its identity. For example, it was suggested in 1949 by Delbruck (quoted in Jablonka and Lamb 1995) that a simple pair of biochemical pathways, each of which produced as an intermediate an inhibitor of the other pathway, could establish a system that could switch between one of two stable states. Actual examples of such systems were found somewhat later in the lac operon of Escherichia coli (Novick and Weiner 1957), and in the lambda phage switch between lysogenic and lytic states (Ptashne 1992). Functionally equivalent models could be envisioned in eukaryotes: The kinds of self-stabilizing inhibitory and stimulatory mechanisms observed in lambda phage are in fact seen in greatly more complex form in higher organisms. In the sea urchin embryo, for example, development proceeds through the establishment and progression of a series of self-stabilizing regulatory networks. However, it is important to recognize an essential difference between the prokaryotic and eukaryotic systems: In the case of the sea urchin, each of the regulatory “modules” is not in a static state, but rather receives from and sends signals to other modules that give rise to the changing, time-dependent phenotype associated with a developing embryo. It should also be noted that although chromatin structure and biochemistry must certainly be involved in the implementation of this program (see Section 5), the system can be modeled entirely in terms of control of gene expression by specific binding of expressed factors to the regulatory regions of relevant genes.
The extent to which nucleus and cytoplasm each contributed to the transmission of a differentiated state in the developing embryo was of course a matter of intense interest and debate; a self-stabilizing biochemical pathway would presumably have to be maintained through cell division. A second kind of epigenetic transmission was clearly shown in Paramecia and other ciliates, in which the ciliary patterns may vary among individuals and are inherited clonally (Beisson and Sonneborn 1965). Altering the cortical pattern by microsurgery results in transmission of a new pattern to succeeding generations. It has been argued that related mechanisms are at work in metazoans, in which the organization of cellular components is influenced by localized cytoplasmic determinants in a way that can be transmitted during cell division (Grimes and Aufderheide 1991).
3. DNA IS THE SAME IN ALL SOMATIC CELLS OF AN ORGANISM
Although chromosome morphology indicated that all somatic cells possessed all of the chromosomes, it could not have been obvious that all somatic cells retained the full complement of DNA present in the fertilized egg. Nor was it even clear that a protein-free DNA molecule could carry genetic information until the work of Avery, MacLeod, and McCarty (Avery et al. 1944), and that of Hershey and Chase (Hershey and Chase 1952), a conclusion strongly reinforced by Watson and Crick’s solution of the structure of DNA in 1953. Work by Briggs and King (Briggs and King 1952) in Rana pipiens and by Laskey and Gurdon (Laskey and Gurdon 1970) in Xenopus had shown that introduction of a nucleus from early embryonic cells into enucleated oocytes could result in development of an embryo. But as late as 1970, Laskey and Gurdon could state that: “It has yet to be proved that somatic cells of an adult animal possess genes other than those necessary for their own growth and differentiation.” In the article containing this statement, they went on to show that to a first approximation, the DNA of a somatic cell nucleus was competent to direct embryogenesis when introduced into an enucleated egg. It was now clear that the program of development, and the specialization of the repertoire of expression seen in somatic cells, must involve signals that are not the result of some deletion or mutation in the germline DNA sequence when it is transmitted to somatic cells. At the same time, other experiments revealed that these signals could confer phenotypic stability over many generations of cell division. Even after undifferentiated Drosophila imaginal disc cells were transplanted and cultured in successive generations of adult flies, they maintained their disc-specific patterns of differentiation when transferred to larvae (Hadorn 1965; McClure and Schubiger 2007).
Of course, there are ways in which the DNA of somatic cells can come to differ from that of the germline with consequences for the cellular phenotype: For example, transposable elements can alter the pattern of expression in somatic cells as demonstrated by the work of Barbara McClintock and other plant geneticists. Similarly, the generation of antibody diversity involves DNA rearrangement in a somatic cell lineage. This rearrangement (or, more precisely, its consequences) can be considered a kind of epigenetic event, consistent with the early observations of position-effect variegation described by Muller (1930). However, much of the work on epigenetics in recent years has focused on systems in which no DNA rearrangements have occurred, and the emphasis has therefore been on modifications to the bases, and to the proteins that are complexed with DNA within the nucleus.
