Abstract
Signal transduction is regulated by protein–protein interactions. In the case of the ErbB family of receptor tyrosine kinases (RTKs), the precise nature of these interactions remains a topic of debate. In this review, we describe state-of-the-art imaging techniques that are providing new details into receptor dynamics, clustering, and interactions. We present the general principles of these techniques, their limitations, and the unique observations they provide about ErbB spatiotemporal organization.
Cutting-edge imaging techniques are providing details into ErbB receptor dynamics in the intact cell membrane. This, combined with structural information from crystallography studies, offers a more complete picture of ErbB function.
Signal transduction is associated with dramatic spatial and temporal changes in membrane protein distribution. Although the biochemical events downstream of membrane receptor activation are often well characterized, the initiating events within the plasma membrane remain unclear. Many cell surface receptors have been shown to redistribute into clusters in response to ligand binding (Metzger 1992). Therefore, correlating membrane receptor activation with dynamics and aggregation state is essential to understanding cell signaling.
The role of receptor aggregation is of particular interest in the case of receptor tyrosine kinases (RTKs). It is generally accepted that ligand binding to the extracellular domain of RTKs induces dimerization, whether ligand- or receptor-mediated (Lemmon and Schlessinger 2010). However, there is evidence that some RTKs exist as oligomers in the absence of ligand, whereas others require higher-order oligomerization for activation (Lemmon and Schlessinger 2010). Understanding the fundamental interactions that regulate RTK signaling still remains an important focus in the field.
Over the past decade, imaging technologies and biological tools have developed to a point such that questions about protein dynamics, clustering, and interactions can now be addressed in living cells (Fig. 1). These techniques reveal information about protein behavior on a spatial and temporal scale that is not provided by traditional biochemical assays. In this review, we will discuss the application of these advanced imaging technologies to the study of the ErbB family of RTKs.
Figure 1.
Summary of imaging techniques for quantifying receptor clustering, dynamics, and interactions.
ErbB SIGNAL TRANSDUCTION
The ErbB family consists of four members: ErbB1 (the classical EGFR, HER1), ErbB2 (HER2), ErbB3 (HER3), and ErbB4 (HER4). In general, these proteins are single-pass transmembrane proteins with an extracellular ligand-binding domain and a cytoplasmic tail containing a tyrosine kinase domain (Yarden and Sliwkowski 2001; Lemmon and Schlessinger 2010). As exceptions, ErbB2 has no known ligand, and ErbB3 has a defective kinase domain. Ligand binding to these transmembrane proteins leads to conformational changes, receptor homo- and hetero-oligomerization, kinase activation, and the transphosphorylation of multiple cytoplasmic tail tyrosines (Schlessinger 2002). These phosphotyrosines, in turn, provide sites for the recruitment and activation of cytoplasmic adaptor proteins, initiating signaling cascades that control numerous cellular processes such as gene expression, cell migration, and cell division. There exists a rich literature of biochemical and structural data describing the mechanisms of ErbB activation. However, despite being a focus of study for more than 25 years, fundamental questions concerning the association states and activation of the ErbB family still remain.
In 2002, Ogiso et al. (2002) solved the X-ray crystal structure of the extracellular domains of the 2:2 epidermal growth factor (EGF):ErbB1 dimer, showing that dimerization of ErbB1 is achieved through a physical interaction between two receptors rather than through the ligand. Comparison with the unliganded or “tethered” structure suggests that on ligand binding, domain I undergoes a large (∼130°) rotation to form a binding pocket for EGF with domain III, known as the “extended” structure. This conformational change subsequently rearranges domain II, exposing the “dimerization arm” to which another EGF-bound ErbB1 in the extended conformation can bind, forming a “back-to-back” dimer. Furthermore, it has also been suggested that ErbB2 is the preferential dimerization partner for ErbB1 because the crystal structure of the ErbB2 ectodomain displays the extended structure (Garrett et al. 2003).
Although the ectodomain crystal structures formed the basis for the model of receptor-driven dimerization, ideas about the size and ligand-occupancy of the ErbB1 signaling complex remain controversial (Sako et al. 2000; Clayton et al. 2005; Lidke et al. 2005; Macdonald and Pike 2008; Chung et al. 2010; Hofman et al. 2010). For example, there have been reports that the 1:2 EGF:ErbB1 dimer should be the predominant signaling species (Macdonald and Pike 2008), whereas other groups have suggested that a transition from dimers to tetramers is the critical step (Clayton et al. 2005). Additionally, there is evidence for “predimers” that form in the absence of ligand (Hofman et al. 2010).
Recent studies have shown that the cytoplasmic kinase domain is also involved in ErbB1 regulation (Zhang et al. 2006; Choi et al. 2007). Crystal structures of the kinase domain (Zhang et al. 2006) have revealed an autoinhibitory conformation, similar to that of Src and cyclin-dependent kinases. The discovery of an allosteric mechanism for ErbB1 kinase activation suggests additional intracellular complexities in the regulation in ErbB signaling. More recently, structural studies of near full-length, purified ErbB1 have shown that activating mutations in the kinase domain can lead to asymmetric dimerization of the kinase domain without the need for ligand or extracellular domain contacts (Wang et al. 2011).
The insights provided by biochemical, biophysical, and structural studies have been invaluable to our understanding of ErbB signaling. However, these studies typically cannot be performed in the context of the intact plasma membrane. With cutting-edge imaging techniques (summarized in Fig. 1), quantitative information of receptor behavior in the cell membrane can be integrated with structural knowledge to gain a more complete mechanistic picture of ErbB function. Importantly, live cell imaging allows for detailed observation of homo- and heterointeractions with high spatiotemporal resolution, as well as the ability to investigate the role of membrane microdomains in regulating receptor function.
