Abstract
Evolutionary hypotheses are correctly interpreted as products of the data they set out to explain, but they are less often recognized as being heavily influenced by other factors. One of these is the history of preceding thought, and here I look back on historically important changes in our thinking about the role of endosymbiosis in the origin of eukaryotic cells. Specifically, the modern emphasis on endosymbiotic explanations for numerous eukaryotic features, including the cell itself (the so-called chimeric hypotheses), can be seen not only as resulting from the advent of molecular and genomic data, but also from the intellectual acceptance of the endosymbiotic origin of mitochondria and plastids. This transformative idea may have unduly affected how other aspects of the eukaryotic cell are explained, in effect priming us to accept endosymbiotic explanations for endogenous processes. Molecular and genomic data, which were originally harnessed to answer questions about cell evolution, now so dominate our thinking that they largely define the question, and the original questions about how eukaryotic cellular architecture evolved have been neglected. This is unfortunate because, as Roger Stanier pointed out, these cellular changes represent life’s “greatest single evolutionary discontinuity,” and on this basis I advocate a return to emphasizing evolutionary cell biology when thinking about the origin of eukaryotes, and suggest that endogenous explanations will prevail when we refocus on the evolution of the cell.
Preceding thought influences how we interpret information. The acceptance of the endosymbiotic origin of mitochondria and plastids may have unduly affected how other aspects of the eukaryotic cell are explained.
The origin and early diversification of eukaryotes are topics that invite big stories because the changes that took place were big ones. The origin of eukaryotes was the biggest change in cell biology since the Last Universal Common Ancestor (LUCA) of all known life—nearly every major aspect of eukaryotic cells is different in some way from the corresponding characters found in bacteria and archaeans, some profoundly so. Not only are they different at the cellular level in possessing a nucleus and endomembrane system, cytoskeleton, and semi-autonomous membrane-bounded organelles (mitochondria and sometimes plastids), for example, but they also possess different means of replicating and expressing genetic information. Moreover, the differences between eukaryotes and other cellular life only increase as you look more closely. At the gross level, eukaryotic genes are organized differently than prokaryotic ones, but the initiation, elongation, and termination of transcription and translation are also different, and even the factors involved in core components of these events are often different. Of course, many aspects of archaeal molecular biology have been found to be shared with eukaryotes (Ouzonis and Sander 1992; Creti et al. 1993; Langer and Zillig 1993; Zillig et al. 1993; Keeling and Doolittle 1995a,b). Notwithstanding these similarities, much about eukaryotes remains unique, and in need of explanation. Unfortunately for evolutionary biologists, this transition has left behind few intermediates (other than possibly Archaea, depending on your point of view), and thus we have a quantum leap in cellular and molecular change to explain, and only a few restraints imposed by direct evidence, which Martin (1999) has nicely described as “… a laborious exercise in deduction and inference.”
Two main ways to look at how such a fundamental transition in evolution took place have emerged. The first of these can be generalized as endogenous or autogenous thinking, in which a large number of small sequential changes took place, generally driven by selection for some ability or property. This is essentially descent with modification. In this case, we have some idea what these changes were, and the challenge is to work out the order of events and come up with explanations as to why one change preceded another or, more powerfully, to suggest where a change could have precipitated other changes. The second view can be generalized as exogenous, or changes driven by endosymbiosis. Here, one looks for a single sweeping cause of many changes that was precipitated by the fusion or merger of different cells (or a cell and a virus, in some cases), producing a cell with new properties. The problem here is to identify the fusion partners and explain what need or benefit drove the partnership and eventual merger, and which extant properties trace back to which partner.
