Abstract
Nuclear reprogramming technology was first established more than 50 years ago. It can rejuvenate somatic cells by erasing the epigenetic memories and reconstructing a new pluripotent order. The recent discovery reviewed here that induced pluripotency can be achieved by a small set of transcription factors has opened up unprecedented opportunities in the pharmaceutical industry, the clinic, and laboratories. This technology allows us to access pathological studies by using patient-specific induced pluripotent stem (iPS) cells. In addition, iPS cells are also expected to be a rising star for regenerative medicine, as sources of transplantation therapy.
A small set of transcription factors can induce pluripotency in differentiated somatic cells. These dedifferentiated cells have opened up unprecedented opportunities for medicine and basic research.
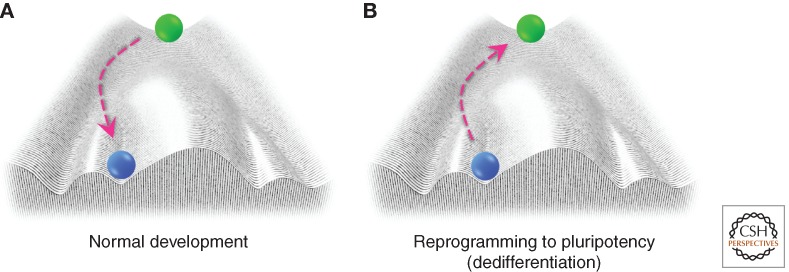
Differentiation has long been thought of as one-way traffic in which a cell can be depicted as a ball rolling down through a “developmental” landscape from an undifferentiated stem or progenitor cell state to a physiologically mature state, as described by Conrad Waddington (Fig. 1A) (Waddington 1957). In fact, all cells roll down this epigenetic landscape into deeper, inescapable valleys, representing the determination of cell fate during development. They continue rolling until they reach their final stable state at the lowest point, functionally equating to their final differentiated state. According to this metaphor, changes in cell fate would be strictly avoided by ridges that do not allow the movement from one valley to another. The discovery of an in vitro method we found for making pluripotent mammalian cells from differentiated somatic cells has added to the body of evidence that this dogma can be reversed. Even more importantly, it has provided an accessible technique to study reprogramming and epigenetics (Takahashi and Yamanaka 2006).
Figure 1.
Cellular reprogramming depicted as a trajectory in Waddington’s epigenetic landscape. (A) A cell’s normal developmental trajectory can be traced starting from a pluripotent cell (green ball) at the top of the hill to its final differentiated state (blue ball), illustrating how epigenetics contributes to cell fate determination during development. (B) A terminally differentiated cell (blue ball) can be reprogrammed back to pluripotency when exposed to a cocktail of transcription factors.
In the past, the inability to transmit genetic information from somatic cells to the next generation was commonly recognized as Weismann’s barrier. Recent discoveries, however, show that cell fate now appears to be far more flexible than previously thought. Based on Waddington’s landscape, rejuvenation refers to the process whereby cells travel back up their maturation path, through the epigenetic landscape, to become more immature cells, and eventually transform to the pluripotent state (Fig. 1B).
The concept of rejuvenation and cellular reprogramming was first proposed by John Gurdon with his landmark experiments producing clones from somatic cells in Xenopus laevis at roughly the same time as Waddington’s doctrine was being advocated (Gurdon et al. 1958). Later, Ian Wilmut and his colleagues reported the successful cloning of a sheep, Dolly, showing that erasure of epigenetic memories that set somatic cell fate is possible even in mammals (Wilmut et al. 1997). The rejuvenation of a cell to the pluripotent state has also been shown by fusing somatic cells with pluripotent stem cells, such as embryonic stem (ES) cells (Tada et al. 2001). These two approaches suggested that fertilized eggs and pluripotent stem cells contain hidden “reprogramming factors” that are able to erase the somatic memories.
Other research countering Waddington’s unidirectional epigenetic landscape model was the work on the conversion of cell fate by defined factors described 25 years ago (Davis et al. 1987). In their groundbreaking studies, Davis et al. performed subtraction experiments using complementary DNA (cDNA), and discovered the myogenic differentiation 1 (MYOD1) gene. Ectopic expression of MYOD1 alone is enough to induce the conversion of fibroblasts to myosin-expressing myoblasts. This pioneering work clearly showed that transcription factor(s) are crucial not only for the maintenance of cellular identity, but also for the determination of cell fate.
Drawing encouragement from these studies, we reasoned and showed that latent pluripotency could be induced in differentiated somatic cells by using a defined cocktail of transcription factors without the need for transfer into an egg (Takahashi and Yamanaka 2006). The cocktail consisted of OCT3/4, SOX2, KLF4, and c-MYC, and was sufficient to revert differentiated somatic cells, including terminally differentiated cells such as T lymphocytes, to a pluripotent fate. The resulting dedifferentiated cells have been designated iPS cells, and they can theoretically be used to generate all cell types in the body, as well as ES cells. This discovery has confirmed the importance of transcription factor networks in cell fate determination, and has definitively affected our understanding of cellular reprogramming.
The efficiency of reprogramming from somatic cells to iPS cells is generally <1%. This suggests that not only are the reprogramming factors important in triggering changes, but so are the subsequent stochastic events required to continue the reprogramming process. Because no major differences in the genomic sequences between the original cells and reprogrammed iPS cells have been observed, changes in the epigenetic status, such as by DNA methylation and histone modification, seem to be the critical events for reprogramming. In fact, the use of small-molecule compounds that inhibit histone deacetylase, thereby increasing overall chromatin acetylation levels, can improve the reprogramming efficiency. During reprogramming, silencing of somatic cell genes and reactivation of pluripotent stem cell–expressed genes have been observed. Although these changes are clearly linked to epigenetic statuses, their mechanisms and driving force are still unclear. The generation of iPS cells provides a good model and tool for understanding the epigenetic changes initiated by transcription factors. In addition, iPS cell technology is currently being used for research both as a source of stem cells for therapeutic use and as a tool for studying pathological processes.
After the landmark experiments that showed MYOD1 as a major commitment determinant for driving the closely related fibroblasts toward a muscle cell fate, the hunt for other factors directing particular cell fates has continued. Other relatively close conversions have been observed, such as from lymphoid to myeloid cells, and from glial cells to neurons, using defined transcription factors. Recent reports have also shown that it is possible to directly convert somatic cells to more distally related differentiated cell types, and even to transcend the germ layer origin (i.e., endoderm, mesoderm, or ectoderm), as illustrated by the conversion of fibroblasts into neurons, hematopoietic cells, cartilage, cardiomyocytes, and hepatocytes. All of these findings have lowered the hurdles of cell fate conversion. Cellular identities are clearly more flexible than previously thought and are largely defined by the epigenetic status of the cell.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
- Davis R, Weintraub H, Lassar AB 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000 [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischber M 1958. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182: 64–65 [DOI] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T 2001. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol 11: 1553–1558 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Waddington CH 1957. The strategy of the genes. Allen & Unwin, London [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385: 810–813 [DOI] [PubMed] [Google Scholar]