DETERMINING CHANGES IN RECEPTOR CLUSTER SIZE USING CONVENTIONAL MICROSCOPY
Receptor clustering and/or oligomerization occur on scales <100 nm, a distance too small to be resolved by conventional, diffraction-limited microscopy, such as confocal or total internal reflection fluorescence (TIRF) microscopy. Furthermore, even at low (physiological) expression levels, the receptors are present at a high enough concentration in the membrane that, on average, there is significantly more than one receptor per diffraction-limited area of the conventional microscope. This makes direct counting of oligomer size (e.g., by brightness) difficult. Nevertheless, several techniques have been developed that use the spatial or temporal information that is present in conventional microscope images or time series to infer the oligomerization state (Kolin and Wiseman 2007; Digman and Gratton 2011).
One method to measure concentration and stoichiometry of complexes is called number and brightness (N&B) (Digman et al. 2009). This imaging technique uses the temporal fluctuations in pixel intensity to determine the average concentration in a pixel as well as the brightness of the fluorophores. The brightness is then correlated with the stoichiometry by comparing to the known brightness of single labels. This works because stochastic fluctuations around the mean follow the relationship that variance/mean2 is proportional to 1/N, in which N is the average number of particles in the area imaged by one pixel. Nagy et al. (2010) used N&B to examine the aggregation of ErbB1 on the plasma membrane of living cells. They found that ErbB1 primarily exists as monomers in low-expressing cells. Yet, even in the absence of ligand, the fraction of ErbB1 homodimers increased with expression level. This result suggests that there is some affinity for the unliganded dimer and that mass action kinetics drives the system toward dimers as receptor concentration increases. In contrast, ErbB2 was found to be in larger clusters, even at low expression. EGF stimulation led to almost complete ErbB1 homodimerization and the subsequent formation of higher-order oligomers that were associated with clathrin structures (Nagy et al. 2010).
Another image analysis method called spatial intensity distribution analysis (SpIDA) has recently been developed (Godin et al. 2011). In this technique, a histogram is generated from the pixel intensities in a single image. The histogram is fit to a model that assumes a random (spatial Poisson) distribution of either monomers or a mixture of monomers and dimers. Because this analysis does not depend on temporal correlation, it can extract data from a single image. Swift et al. (2011) applied SpIDA to simultaneously monitor ErbB1 dimerization and internalization (concentration at the membrane) in response to transactivation via G-protein coupled receptors (GPCR). They found a differential response depending on the specific GPCR involved; all GPCR transactivation induced ErbB1 dimerization, yet not all of the GPCRs induced rapid receptor internalization.
These techniques are useful in their ability to extract clustering information from intact and living cells without the need for specialized instrumentation. Particularly useful is the ability to follow changes in live cells over time to provide dynamic information. However, it is important to note that these techniques critically rely on the assumption that the true dynamics and clustering behavior follow the model on which the parameter estimation is derived, and therefore the results are highly dependent on the models assumed. The model most often assumed was a mixture of oligomers (or just monomers and dimers) undergoing free Brownian motion on the cell membrane. Often, a range of alternative hypotheses was not tested. For example, in both N&B and SpIDA, it may be very difficult to distinguish between dimers and a collection of higher-order oligomers. More complicated behavior such as transient immobilization, corralled diffusion, or movement of proteins transiently confined to lipid rafts or other heterogeneous membrane structures render the interpretation of results more difficult. Finally, these techniques alone cannot distinguish protein proximity from functional protein–protein interactions.
HIGH-RESOLUTION IMAGING OF PROTEIN DISTRIBUTION
There are a number of techniques that can be used to directly map the distribution of receptors with high spatial resolution. These include electron microscopy (EM), near-field scanning optical microscopy (NSOM), and super-resolution (SR) imaging. Each of these can provide a nanometer-scale image of protein distribution.
NSOM is capable of mapping out the nanoscale organization of the membrane by scanning an optical fiber across the surface of the cell such that excitation and fluorescence collection only occurs near the tip (Betzig et al. 1986). Nagy et al. (1999) used NSOM to show the presence of ErbB2 clusters on the surface of SKBR3 cells and that the cluster size was associated with activation. Furthermore, they showed that ErbB2 cluster size increased with EGF stimulation of ErbB1. More recently, NSOM was used to show that ErbB1 exists in clusters on the surface of HeLa cells (Abulrob et al. 2010). In contrast to the changes seen for ErbB2, ErbB1 cluster size was not observed to change with ligand activation, although cluster size did decrease with ErbB1 kinase domain inhibitors. Using two-color imaging, Abulrob et al. also showed that ErbB1 could be found in three states: colocalized with caveolae, colocalized with lipid rafts, or not associated with either compartment.