In all likelihood, neither of these two ways of looking at eukaryotic origins is entirely correct; the truth probably lies between the two extremes. But here I wish to review not so much what is right or wrong, but how the history of ideas plays an important role in how we construct and evaluate these hypotheses. In all hypotheses for the origin of eukaryotes, the order of events matters; this is true both in the hypotheses about evolutionary history, but also in how the history of science influences hypotheses themselves. This effect is not limited to the origin of eukaryotes by any means. Philosophers have discussed in detail how history distorts our thinking in many fields, just as psychologists are well aware of cognitive processes that affect how we interpret information. However, most scientists working in any particular field (including myself) have little awareness of these ideas, and they are seldom foremost in our minds when we interpret our data, almost as though we are ourselves experimental subjects but never get the diagnosis needed to be more self-aware of the reflexive nature of scientific theories.
HISTORICAL VIEWS OF THE ORIGIN OF EUKARYOTES
Although the basic concepts are older (Chatton 1925, 1938; Sapp 2005), our modern ideas of the differences between prokaryotic and eukaryotic cells are relatively new, primarily informed as they were by the technical inventions of electron microscopy (Robinow 1956, 1962; Stanier and Van Niel 1962), and more recently molecular biology (Woese 1987, 1994; Woese et al. 1990; Olsen et al. 1994). The first attempts to comprehensively explain these differences were synthetic, and primarily endogenous. That is to say, these explanations generally arose from a desire to explain the whole process of how eukaryotic cells evolved from prokaryotic cells, and primarily relied on the dominant mode of evolution accepted at the time, gradual and incremental accumulation of mutational changes, each mostly imparting a small effect (e.g., Dodge 1965; Stanier 1970; Cavalier-Smith 1978, 1987a,b; Stewart and Mattox 1980). A few lines of reasoning dominated this debate; one was to simply identify a plausible series of events that could lead from a generic prokaryotic form to a generic eukaryotic one (more on this below), but others attempted to identify some link between a “special” group of prokaryotes and eukaryotes or, similarly, to identify a particular group or groups of eukaryotes as being primitive and therefore closer to prokaryotes as a whole.
An example of a special relationship is the hypothesis that red algal eukaryotes arose from cyanobacteria and were thus the first lineage of eukaryote (Dougherty and Allen 1960). This was based on an observed connection between the two groups’ photosynthetic pigments associated with phycobilisomes, as well as the lack of flagella in red algae. This was, of course, misleading, because at the time the idea of an endosymbiotic origin of plastids from cyanobacteria was not widely accepted, and the hypothesis died with the recognition that the plastid was a cyanobacterium (Bonen and Doolittle 1975, 1976).
Identification of putatively “primitive” eukaryotes has led to several ideas. Two recent examples are Cavalier-Smith’s proposal that euglenids are the deepest-branching lineage, primarily because of the absence of proteins associated with mitochondrial import (Cavalier-Smith 2010), or Martin’s proposal that fungi represent the earliest lineage because of their metabolic properties (Martin et al. 2003). It is too soon to say what kind of impact these ideas will have in the long term, but looking at two older ideas shows the range of possibilities: the Mesokaryotes and Archezoa. Mesokaryotes were dinoflagellates, and they were proposed to represent the most ancient lineage of eukaryotes that arose from some unidentified bacterial lineage because of their unusual nuclei (Dodge 1965). Dinoflagellates lack histones and conventional chromatin, and instead have permanently condensed DNA bound to nonhistone proteins (Dodge 1965; Leadbeater and Dodge 1967; Li 1983; Rizzo and Morris 1983; Rizzo 1991). Based on this, they were proposed to be primitively prehistone and prenucleosome (Dodge 1965; Rizzo and Nooden 1972; Hamkalo and Rattner 1977; Rizzo and Morris 1983). This theory died with the recognition that dinoflagellates are not an ancient lineage. They branch within the alveolates, whose other members have normal nuclei, and thus these characters are reinterpreted as derived, and their chromosomes are now interpreted as being highly derived (Gornik et al. 2012; Talbert and Henikoff 2012). The Mesokaryote hypothesis has had little impact since it was overturned, but the impact of the Archezoa hypothesis has been more significant, surviving well beyond the hypothesis itself. The Archezoa hypothesis was based on the idea that