4. THE ROLE OF DNA METHYLATION
X-chromosome inactivation in the mouse provided an early model of this kind of epigenetic mechanism that involved no DNA rearrangement (Ohno et al. 1959; Lyon 1961). The silenced X chromosome was clearly chosen at random and then clonally inherited in somatic cells, and there was no evidence of changes in the DNA sequence itself. Riggs (1975) and Holliday and Pugh (1975) proposed that DNA methylation could act as an epigenetic mark, in part, to account for this kind of inactivation. The key elements in this model were the ideas that sites of DNA methylation were palindromic, and that distinct enzymes were responsible for methylation of unmodified DNA and DNA already methylated on one strand. It was postulated that the first DNA methylation event would be much more difficult than the second; once the first strand was modified, however, the complementary strand would quickly be modified at the same palindromic site. A DNA methylation mark present on a parental strand would be copied on the daughter strand following replication resulting in faithful transmission of the methylated state to the next generation. Shortly thereafter, Bird took advantage of the fact that the principal target of methylation in animals is the sequence CpG (Doskocil and Sorm 1962) to introduce the use of methylation-sensitive restriction enzymes as a way of detecting the DNA methylation state. Subsequent studies (Bird and Southern 1978; Bird 1978) then showed that endogenous CpG sites were either completely unmethylated or completely methylated. The predictions of the model were thus confirmed, establishing a mechanism for epigenetic transmission of the methylation mark through semiconservative propagation of the methylation pattern.
In the years following these discoveries, a great deal of attention has been focused on endogenous patterns of DNA methylation, the possible transmission of these patterns through the germline, the role of DNA methylation in silencing gene expression, possible mechanisms for initiation or inhibition of methylation at a fully unmethylated site, and the identification of the enzymes responsible for de novo methylation and for maintenance of methylation on already methylated sites. There also has been intense interest in possible mechanisms by which the methyl group at methylated cytosine residues (or 5mC itself) is removed, something that occurs early in development and in the germline. Evidently, the extent to which DNA methylation can be an epigenetic mark preserved through the germline is determined by which sites survive the demethylation events. Although much of the DNA methylation seen in vertebrates is associated with repetitive and retroviral sequences and may serve to maintain these sequences in a permanently silent state, there can be no question that in many cases this modification provides the basis for epigenetic transmission of the gene activity status. This is most clearly demonstrated at imprinted loci (Cattanach and Kirk 1985) such as the mouse or human Igf2/H19 locus, in which one allele is marked by DNA methylation, which in turn controls expression from both genes (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000).
At the same time, it was clear that this could not be the only mechanism for epigenetic transmission of information. For example, as noted in Section 2, position-effect variegation had been observed many years earlier in Drosophila, an organism that has extremely low levels of DNA methylation. Furthermore, in subsequent years, Drosophila geneticists had identified the Polycomb and Trithorax groups of genes, which appeared to be involved in permanently “locking in” the state of activity, either off or on, respectively, of clusters of genes during development. The fact that these states were stably transmitted during cell division suggested an underlying epigenetic mechanism.
5. THE ROLE OF CHROMATIN
It had been recognized for many years that the proteins bound to DNA in the eukaryotic nucleus, especially the histones, might be involved in modifying the properties of DNA. Well before most of the work on DNA methylation began, Stedman and Stedman (1950) proposed that the histones could act as general repressors of gene expression. They argued that because all somatic cells of an organism had the same number of chromosomes, they had the same genetic complement (although this was not demonstrated until some years later, as noted in Section 3). Understanding the subtlety of histone modifications was far in the future though, so Stedman and Stedman operated on the assumption that different kinds of cells in an organism must have different kinds of histones to generate the observed differences in phenotype. Histones can indeed reduce levels of transcript far below those commonly observed for inactive genes in prokaryotes. Subsequent work addressed the capacity of chromatin to serve as a template for transcription, and asked whether that capacity was restricted in a cell-type-specific manner. Other results suggested that only a small fraction of DNA, packaged as chromatin, was accessible to enzyme probes (Cedar and Felsenfeld 1973). Nonetheless, there was a period in which it was commonly believed that the histones were suppressor proteins that passively silenced gene expression. In this view, activating a gene simply meant stripping off the histones; once that was done, it was thought transcription would proceed pretty much as it did in prokaryotes. There was, however, some evidence that extended regions of open DNA did not exist in eukaryotic cells (Clark and Felsenfeld 1971), focusing attention on promoters and other specific regulatory sites. Furthermore, even if the naked DNA model was correct, it was not clear how the decision would be made as to which histone-covered regions should be cleared.