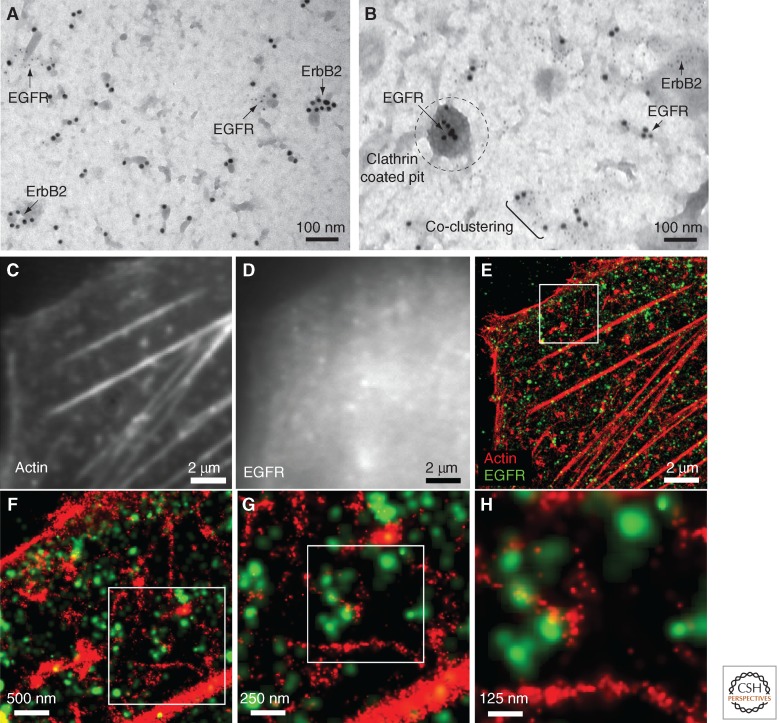
Using EM, Wilson and colleagues have also shown that resting ErbB receptors are found in clusters on the cell surface (Fig. 2A,B) and these clusters did not increase in size on stimulation (Yang et al. 2007). However, they did note a large increase in ErbB3 cluster size in response to heregulin stimulation. By labeling with two sizes of gold particles, they found that pairs of family members (ErbB1/ErbB2, ErbB2/ErbB3, ErbB1/ErbB3) showed very little coclustering, even after ligand addition. This unexpected segregation of receptors suggests a possible mechanism through which membrane microdomains may regulate heterointeractions.
Figure 2.
High-resolution imaging of receptor distribution. (A,B) Transmission electron microscopy images of ErbB1/ErbB2 distribution on SKBR3 membrane sheets. Dual labeling (10-nm and 5-nm gold) allows for mapping of ErbB1 and ErbB2 proximity. Note the localization of ErbB1 to a clathrin-coated pit in (B). Images courtesy of Bridget Wilson created from data based on Yang et al. (2007). (C)–(H) Two-color stochastic optical reconstruction microscopy (STORM) imaging of ErbB1 with respect to actin on Chinese hamster ovary cells stably expressing ErbB1. Diffraction-limited images of actin (C) and ErbB1 (D) as imaged on the basal cell surface using TIRF microscopy. Super-resolution reconstruction overlay (E) of actin (red, phalloidin-AlexaFluor647) and ErbB1 (green, anti-ErbB1 antibody conjugated to Cy3b) showing the subdiffraction resolution attained using STORM and the localization of ErbB1 within the actin cytoskeleton. The reconstruction overlay is shown at increasing zoom (white boxes) to highlight imaging resolution (F,G,H).
Emerging super-resolution techniques in fluorescence microscopy are allowing for imaging below the diffraction limit of the light microscope (Fig. 2C–H), providing 100 nm or better resolution with far-field microscopy (Leung and Chou 2011; Lidke and Lidke 2012). These techniques include photoactivation localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM), stimulated emission depletion (STED), and structured illumination microscopy (SIM) (Hell 2007; van de Linde et al. 2008; Huang et al. 2009; Patterson et al. 2010; Lidke and Lidke 2012). The single-molecule localization techniques, such as PALM and STORM, are powerful ways to map protein distribution, but have suffered from difficulties in quantifying protein numbers. Because localization microscopy typically relies on the sequential localization of individual fluorophores, repeated observation of the same fluorophore can lead to overcounting and apparent clustering of proteins. In addition, unlabeled, photobleached, or nonfunctional probes can lead to undercounting (Renz et al. 2012). Therefore, although information regarding protein distribution may be easily obtained, development of quantitative methods to identify protein clustering with precise protein numbers has remained a challenge within the field. There are many groups working to improve the quantitative analysis of superresolution images. Veatch and colleagues have used pair correlation analysis to determine cluster size from super-resolution images (Sengupta et al. 2011; Veatch et al. 2012). However, this method reports only the average cluster size assuming homogeneous clusters. Needham et al. (2013) have developed a super-resolution imaging method to identify receptor separation down to 10 nm using fluorophore localization and photobleaching. They find five discrete inter-ErbB1 receptor separations from 8–57 nm, in the absence of ligand. They postulate that this distribution is due to alignment of resting ErbB1 along cortical actin filaments, forming polymers of up to 10 receptors (Needham et al. 2013). We note that, as with many antibody labeling protocols, in this study the cells were labeled at 4°C before fixation.
It is important to emphasize that although these studies can reveal high-resolution information about protein proximity and distributions, they cannot distinguish clustering from functional protein interactions. Nevertheless, information on cluster distributions, receptor coclustering, and colocalization with membrane compartments has provided a unique view of membrane organization. Importantly, the observed clustering of receptors, particularly in the absence of ligand, has reinforced the importance of microdomains in organizing receptors on the cell surface.