several lineages were premitochondrial (Cavalier-Smith 1983, 1993). One reason why this hypothesis had such impact was that the initial phylogenetic data appeared to support it because representatives of Archezoa tended to branch deeply in SSU rRNA and early protein trees (Vossbrinck et al. 1987; Sogin et al. 1989; Sogin 1991; Hinkle and Sogin 1993; Hashimoto et al. 1994, 1995; Hashimoto and Hasegawa 1996; Kamaishi et al. 1996). However, this hypothesis also died with the discovery of mitochondrion-derived genes and eventually mitochondrion-related organelles (MROs, e.g., mitosomes and hydrogenosomes) in all archezoan lineages (for review, see Roger 1999; Williams and Keeling 2003) and concurrent revisions to our understanding of the tree of eukaryotes, which showed no specifically deep position for these lineages, and that some were obviously related to mitochondrion-bearing sisters (Hampl et al. 2009). Interestingly, however, even though the idea that the four lineages originally proposed to be Archezoa is scientifically dead, it does continue to have a notable effect on our thinking about those lineages. They have, in general, and the diplomonad Giardia, in particular, maintained a stubborn reputation as being ancient or primitive eukaryotes (Morrison et al. 2007; Gourguechon et al. 2013). This is no longer specifically tied to their lack of mitochondria, but it is doubtful that this perception would have gained such popularity without the Archezoa hypothesis. This therefore represents an interesting case in which an implication of a hypothesis outlives the hypothesis itself, a topic that is discussed in detail below.
COGNITIVE INERTIA AND THE TRIUMPH OF MODERN ENDOSYMBIOTIC THEORIES
Although there were, of course, alternative views on eukaryotic origins (Goksoyr 1966), they did not really take hold of mainstream scientific thinking until the now-famous and sometimes bitter struggle over the endogenous-versus-endosymbiotic origin of mitochondria and plastids, which primarily took place through the 1960s and 1970s (Gibor and Granick 1964; Klein and Cronquist 1967; Sagan 1967; Margulis 1970, 1981; Raff and Mahler 1972, 1975; Uzzell and Spolsky 1974; Mahler and Raff 1975; Taylor 1979; Cavalier-Smith 1987b). Ideas about the endosymbiotic origin of plastids, in particular, date back considerably further (for review, see Sapp 1994), but at this point in time, they were not widely accepted among cell biologists, even when a good deal of data on their semi-autonomous nature had accumulated. Ultimately, however, the data in favor of the endosymbiotic theory were simply overwhelming, so that, in 1982, Gray and Doolittle were able to ask, “Has the endosymbiosis hypothesis been proven?” (Gray and Doolittle 1982).
This marked a sea change in thinking, and not just related to the acceptance of the endosymbiotic origin of mitochondria and plastids, but also relating to how we think about cellular evolution in general. To some, this was simply an awakening to the truth, for example, Margulis’s new field of endocytobiology (Margulis 1990). However, another perspective is that we have taken a principle that aptly applied to mitochondria and plastids and pushed it beyond its useful limits. The case for an endosymbiotic origin of eukaryotic flagella (or undulopodia) is a good illustration. This hypothesis was first proposed by Margulis based on the fundamental differences between bacterial and eukaryotic flagella and on perceived similarities between eukaryotic flagella and spirochete bacteria that adhere to the surface of certain protists (Sagan 1967; Margulis 1970, 1981), and it sought to follow in the footsteps of plastids and mitochondria but failed simply because it was wrong. Unlike plastids and mitochondria, the more we learned about spirochetes and flagella, the less they had in common; no direct link has ever been established, in particular, at the molecular level (e.g., Johnson and Dutcher 1991). Similar stories were told and eventually disproven or largely forgotten for the endoplasmic reticulum, glycosomes, peroxisomes, cytoskeleton, and even the nucleus itself (Cavalier-Smith 1987b, 1997; Gupta and Golding 1993; Gupta and Singh 1994; Lake and Rivera 1994; Golding and Gupta 1995; Gupta 1999; Li and Wu 2005). In his trenchant review of these theories, and the nucleus in particular, Martin (1999) points out that these hypotheses share a fundamental characteristic that distinguishes them from the hypotheses for an endosymbiotic origin of mitochondria and plastids, whereas the latter were proposed to explain specific similarities between the organelles and particular free-living cells, the former were put forward to explain traits that were complex, but either lacked such a connection to specific free-living cells or were based on superficial similarities that did not stand up to more detailed comparison.