The resolution of this problem began as early as 1964, when Allfrey and Mirsky (1964) speculated that histone acetylation might be correlated with gene activation, and that “active” chromatin might not necessarily be stripped of histones. In the ensuing decade, there was great interest in examining the relationship between histone modifications and gene expression. Modifications other than acetylation (methylation and phosphorylation) were identified, but their functional significance was unclear. It became much easier to address this problem after the discovery by Kornberg and Thomas (1974) that DNA was packaged into the nucleosome, the fundamental histone-containing chromatin subunit. The determination of the crystal structure of the nucleosome, first at 7-Å and then at 2.8-Å resolution, also provided important structural information, particularly evidence for the extension of the histone amino-terminal tails beyond the DNA–protein octamer core, making evident their accessibility to modification (Richmond et al. 1984; Luger et al. 1997). Beginning in 1980 and extending for some years, Grunstein and his collaborators (Wallis et al. 1980; Durrin et al. 1991), applying yeast genetic analysis, were able to show that the histone amino-terminal tails were essential for regulation of gene expression, and for the establishment of silent chromatin domains.
The ultimate connection to detailed mechanisms began with the critical demonstration by Allis and coworkers (Brownell et al. 1996) that a histone acetyltransferase from Tetrahymena was homologous to the yeast transcriptional regulatory protein Gcn5, providing direct evidence that histone acetylation was connected to control of gene expression. Complementary evidence came from a study showing that a mammalian histone deacetylase was related to the yeast repressive transcriptional regulator Rpd3p (Taunton et al. 1996). Since then there has been an explosion in the discovery of histone modifications, as well as a reevaluation of the roles of those that were known previously.
This still did not answer the question of how the sites for modification were chosen in vivo. It had been shown, for example (Pazin et al. 1994), that Gal4-VP16 could activate transcription from a reconstituted chromatin template in an ATP-dependent manner. Activation was accompanied by repositioning of nucleosomes, and it was suggested that this was the critical event in making the promoter accessible. A fuller understanding of the significance of these findings required the identification of ATP-dependent nucleosome remodeling complexes such as SWI/SNF and NURF (Peterson and Herskowitz 1992; Tsukiyama and Wu 1995), and the realization that both histone modification and nucleosome remodeling were involved in preparing the chromatin template for transcription. More recent results from many laboratories reveal that individual sites vary in the order and identity of the enzymes that performed these steps. It should, however, be clear that the initial determinants of specific gene activity during normal development must involve regulatory factors that recognize and bind to particular DNA sequences at enhancers, promoters, and other control sites. These factors have typically been proteins with DNA sequence-specific binding domains coupled to domains that recruit cofactors, directly or indirectly affecting gene expression, including, in many cases, histone modification or nucleosome remodeling complexes. The first direct evidence for the primacy of DNA-binding factors came from the work of Weintraub (Davis et al. 1987; Tapscott et al. 1988; Weintraub et al. 1989) and his collaborators who showed that overexpression of the protein MyoD in fibroblasts and other tissues induced conversion to myoblasts. It is now understood that such sequence-specific binding events establish the initial states of regulation; the epigenetic mechanisms that follow provide ways of maintaining those initial states once they are established. Of course, disruption of such established epigenetic patterns can alter phenotypes.
Complexes that modify or remodel histones can be delivered in a site-specific way, not only by proteins, but also by RNA. Recently, it has become clear that certain kinds of noncoding RNAs are also capable of localized binding coupled to recruitment of regulatory complexes (Chu et al. 2011). For example, in the case of HOTAIR (Rinn et al. 2007) and Kcnq1ot1 (Pandey et al. 2008; Mohammad et al. 2010), the RNAs tend to associate with DNA fairly near their own sites of synthesis, and bring with them the histone modifying Polycomb complex PRC2 (see below), bound to a specific sequence on each RNA. Kcnq1ot1 can recruit a DNA methyltransferase as well (Mohammad et al. 2010). Both HOTAIR and the noncoding RNA telomerase RNA component have been shown to bind directly to distinct motifs on DNA, providing targeting specificity (Chu et al. 2012).