ENSEMBLE PROXIMITY ASSAYS TO DETERMINE PROTEIN INTERACTIONS
To study protein–protein interactions in the ErbB family, a number of groups have used Förster (or fluorescence) resonance energy transfer (FRET). FRET can determine the proximity of fluorophore-labeled proteins based on the level of excited state energy transferred from one fluorophore (donor) to a second fluorophore (acceptor), within a range of ∼2–10 nm (Jares-Erijman and Jovin 2003). The classical FRET studies by Gadella and Jovin confirmed that ligand-mediated interaction of receptors within the plasma membrane is a key initiating step in ErbB signaling (Gadella and Jovin 1995). Energy transfer between proximal, ligand-bound receptors was determined by donor photobleaching FRET. From these measurements, it was concluded that the ligand-induced dimeric association of receptors occurs within minutes of incubation with EGF and that the high-affinity receptors are found as preformed dimers. In these experiments, they also revealed a dependence of dimer formation on temperature, with a shift from 13% dimers at 4°C to 69% at 37°C. This result emphasizes the importance of performing experiments at physiological temperatures.
Since those early experiments, other groups have used FRET to examine ErbB1 interactions that have actually led to variations on the model of ErbB1 activation. For example, Martin-Fernandez et al. (2002) showed that ligand binding causes a rapid and transient structural transition followed by oligomer “dissociation” on a longer timescale, possibly explained by negative cooperativity of EGF-ErbB1 binding/dimerization characterized by others based on Scatchard plots (Macdonald and Pike 2008). In contrast, Clayton and colleagues have used FRET in combination with image correlation spectroscopy (ICS) measurements to suggest that a dimer-to-tetramer transition is the key event in ErbB1 activation (Clayton et al. 2005).
Energy transfer between identical fluorophores, known as homo-FRET, has been used to quantify the degree of protein clustering based on the degree of emission polarization or anisotropy (Lidke et al. 2003; Bader et al. 2007, 2009, 2011; Hofman et al. 2010). Similar to conventional hetero-FRET, homo-FRET involves the transfer of energy from the excited state of one fluorophore molecule to a proximal fluorophore (or fluorophores), in which the fluorescence emission of the latter is essentially depolarized with respect to the original excitation dipole. Thus, energy transfer between identical fluorophores results in a decrease in the steady-state fluorescence anisotropy or time-resolved anisotropy decay and can be used to quantify cluster size of fluorescent molecules. Homo-FRET systems are ideal for studying the dimerization and oligomerization events in like molecules without the need for distinct donor/acceptor labels in appropriate ratios.
Using microscopy-based fluorescence anisotropy measurements of ErbB1-green fluorescent protein (GFP), Hofman et al. (2010) found that ErbB1 forms clusters in the absence of ligand as measured by a decrease in fluorescence anisotropy. Converting fluorescence anisotropy values into cluster sizes (Bader et al. 2009), it was predicted that ∼40% of ErbB1-GFP exists as predimers in the absence of ligand and the percentage is not dependent on kinase activity. The investigators concluded that predimerization is likely driven through protein interactions within the cytosolic domain, which is consistent with the putative autoinhibitory symmetric dimer as observed crystallographically from the kinase domain crystal structure (Jura et al. 2009). Higher receptor clustering was dependent on kinase activity, suggesting that high-order oligomers are stabilized through recruitment of cytoplasmic adaptor proteins. Furthermore, because anisotropy is calculated spatially, the investigators show that ErbB1 remains clustered within early endosomes, suggesting that signaling may continue through the process of endocytosis.
An alternative method for visualizing protein interactions involves conjugating nonfluorescent, complementary amino- and carboxy-terminal fragments of a fluorescent protein to multiple proteins of interest. Although neither fragment in isolation shows fluorescent properties, the complementary fragments form a complete, photoactive fluorophore when their respective conjugated proteins are in close proximity. This method, known as BiFC, bimolecular fluorescence complementation (Kerppola 2008), allows for visualization of protein interactions occurring at the nanometer scale and localization of those interactions within the diffraction limit. Conjugation and coexpression of complementary fluorescent protein fragments to ErbB1 showed the formation of ligand-independent interactions at the cell surface, which internalized on the addition of EGF. They found that the overall fluorescence intensity did not change in response to ligand, suggesting that most receptors exist in preformed dimers in the resting cell. It is important to note, however, that the favorable interaction between complementary fluorescent protein fragments may bias quantified results toward the receptor complexed state. BiFC was further used to show the presence of ErbB2 and ErbB4 homodimers at the cell surface, as well as ErbB3 homodimers within the nucleus. As with FRET, BiFC is particularly useful in studying the interaction of different protein species. As such, heterodimers within the ErbB family were also identified, as was their subcellular localization (plasma membrane, nucleus, or cytosolic). The investigators’ conclusions that ligand-independent dimerization occurs primarily in the endoplasmic reticulum highlights the importance of microscopy-based methods for spatially characterizing protein interactions (Tao and Maruyama 2008).
Pike and colleagues have recently generated a complementation system using fragments of firefly luciferase (Luker et al. 2004; Macdonald-Obermann et al. 2012). With this clever system, they were able to determine not only the formation of ErbB1/ErbB2 homo- and heterodimers, but also the sequence of activation between the kinase domains. They showed that ErB1/ErbB2 heterodimerization is dependent on EGF binding to ErbB1, and that ErbB1 phosphorylates ErbB2 within the heterodimer. These results support the asymmetric kinase domain dimer structural model and introduce the idea that kinase domain interactions are directional.
The ability to express proteins of interest in a variety of cellular backgrounds, along with the ability to genetically encode fluorescent tags on proteins, has revolutionized the field of cellular imaging. However, with any heterologous expression system, it is important to consider that overexpression may cause improper posttranslational modification (such as receptor glycosylation) and subsequent misfolding, which can lead to aggregation within intracellular compartments. Further complexities in protein folding and processing may occur on the addition of GFP (or GFP fragments); therefore, the assessment of protein folding, trafficking, and function of GFP-tagged proteins is critical in such experimental systems.