This is an interesting observation; if these hypotheses did not take their cues from specific details of free-living cells, then it is worth asking why were they proposed? The process of proposing a scientific hypothesis and testing it with data is well known to everyone practicing research, even if they are not the only ways to do so, but the mechanism by which we accomplish this is itself an imperfect product of evolution and doubtless arose for some entirely different cognitive purpose. How we as individuals assemble data into a hypothesis, or more importantly how we interpret data in the context of existing hypotheses, are things about which we are not generally self-aware, so it might be fun to consider how some of the hypotheses for how we think impact what we think.
At the risk of treading way beyond my expertise, it is worth introducing a few relevant concepts from Piaget’s schema theory, which is a general model for how humans interpret new information in the context of preexisting frameworks that have been built around existing ideas (Piaget 1953). It is important to stress that a schemata will not reflect all available information, but just those pieces that easily fit into the framework that is already in place (i.e., things that agree with what we already believe). People are remarkably good at ignoring or even altering information that does not fit with preconceived structures, and we are more likely to remember those facts that fit these structures than we are to remember other facts that are inconsistent with them. Two specific and related behaviors that we can likely all recognize (at least in others if not ourselves) relate to our reliance on these constructs. One is known as disconfirmation bias, which is our propensity to require a higher standard of evidence to disprove an accepted structure than was required to form the structure in the first place. The second is cognitive inertia, which relates to the general tendency for our beliefs to survive longer than the evidence that supported them (the latter is more often applied to the world of business, but applies equally well to science, e.g., the vague notion that Giardia is ancient).
With these tendencies in mind, it is tempting to speculate that the timing of this rush of sometimes ill-founded endosymbiotic explanations for various components of the eukaryotic cells was not a coincidence but, rather, was a predictable outcome of the intellectual victory of the endosymbiotic origin of mitochondria and plastids over the endogenous explanation that had previously dominated our thinking. The endosymbiotic theory created a new framework for interpreting information, which is advantageous if the information really fits into this framework (e.g., much of the data relating to the origin of mitochondria and plastids), but misleading when information is forced into the framework artificially, or ignored when it disagrees with the framework (e.g., proposals suggesting an endosymbiotic origin of the nucleus that ignored the fact that the nucleus is not bounded by a double membrane as are mitochondria and plastids).
ARE CHIMERAS REAL, OR MONSTERS FROM A MYTHICAL ENDOSYMBIOTIC PANTHEON?
These theories for endosymbiotic origins of singular structures like flagella, peroxisomes, and the nucleus grew out of a desire to explain a specific, complex feature of eukaryotic cells. But a second, overlapping class of new theories that we might loosely define as chimeric theories also followed, and these sought to explain the very origin of eukaryotic cells in an endosymbiotic framework. One important distinction to make clear is that, for the most part, I am not referring here to theories that expand on the contribution of the mitochondrion at the origin of eukaryotes (this distinction is discussed in Koonin 2010 and Katz 2012) but, rather, theories that propose a novel fusion between two cells giving rise to the nucleocytoplasmic component of eukaryotes. The demonstration that no known extant eukaryotic lineage primitively lacks mitochondria has led to suggestions that mitochondrial origins might coincide with the origin of eukaryotes as a whole. Koonin (2010) has usefully defined theories on mitochondrial origins as archezoan, which are those that propose that some protoeukaryote evolved, then took up mitochondria, and symbiogenic, which are those that propose that mitochondria were taken up by a cell we would not recognize as a eukaryote and that this symbiotic merger was the trigger for the evolution of eukaryotes. The bases for these symbiogenic hypotheses (some of which are listed in Table 1) have been reviewed in detail elsewhere (e.g., Katz 1998, 2002, 2012; Martin 1999; Koonin 2010) and are not reviewed here.