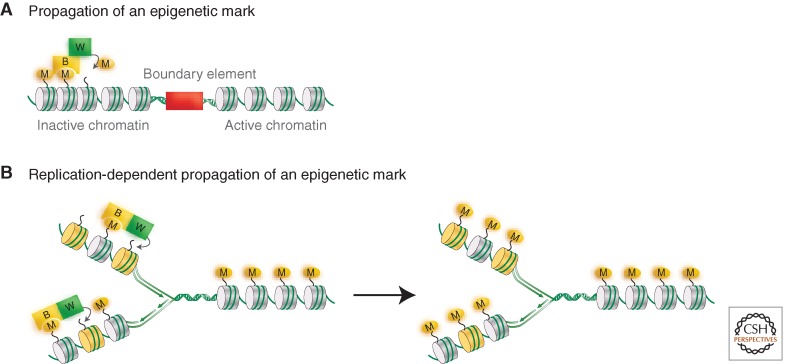
It was not clear how information about the state of activity could, using these mechanisms, be transmitted through cell division; their role in epigenetic transmission of information was thus unclear. The next important step came from the realization that modified histones could recruit proteins in a modification-specific way that could, in turn, affect the local structural and functional states of chromatin. It was found, for example, that methylation of histone H3 at lysine 9 resulted in the recruitment of the heterochromatin protein HP1 (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001). Furthermore, HP1 could recruit the enzyme (Suv39 H1) that is responsible for that methylation. This led to a model for the propagation of the silenced chromatin state along a region, through a processive mechanism (Fig. 1A). Equally important, it provided a reasonable explanation of how that state could be transmitted and survive the DNA replication cycle (Fig. 1B). Recent attention has focused on the Polycomb group proteins (Margueron and Reinberg 2011), and particularly the PRC2 complex that contains a component protein, Ezh2/E(Z), that methylates histone H3 at residue K27, a mark associated with heterochromatin. Analogously to the H3 lysine 9-related mechanism, the PRC2 complex binds to H3K27me3 (Hansen et al. 2008). This involves another member of the PRC2 complex, Eed/ESC, that contains a domain that interacts with methylated H3K27, and this interaction in turn stimulates the methyltransferase activity of Ezh2 (Margueron et al. 2008; Margueron et al. 2009). The arrangement suggests the same kind of propagation mechanism proposed for H3K9 methylation as illustrated in Figure 1A. It still remains to be determined whether these mechanisms that can account for spreading of a histone modification down a polynucleosome chain also function during mitosis.
Figure 1.
Propagation of epigenetic marks. (A) A general mechanism for propagating a histone modification such as H3K9 methylation typically found in heterochromatic regions. The modified histone tail (M) interacts with a protein binder (B) that has a binding site specific for that modification. B also has a specific interaction site with an enzyme “writer” (W) that carries out the same histone modification on an adjacent nucleosome (gray cylinder). Spreading of the histone mark will continue until the modifying machinery reaches a boundary element, delineating the boundary between heterochromatin and euchromatin. (B) A general mechanism for maintaining a histone modification during replication. Newly deposited nucleosomes (yellow), which may incorporate histone variants, are interspersed with parental nucleosomes (shaded in gray) following DNA replication. The modified histone tail (M) on the parental nucleosome interacts with a protein binder (B). As in A, B interacts with a “writer” (W), which catalyzes a histone modification on the histone tail from an adjacent daughter nucleosome.
Despite these results, the issue of the role of histone modifications in epigenetic processes continues to be a source of confusion. It is clear that although the term “epigenetic modification” is frequently used, a given histone modification occurring at a given site in the genome may not necessarily be part of an epigenetic mechanism, but simply part of a biochemical process such as gene expression or DNA strand breakage repair.