DIRECT OBSERVATION OF PROTEIN–PROTEIN INTERACTIONS: SINGLE-MOLECULE STUDIES
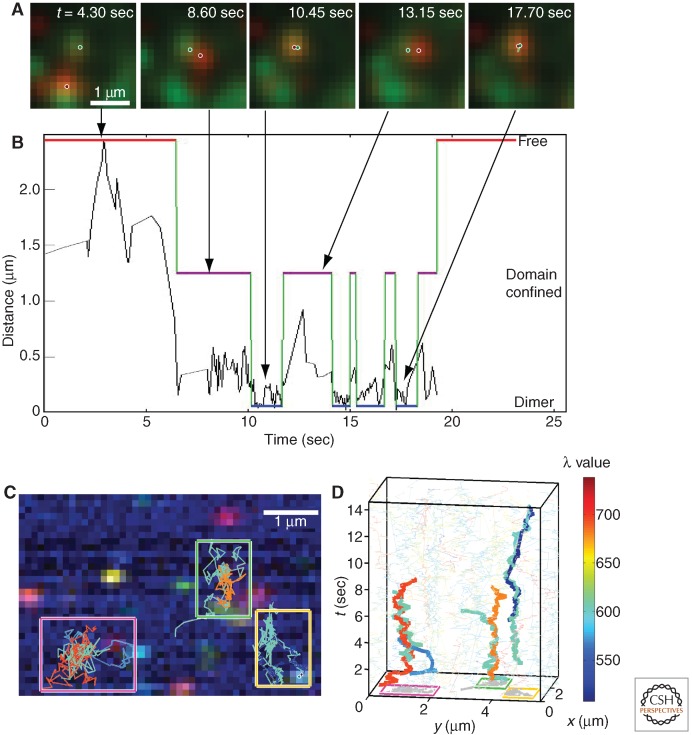
Evaluation of dimerization and higher-order clustering of ErbB1 using FRET and complementation assays are typically limited to time- and ensemble-averaged techniques. Moreover, the timescale with which FRET and BiFC data are collected greatly exceeds that of protein dynamics. Recent advances in single-molecule imaging allow for the localization of individual receptor molecules on the surface of living cells with high spatial and temporal resolution (Schmidt et al. 1996; Weiss 1999). These single-particle tracking (SPT) techniques involve the subdiffraction localization of a subset of membrane receptors at the millisecond timescale and have been most often been used to characterize diffusion of receptor molecules within the plasma membrane (Kusumi et al. 1993; Orr et al. 2005; Chung et al. 2010; Subach et al. 2010). SPT has recently proven to be a powerful technique for visualizing protein interactions by directly observing dimerization at the single-molecule level (Fig. 3A) (Sako et al. 2000; Low-Nam et al. 2011).
Figure 3.
SPT captures dimerization of EGF-bound ErbB1. (A) Images from a time series in which EGF-QD585 (green) and EGF-QD655 (red) are simultaneously tracked. The pixelated image shows the raw data, and the circles indicate the subdiffraction localization of individual receptor molecules. (B) Plot showing the separation over time of the two receptors in (A). Particle trajectories are analyzed using a three-state hidden Markov model based on interparticle distance to distinguish between free (red line), confined (purple line), and dimeric (blue line) receptor populations. In this example, the EGF-bound receptors are seen to undergo repeated (four) dimerization events (see also Low-Nam et al. 2011). (C) Hyperspectral imaging of eight spectrally distinct QDs bound to EGF. Pseudocoloring of the data is generated based on each quantum dot’s (QD) peak spectral wavelength (λ). (D) Three-dimensional particle trajectories (x,y,t) of the corresponding boxed regions in (C). Correlated motion of dimerized receptors can be observed. Color map (right) indicates QD emission peak. (Images from Cutler et al. 2013; reprinted, with permission, from the investigators.)
Using small organic dyes, Sako and colleagues provided the first microscopy data showing direct dimerization of EGF-bound ErbB1 within the plasma membrane (Sako et al. 2000). By labeling A431 cells with a low concentration of fluorescent EGF, they reported diffusion and dimerization of two ligand-bound ErbB1 monomers. Interestingly, in their one-color system, in which dimers are classified based on relative intensity, they more commonly observed a sudden increase in fluorescence of a single ligand-bound ErbB1 on the cell surface, followed by a two-step bleaching process. This suggests either ligand binding to predimerized receptors, or that ligand binding to one ErbB1 results in a dimerization event that further increases the affinity for another EGF molecule to bind. However, the latter model is in contrast to existing models of negative cooperativity based on Scatchard plot and structural data, in which binding of one EGF molecule causes lower affinity ligand binding for the second EGF molecule via structural constraints (Macdonald and Pike 2008). Additionally, two-color tracking and single-pair FRET was performed using the Cy3- and Cy5-labeled EGF showing energy transfer in colocalized receptors, confirming dimerization at the nanometer scale. An important result from this work was that doubly liganded receptor dimers were also seen to label with an antibody that recognizes phosphorylated ErbB1, consistent with the hypothesis that dimerization is sufficient for activation.