Table 1.
Summary of chimeric theories for the origin of eukaryotes
| Hypothesis | Partners (and what it aims to explain) | References |
|---|---|---|
| Hartman (1984) | Archaean and bacterium and RNA organism (RNA metabolism) | Hartman 1984 |
| Zillig (1989–1991) | Ancestral bacterium and ancestral archaean (conflicting characters) | Zillig et al. 1989; Zillig 1991 |
| Sogin (1991) | Protoeukaryote and archaean (conflicting roots of protein and rRNA trees) | Sogin 1991a |
| Gupta and Golding (1993–1996) | Gram-negative bacterium and archaean (conflicting phylogenies) | Gupta and Golding 1993; Gupta 1995, 1999; Golding and Gupta 1995 |
| Lake and Rivera (1994) | Nuclear endosymbiosis | Lake and Rivera 1994 |
| Hydrogen hypothesis (Martin and Muller 1998) | Methanogenic archaean and α-protebacterium (metabolism) | Martin and Muller 1998 |
| Syntropy hypothesis (Moreira and Lopez-Garcia 1998–2006) | Methanogenic archaean and δ-protebacterium (metabolism) | Moreira and Lopez-Garcia 1998 |
| Clostridium–Sulfolobus nexus (Karlin 1999) | Sulfolobus-like archaean and Clostridium-like bacterium (dinucleotide signatures and metabolism) | Karlin et al. 1999 |
| Karyomastigont hypothesis (Margulis 2000) | Thermoplasma-like archaean and spirochete bacterium (cellular traits) | Margulis et al. 2000 |
| Takemura (2001) | Archaean and orthopoxvirus (α DNA polymerase) | Takemura 2001 |
| Ring of life (Rivera and Lake 2004) | Eocyte and proteobacterium (conflicting phylogenies) | Rivera and Lake 2004 |
| Viral eukaryogensis hypothesis (Bell 2006) | Archaean and virus (meiosis and mitosis) | Bell 2006 |
| Forterre (2006–2011) | Thaumarchaeon and Planctomycetes-Verrucomicrobia-Chlamydiae-like bacterium (DNA) | Forterre 2006, 2011 |
The majority of chimeric theories are fundamentally different from both the endogenous theories for the origin of eukaryotes and the endosymbiotic theories for individual organelles. One important difference is that they mostly seek to explain conflicting molecular data and typically use cells as vehicles to explain phylogenetic patterns. Because of this, they are not synthetic in the sense that they do not seek to comprehensively explain the many differences between eukaryotic and prokaryotic cells, but mostly seek to explain some general observations about the genes encoded in their genomes. Sometimes they delve into differences at the cellular level (at times perilously; several suggest an endosymbiotic origin of the nucleus, as discussed above and in Martin 1999), but typically they are driven by molecular observations, and the cellular implications of the explanations are less well explored. The dominance of molecular data in these theories is also important in their history because, unlike our knowledge of cytological differences between prokaryotes and eukaryotes, which is changing rather slowly, our access to molecular data has increased dramatically since the 1980s and is changing comparatively quickly. As a result, early theories emerged from the analysis of a single gene, or conflict between a gene and the then established SSU rRNA phylogeny (e.g., Zillig and RNA polymerase, Sogin and the root, or Gupta and HSP70) (Table 1). Sometimes these were expanded to take in other characteristics or genes (Golding and Gupta 1995; Gupta 1995), and sometimes they were not. Eventually these grew into analyses of many genes (for review, see Brown and Doolittle 1997), and ultimately whole genomes (Table 1) (e.g., Rivera et al. 1998; Rivera and Lake 2004), but they tend to stick to genes. Obvious exceptions to these generalities are the syntrophy and hydrogen hypotheses, which focus on biological reasons for the partnership rather than seeking to explain a specific incongruence in molecular data (Martin and Muller 1998; Moreira and Lopez-Garcia 1998), and are included in Table 1 despite this critical difference because they are theories that have had impacts on our thinking and should not be ignored.