Different kinds of propagation mechanisms have been suggested that depend on variant histones rather than modified histones (Ahmad and Henikoff 2002; McKittrick et al. 2004). Histone H3 is incorporated into chromatin only during DNA replication. In contrast, the histone variant H3.3, which differs from H3 by four amino acids, is incorporated into nucleosomes in a replication-independent manner, and it tends to accumulate in active chromatin, in which it is enriched in the “active” histone modifications (McKittrick et al. 2004). It has been proposed that the presence of H3.3 is sufficient to maintain the active state and that after replication, although it would be diluted twofold, enough H3.3 would remain to maintain the active state. The consequent transcription would result in replacement of H3-containing nucleosomes with H3.3, thus perpetuating the active state in the next generation. Results obtained by Ng and Gurdon (Ng and Gurdon 2005; Ng and Gurdon 2008a; Ng and Gurdon 2008b) strongly support such a model. In nuclear transplant experiments, they showed that when nuclei from cells expressing endodermal genes are transplanted into enucleated Xenopus eggs there is considerable expression of those genes in animal pole cells (which should not express them) from the resulting embryos. The extent of this epigenetic defect is controlled by the abundance of histone H3.3: Decreased H3.3 leads to a diminution in the fraction of cells expressing the endodermal genes, whereas increased H3.3 expression results in a larger fraction of animal pole cells with aberrant endodermal gene expression. Other histone variants can help confer stability on a silenced epigenetic state. The variant macroH2A (mH2A) is associated with the irreversible inactivation of the mouse X chromosome. Incorporation of this variant helps confer resistance to reprogramming of the inactive X in nuclear transfer experiments (Pasque et al. 2011).
It has been proposed that the definition of an epigenetic mechanism should include, in addition to the property of being maintained through cell division, a requirement for an initial signal, such as expression of a transcription factor, that is not needed once the new state is established (Berger et al. 2009). Behavior of this kind has been described for the glucocorticoid response element in which transient binding of glucocorticoid receptor (GR) to some sites leads to nucleosome remodeling that makes it possible for a modified estrogen receptor molecule to bind after the GR has departed (Voss et al. 2011). It should be kept in mind that most eukaryotic transcription factors do not have long residence times at their binding sites, but turn over rapidly. Certain kinds of chromatin modifications could, in principle, provide a mechanism to integrate signals from multiple transcription factors (Struhl 1999).
6. ALL MECHANISMS ARE INTERRELATED
These models finally begin to complete the connection between modified or variant histones, specific gene activation, and epigenetics, although there is much more to be done. Although we have some ideas about how the heterochromatic state may be maintained, they do not explain how silencing chromatin structures are established. Much of the evidence for such mechanisms has come from work on the silencing of mating-type locus and centromeric sequences in Schizosaccharomyces pombe. Formation of heterochromatin involves the production of RNA transcripts, particularly from repeated sequences, that are processed into small RNAs through the action of proteins such as Dicer, Argonaute, and RNA-dependent RNA polymerase (Zofall and Grewal 2006). These RNAs are subsequently recruited to the homologous DNA sites as part of complexes that will eventually include enzymes that deliver “silencing” histone modifications, thus initiating the formation of heterochromatin. There is also evidence that the same mechanisms are required for maintenance of at least some heterochromatic regions in plants and vertebrates.
We now know of countless examples of epigenetic mechanisms at work in the organism. In addition to the allele-specific and random X-chromosome inactivation described in Section 5 and similar allele-specific expression at many more imprinted loci, there are epigenetic phenomena involved in antibody expression in which the rearrangement of the immunoglobulin genes on one chromosome is selectively inhibited. In Drosophila, the Polycomb group genes are responsible for establishing a silenced chromatin domain that is maintained through all subsequent cell divisions (Grossniklaus and Paro 2014). Epigenetic changes are also responsible for paramutation in plants, in which one allele can cause a heritable change in expression of the homologous allele (Brink 1956; Stam et al. 2002). This is an example of an epigenetic state that is inherited meiotically as well as mitotically, a phenomenon documented in plants and recently in animals (Rassoulzadegan et al. 2006). In addition, the condensed chromatin structure characteristic of centromeres in organisms as diverse as flies and humans has been shown to be transmissible through centromere-associated proteins rather than DNA sequence. In all of these cases, the DNA sequence remains intact, but its capacity for expression is suppressed. This is likely in all cases to be mediated by DNA methylation, histone modification, presence of a histone variant, or all three; in some cases, we already know that to be true. Perhaps the X chromosome, which inspired early ideas about the role of DNA methylation in epigenetic signaling, is the best example of how all of these mechanisms are interrelated and function together to achieve epigenetic regulation. Recent studies show that silencing of the inactive X chromosome involves, in addition to DNA methylation, specific silencing histone modifications, Polycomb group proteins, noncoding RNAs, and histone variants (Lee 2011). All of these are likely to be involved in transmission of the silenced state during cell division.