The previous work (Sako et al. 2000) revealed many insights about ErbB1 interactions at the single-molecule level. However, they were limited in the duration of the measurements by photobleaching of the organic fluorophores used and only tracked the EGF-bound receptors. In our own work, we have used highly photostable quantum dot (QD) probes to monitor diffusion and dimerization of both liganded and unliganded receptors over longer timescales (Lidke et al. 2004, 2005; Low-Nam et al. 2011). Two-color single-QD tracking of EGF-bound QDs was first used to determine that dimerization was sufficient to initiate retrograde transport of ErbB1 along cellular filopodia (Lidke et al. 2005). Because retrograde transport was shown to be dependent on phosphorylation of ErbB1, the transport of dimers was consistent with the dimer being the minimal signaling unit of ErbB1. More recently, we have used single-QD tracking to quantify ErbB1 homodimerization kinetics at the single-molecule level on the apical surface of living cells (Low-Nam et al. 2011). Direct observation of dimerization using two-color QD tracking revealed a fundamental relationship between receptor ligand occupancy and dimerization. Using a three-state hidden Markov model (Fig. 3B), dimerization events were identified and the dimer off rates were calculated. Interactions between unliganded receptors were observed, but were transient, whereas two ligand-bound receptors formed long-lived dimers. In contrast to Sako et al. (2000), we often observed the coming together of independent receptors, rather than the spontaneous recruitment of the second ligand. The 2:2 EGF:ErbB1 dimers showed a marked reduction in mobility that was depended on kinase activity. Interestingly, a 1:2 EGF:ErbB1 dimer was found to have an intermediate dimer lifetime, but did not show reduced mobility, suggesting that these less stable interactions are not as efficient at initiating signaling events. This work also indicated that the actin cytoskeleton promotes dimer formation by coconfining receptors. Taken together, these results support the model suggested by the crystal structure that ligand binding to the extracellular domain leads to a conformational change that promotes dimerization and receptor activation.
Although evidence for functional ErbB1 homodimers has been provided from single-molecule studies, there is still the question of whether higher-order oligomers occur and what their significance may be. Recent advances in multicolor imaging are making it possible to explore these questions of higher-order oligomers (Arnspang et al. 2012; Cutler et al. 2013). In particular, a novel high-speed hyperspectral microscope developed in the laboratory of Keith Lidke allows for simultaneous single particle tracking of up to eight spectrally distinct QDs at video rate (Fig. 3C). Using this instrument, we have consistently observed the formation of ErbB1 homodimers by tracking QD-EGF (Fig. 3D) (Cutler et al. 2013). Notably, we have not yet observed higher-order interactions. This instrument also extends our ability to examine oligomerization with respect to the membrane landscape (Cutler et al. 2013). Previous work using simultaneous imaging of GFP-actin and single-QD tracking of the immunoreceptor FcεRI showed that membrane receptor motion can be restricted by the membrane-proximal actin bundles (Andrews et al. 2008). The ability to perform multicolor SPT with markers of membrane domains and cytoskeletal proteins will yield new information about receptor mobility and oligomerization within the context of the cellular and membrane architecture.
SPT provides information on the stochastic behavior of individual proteins that is lost in ensemble techniques. However, SPT is limited to sparse labeling densities such that only a subset of proteins is labeled at one time, making it difficult to determine the oligomer distribution accurately. Despite this limitation, SPT is a critical tool that provides direct observation of protein dynamics and interactions within the native cell membrane that cannot be achieved by any other method.
IMAGING RECEPTOR ACTIVATION AND DOWNSTREAM SIGNALING
Microscopy-based characterization of ErbB1 signaling has focused primarily on events in the membrane. Direct imaging of downstream signaling events in live cells is difficult, owing largely to the challenge of delivering fluorophores to the cytosol. Antibody labeling of cytoplasmic proteins in live cells requires mild membrane permeabilization (Sako et al. 2000), and it is unknown whether such membrane perturbations affect ErbB mobility, interactions, or signal initiation. An alternative to antibody labeling is the use of FRET biosensors in which FRET via GFP spectral variants are used to detect intracellular signaling events such as protein recruitment, posttranslational modifications, and/or structural changes (Hahn and Toutchkine 2002). FRET-based methods have been used in several recent studies to address questions of intracellular events in ErbB signaling. Sorkin and colleagues characterized the recruitment of Grb2 to activated ErbB1 by coexpression of Erb1-CFP donor and Grb2-YFP acceptor molecules (Sorkin et al. 2000). FRET between three spectral variants of GFP (3-FRET) has been used to visualize the simultaneous recruitment of multiple downstream factors in live cells. Galperin et al. (2004) used 3-FRET to determine the corecruitment of Grb2 and Cbl to activated ErbB1 through direct and indirect interactions, respectively. BiFC has also been used to show STAT5 recruitment to ErbB1 (Chu et al. 2009). An alternative method has been developed to study protein phosphorylation using a FRET biosensor, in which intramolecular FRET is dependent on the protein phosphorylation state (Sato and Umezawa 2004). Coupling these signaling readouts to receptor dynamics and oligomerization will be an import future goal.
CONCLUDING REMARKS
Efficient signal transduction along the ErbB signaling pathways involves the formation of stable interactions between inherently dynamic proteins. Our current understanding of ErbB structure–function relationships has relied heavily on the detailed structural information of crystallography studies. Crystallography, however, cannot provide the dynamic information that is clearly important in regulating protein functions and requires isolating the protein from its physiological environment. Advanced techniques in high-resolution imaging are being used to complement the structural models by providing information on receptor dynamics, including the direct observation of receptor dimerization. The ability to measure protein behavior in living cells is making it possible to directly test the existence of structural predictions and their functional consequences.
Despite their advantages, these new technologies have not provided a definitive mechanism for ErbB activation. As shown in this review, there exist imaging results that are consistent with seemingly disparate ideas. For example, FRET and SPT have suggested both the existence of ligand-independent dimerization (Chung et al. 2010; Hofman et al. 2010) and ligand-dependent dimer stabilization (Gadella and Jovin 1995; Lidke et al. 2005; Low-Nam et al. 2011). Even the fundamental signaling unit of the receptor is debated, with some data supporting the concept that dimerization is key (Sako et al. 2000; Lidke et al. 2005; Low-Nam et al. 2011), and other results suggesting that a dimer-to-tetramer transition is the critical step (Clayton et al. 2005). This confusion may be partially due to variations in sample treatment (fixed vs. live cells, imaging temperature, steric hindrance due to labeling, protein overexpression, cell type, etc.) or the choice of models used to quantify and interpret data. Moreover, each technique has its own limitations that must be taken into consideration (Fig. 1). Or, perhaps, each of these mechanisms plays some role under the appropriate physiological conditions. A future challenge in the field will be to determine whether all these observations can be unified into one comprehensive model for ErbB activation. Ultimately, it will be the integration of structural, biochemical, and biophysical results that provide a complete picture of ErbB signal transduction.
ACKNOWLEDGMENTS
The work is supported by National Science Foundation (NSF) CAREER Award MCB-0845062 to D.S.L., NSF CAREER Award #0954836 to K.A.L., and the New Mexico Spatiotemporal Modeling Center (National Institutes of Health P50GM085273).
Footnotes
Editors: Joseph Schlessinger and Mark A. Lemmon
Additional Perspectives on Signaling by Receptor Tyrosine Kinases available at www.cshperspectives.org
REFERENCES
- Abulrob A, Lu Z, Baumann E, Vobornik D, Taylor R, Stanimirovic D, Johnston LJ 2010. Nanoscale imaging of epidermal growth factor receptor clustering: Effects of inhibitors. J Biol Chem 285: 3145–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NL, Lidke KA, Pfeiffer JR, Burns AR, Wilson BS, Oliver JM, Lidke DS 2008. Actin restricts FcepsilonRI diffusion and facilitates antigen-induced receptor immobilization. Nat Cell Biol 10: 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnspang EC, Brewer JR, Lagerholm BC 2012. Multi-color single particle tracking with quantum dots. PLoS ONE 7: e48521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AN, Hofman EG, van Bergen En Henegouwen PM, Gerritsen HC 2007. Imaging of protein cluster sizes by means of confocal time-gated fluorescence anisotropy microscopy. Opt Express 15: 6934–6945 [DOI] [PubMed] [Google Scholar]
- Bader AN, Hofman EG, Voortman J, en Henegouwen PM, Gerritsen HC 2009. Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophys J 97: 2613–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AN, Hoetzl S, Hofman EG, Voortman J, van Bergen en Henegouwen PM, van Meer G, Gerritsen HC 2011. Homo-FRET imaging as a tool to quantify protein and lipid clustering. Chemphyschem 12: 475–483 [DOI] [PubMed] [Google Scholar]
- Betzig E, Lewis A, Harootunian A, Isaacson M, Kratschmer E 1986. Near field scanning optical microscopy (NSOM): Development and biophysical applications. Biophys J 49: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Mendrola JM, Lemmon MA 2007. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene 26: 1567–1576 [DOI] [PubMed] [Google Scholar]
- Chu J, Zhang Z, Zheng Y, Yang J, Qin L, Lu J, Huang ZL, Zeng S, Luo Q 2009. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens Bioelectron 25: 234–239 [DOI] [PubMed] [Google Scholar]
- Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I 2010. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 464: 783–787 [DOI] [PubMed] [Google Scholar]
- Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW 2005. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem 280: 30392–30399 [DOI] [PubMed] [Google Scholar]
- Cutler PJ, Malik MD, Liu S, Byars JM, Lidke DS, Lidke KA 2013. Multi-color quantum dot tracking using a high-speed hyperspectral line-scanning microscope. PLoS ONE 8: e64320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Gratton E 2011. Lessons in fluctuation correlation spectroscopy. Annu Rev Phys Chem 62: 645–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Wiseman PW, Choi C, Horwitz AR, Gratton E 2009. Stoichiometry of molecular complexes at adhesions in living cells. Proc Natl Acad Sci 106: 2170–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadella TW Jr, Jovin TM 1995. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J Cell Biol 129: 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin E, Verkhusha VV, Sorkin A 2004. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods 1: 209–217 [DOI] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, et al. 2003. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell 11: 495–505 [DOI] [PubMed] [Google Scholar]
- Godin AG, Costantino S, Lorenzo LE, Swift JL, Sergeev M, Ribeiro-da-Silva A, De Koninck Y, Wiseman PW 2011. Revealing protein oligomerization and densities in situ using spatial intensity distribution analysis. Proc Natl Acad Sci 108: 7010–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn K, Toutchkine A 2002. Live-cell fluorescent biosensors for activated signaling proteins. Curr Opin Cell Biol 14: 167–172 [DOI] [PubMed] [Google Scholar]
- Hell SW 2007. Far-field optical nanoscopy. Science 316: 1153–1158 [DOI] [PubMed] [Google Scholar]
- Hofman EG, Bader AN, Voortman J, van den Heuvel DJ, Sigismund S, Verkleij AJ, Gerritsen HC, van Bergen en Henegouwen PM 2010. Ligand-induced EGF receptor oligomerization is kinase-dependent and enhances internalization. J Biol Chem 285: 39481–39489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang X 2009. Super-resolution fluorescence microscopy. Annu Rev Biochem 78: 993–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman EA, Jovin TM 2003. FRET imaging. Nat Biotechnol 21: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J 2009. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci 106: 21608–21613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys 37: 465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolin DL, Wiseman PW 2007. Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem Biophys 49: 141–164 [DOI] [PubMed] [Google Scholar]