In recent years, the study of epigenetics has focused on defining mechanisms of transmission of information not encoded in DNA. Perhaps it is appropriate to reconsider the original use, 70 years ago, of the term epigenetics to describe the then poorly understood processes leading from fertilized zygote to organism. We now know a lot about these processes, thanks to recent results in embryonic stem cells, which show how expression of a few critical factors can establish a self-stabilizing pluripotent state. That state can be transmitted through cell division by what can be thought of as an epigenetic mechanism (according to present definitions). The state can also be perturbed, leading to different epigenetically maintained patterns of expression corresponding to different paths of differentiation into distinct cell types. In an elegant elaboration of these results and of the earlier nuclear transplant studies, somatic cells can be reprogrammed to pluripotency (Yamanaka and Blau 2010). There will be a proliferation of details for many years to come, but we now think we know more or less how this works.
Although this has been presented as a sequential story, it should more properly be viewed as a series of parallel and overlapping attempts to define and explain epigenetic phenomena. The definition of the term epigenetics has changed, but the questions about mechanisms of development raised by earlier generations of scientists are now once again the center of attention. Contemporary epigenetics still addresses those central questions. More than 80 years have passed since Muller described what is now called position-effect variegation. It is gratifying to trace the slow progress from observation of phenotypes through elegant genetic studies to the recent analysis and resolution of mechanisms at the molecular level, and especially to the synthesis of all this information in the analysis of the progression from pluripotent stem cell to individual differentiated states. With this knowledge has come the understanding that epigenetic mechanisms are, in fact, responsible for a considerable part of the phenotype of complex organisms.
ACKNOWLEDGMENTS
I am grateful to Dr. John Gurdon for illuminating exchanges, comments, and advice. This work was supported by the intramural research program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahmad K, Henikoff S 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Mirsky AE 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci 51: 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery OT, MacLeod CM, McCarty M 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med 79: 137–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Beisson J, Sonneborn TM 1965. Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. Proc Natl Acad Sci 53: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A 2009. An operational definition of epigenetics. Genes Dev 23: 781–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP 1978. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol 118: 49–60 [DOI] [PubMed] [Google Scholar]
- Bird AP, Southern EM 1978. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol 118: 27–47 [DOI] [PubMed] [Google Scholar]
- Briggs R, King TJ 1952. Transplantation of living nuclei from blastula cells into enucleated frogs' eggs. Proc Natl Acad Sci 38: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink RA 1956. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41: 872–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD 1996. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851 [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Kirk M 1985. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 315: 496–498 [DOI] [PubMed] [Google Scholar]
- Cedar H, Felsenfeld G 1973. Transcription of chromatin in vitro. J Mol Biol 77: 237–254 [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Quinn J, Chang HY 2012. Chromatin isolation by RNA purification (ChIRP). J Vis Exp 61 pii: 3912 10.3791/3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RJ, Felsenfeld G 1971. Structure of chromatin. Nat New Biol 229: 101–106 [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000 [DOI] [PubMed] [Google Scholar]
- Doskocil J, Sorm F 1962. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta 55: 953–959 [DOI] [PubMed] [Google Scholar]
- Durrin LK, Mann RK, Kayne PS, Grunstein M 1991. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell 65: 1023–1031 [DOI] [PubMed] [Google Scholar]
- Grimes GW, Aufderheide KJ 1991. Cellular aspects of pattern formation: The problem of assembly. Monogr Dev Biol 22: 1–94 [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R 2014. Transcriptional silencing by Polycomb group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn E 1965. Problems of determination and transdetermination. Brookhaven Symp Biol 18: 148–161 [Google Scholar]
- Hannah A 1951. Localization and function of heterochromatin in Drosophila melanogaster. Adv Genet 4: 87–125 [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Hershey AD, Chase M 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol 36: 39–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE 1975. DNA modification mechanisms and gene activity during development. Science 187: 226–232 [PubMed] [Google Scholar]
- Jablonka E, Lamb MJ 1995. Epigenetic inheritance and evolution. Oxford University Press, New York [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Thomas JO 1974. Chromatin structure; oligomers of the histones. Science 184: 865–868 [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Laskey RA, Gurdon JB 1970. Genetic content of adult somatic cells tested by nuclear transplantation from cultured cells. Nature 228: 1332–1334 [DOI] [PubMed] [Google Scholar]
- Lee JT 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 12: 815–826 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Lyon MF 1961. Gene action in the X-chromosome of the mouse. Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D 2008. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1965. The control of gene action in maize. Brookhaven Symp Biol 18: 162–184 [Google Scholar]
- McClure KD, Schubiger G 2007. Transdetermination: Drosophila imaginal disc cells exhibit stem cell-like potency. Int J Biochem Cell Biol 39: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E, Gafken PR, Ahmad K, Henikoff S 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci 101: 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C 2010. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137: 2493–2499 [DOI] [PubMed] [Google Scholar]
- Morgan TH 1911. An attempt to analyze the constitution of the chromosomes on the basis of sex-linked inheritance in Drosophila. J Exp Zool 11: 365–414 [Google Scholar]
- Muller HJ 1930. Types of visible variations induced by X-rays in Drosophila. J Genet 22: 299–334 [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB 2005. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. Proc Natl Acad Sci 102: 1957–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB 2008a. Epigenetic inheritance of cell differentiation status. Cell Cycle 7: 1173–1177 [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB 2008b. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10: 102–109 [DOI] [PubMed] [Google Scholar]
- Novick A, Weiner M 1957. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci 43: 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Kaplan WD, Kinosita R 1959. Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp Cell Res 18: 415–418 [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Pasque V, Halley-Stott RP, Gillich A, Garrett N, Gurdon JB 2011. Epigenetic stability of repressed states involving the histone variant macroH2A revealed by nuclear transfer to Xenopus oocytes. Nucleus 2: 533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kamakaka RT, Kadonaga JT 1994. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science 266: 2007–2011 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I 1992. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68: 573–583 [DOI] [PubMed] [Google Scholar]
- Ptashne M 1992. A genetic switch: Phage λ and higher organisms, 2nd ed. Cell Press and Blackwell Scientific, Cambridge, MA [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F 2006. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 441: 469–474 [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A 1984. Structure of the nucleosome core particle at 7 A resolution. Nature 311: 532–537 [DOI] [PubMed] [Google Scholar]
- Riggs AD 1975. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 14: 9–25 [DOI] [PubMed] [Google Scholar]
- Riggs AD, Porter TN 1996. Overview of epigenetic mechanisms. In Epigenetic mechanisms of gene regulation (ed. Russo VEA, Martienssen R, Riggs AD), pp. 29–45 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Riggs AD, Martienssen RA, Russo VEA 1996. Introduction. In Epigenetic mechanisms of gene regulation (ed. Russo VEA, et al. ), pp. 1–4 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, Belele C, Dorweiler JE, Chandler VL 2002. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev 16: 1906–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman E, Stedman E 1950. Cell specificity of histones. Nature 166: 780–781 [DOI] [PubMed] [Google Scholar]
- Struhl K 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1–4 [DOI] [PubMed] [Google Scholar]
- Sturtevant AH 1913. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J Exp Zool 14: 43–59 [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB 1988. MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242: 405–411 [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408–411 [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH 1953. Epigenetics and evolution. Symp Soc Exp Biol 7: 186–199 [Google Scholar]
- Wallis JW, Hereford L, Grunstein M 1980. Histone H2B genes of yeast encode two different proteins. Cell 22: 799–805 [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci 86: 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM 2010. Nuclear reprogramming to a pluripotent state by three approaches. Nature 465: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SI 2006. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol 71: 487–496 [DOI] [PubMed] [Google Scholar]