- Kusumi A, Sako Y, Yamamoto M 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J 65: 2021–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung BO, Chou KC 2011. Review of super-resolution fluorescence microscopy for biology. Appl Spectrosc 65: 967–980 [DOI] [PubMed] [Google Scholar]
- Lidke DS, Lidke KA 2012. Advances in high-resolution imaging—Techniques for three-dimensional imaging of cellular structures. J Cell Sci 125: 2571–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Nagy P, Barisas BG, Heintzmann R, Post JN, Lidke KA, Clayton AH, Arndt-Jovin DJ, Jovin TM 2003. Imaging molecular interactions in cells by dynamic and static fluorescence anisotropy (rFLIM and emFRET). Biochem Soc Trans 31: 1020–1027 [DOI] [PubMed] [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM 2004. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol 22: 198–203 [DOI] [PubMed] [Google Scholar]
- Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ 2005. Reaching out for signals: Filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol 170: 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PM, Wilson BS, Lidke DS 2011. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat Struct Mol Biol 18: 1244–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D 2004. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci 101: 12288–12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JL, Pike LJ 2008. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc Natl Acad Sci 105: 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ 2012. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc Natl Acad Sci 109: 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Fernandez M, Clarke DT, Tobin MJ, Jones SV, Jones GR 2002. Preformed oligomeric epidermal growth factor receptors undergo an ectodomain structure change during signaling. Biophys J 82: 2415–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger H 1992. Transmembrane signaling: The joy of aggregation. J Immunol 149: 1477–1487 [PubMed] [Google Scholar]
- Nagy P, Jenei A, Kirsch AK, Szollosi J, Damjanovich S, Jovin TM 1999. Activation-dependent clustering of the erbB2 receptor tyrosine kinase detected by scanning near-field optical microscopy. J Cell Sci 112: 1733–1741 [DOI] [PubMed] [Google Scholar]
- Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ 2010. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci 107: 16524–16529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham SR, Hirsch M, Rolfe DJ, Clarke DT, Zanetti-Domingues LC, Wareham R, Martin-Fernandez ML 2013. Measuring EGFR separations on cells with approximately 10 nm resolution via fluorophore localization imaging with photobleaching. PLoS ONE 8: e62331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, et al. 2002. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110: 775–787 [DOI] [PubMed] [Google Scholar]
- Orr G, Hu D, Ozcelik S, Opresko LK, Wiley HS, Colson SD 2005. Cholesterol dictates the freedom of EGF receptors and HER2 in the plane of the membrane. Biophys J 89: 1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G, Davidson M, Manley S, Lippincott-Schwartz J 2010. Superresolution imaging using single-molecule localization. Ann Rev Phys Chem 61: 345–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M, Daniels BR, Vamosi G, Arias IM, Lippincott-Schwartz J 2012. Plasticity of the asialoglycoprotein receptor deciphered by ensemble FRET imaging and single-molecule counting PALM imaging. Proc Natl Acad Sci 109: E2989– E2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Minoghchi S, Yanagida T 2000. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol 2: 168–172 [DOI] [PubMed] [Google Scholar]
- Sato M, Umezawa Y 2004. Imaging protein phosphorylation by fluorescence in single living cells. Methods 32: 451–455 [DOI] [PubMed] [Google Scholar]
- Schlessinger J 2002. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110: 669–672 [DOI] [PubMed] [Google Scholar]
- Schmidt T, Schutz GJ, Baumgartner W, Gruber HJ, Schindler H 1996. Imaging of single molecule diffusion. Proc Natl Acad Sci 93: 2926–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Jovanovic-Talisman T, Skoko D, Renz M, Veatch SL, Lippincott-Schwartz J 2011. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods 8: 969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, McClure M, Huang F, Carter R 2000. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol 10: 1395–1398 [DOI] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Renz M, Lippincott-Schwartz J, Verkhusha VV 2010. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J Am Chem Soc 132: 6481–6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift JL, Godin AG, Dore K, Freland L, Bouchard N, Nimmo C, Sergeev M, De Koninck Y, Wiseman PW, Beaulieu JM 2011. Quantification of receptor tyrosine kinase transactivation through direct dimerization and surface density measurements in single cells. Proc Natl Acad Sci 108: 7016–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao RH, Maruyama IN 2008. All EGF(ErbB) receptors have preformed homo- and heterodimeric structures in living cells. J Cell Sci 121: 3207–3217 [DOI] [PubMed] [Google Scholar]
- van de Linde S, Kasper R, Heilemann M, Sauer M 2008. Photoswitching microscopy with standard fluorophores. Appl Phys B 93: 725–731 [Google Scholar]
- Veatch SL, Machta BB, Shelby SA, Chiang EN, Holowka DA, Baird BA 2012. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE 7: e31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, Cole PA 2011. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol 18: 1388–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S 1999. Fluorescence spectroscopy of single biomolecules. Science 283: 1676–1683 [DOI] [PubMed] [Google Scholar]
- Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, Williams L, Oliver JM, Wilson BS 2007. Mapping ErbB receptors on breast cancer cell membranes during signal transduction. J Cell Sci 120: 2763–2773 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX 2001. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137 [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125: 1137–1149 [DOI] [PubMed] [Google Scholar]