At the same time as molecular databases grew, the chimeric theories also became more specific. Early versions tended to suggest vague partnerships between archaea and bacteria or eventually Gram-positive bacteria because the diversity of available data was so poor (Sogin 1991; Zillig 1991; Gupta and Golding 1993). Subsequent versions tended to be more precise (e.g., proteobacteria or Thermoplasma-like organism) (Moreira and Lopez-Garcia 1998; Rivera and Lake 2004), but even now genome-wide analyses are not leading to a great deal of specificity, and theories with the most exactly specified partners tend to do so based on modern analogs rather than an overwhelming signal from genomic data (e.g., Martin and Muller 1998; Moreira and Lopez-Garcia 1998). This is excellent in terms of testability but problematic in the sense that we must assume that the metabolic properties of modern bacteria reflect well those of their ancient relatives, which maybe untrue given the timescales involved.
As can be seen from Table 1, these theories came to several different conclusions based on different observed conflicts between different kinds of data; thus, it is not really possible to summarize this as a polemic like the endosymbiotic origin of mitochondria and plastids, with chimeric theories versus endogenous theories, because neither is a homogeneous collection of consistent proposals. Instead, both kinds of theories are collections of independent and generally incompatible proposals, in the case of chimeric theories most often based on nonoverlapping data that led to speculation for chimerism between different partners. The fact that many people have proposed chimeric theories does not lend support to the notion as a whole; because the theories tend to propose different chimeric events, each should be weighed on its merits individually. In addition to this issue of consistency (who ate whom), there is also the issue of rationale (why). The theories were mostly proposed to explain different inconsistencies in the data (e.g., phylogenies of different genes), many of which have either disappeared with addition of more data (e.g., Sogin 1991; Zillig 1991) or have not been revisited since additional data on the issue became available, or since our understanding of horizontal gene transfer has matured somewhat.
Despite the inconsistencies between different chimeric theories, it is easy to fall into the trap of summarizing them on aggregate because we often think about the progress of scientific ideas according to changing trends in what gets published. It is difficult to publish papers stating the continued persistence of a decades-old idea, even if the idea remains widely accepted, but remarkably easy to publish papers proposing ideas that are new. Especially when higher-profile papers tend to make more radical claims (that are on average more likely to be wrong), it is impossible to tally up the data in favor of one or the other kind of hypothesis by what is being published. Indeed, it is possible that the vast majority of specialists in a field believe a set of ideas that has not been explicitly detailed in a publication for many years (because it is widely held and unchanging), but it is conversely possible that the repeated statement that an idea is widely held because of the number of papers on this topic can actually lend credibility to the idea, deserved or not.
DOES THE ORDER OF DISCOVERIES AFFECT OUR INTERPRETATION OF THEM?
Looking back at the history of ideas for the origin of eukaryotes, there are a couple of potentially interesting correlations, but distinguishing between correlation and cause is not straightforward. The rise in popularity of chimeric theories did correlate with the acceptance of the endosymbiotic origin of mitochondria and plastids, but also correlates with the rise of molecular biology, and these are the data from which they were predominantly derived. Thus, it maybe more complicated than stating that one gave rise to the other directly, because on one hand is a new source of information, while on the other is a new way to interpret information. Both are extremely influential, but for the most part we are only conscious of the former, and it is very difficult to untangle their relative contributions to the way information is organized and perceived.
Some insights might come from looking at the consistency of interpretation of similar kinds of data. For the sake of a thought experiment (and because these were dominant structures in our thinking when the conclusions I am about to review were drawn), accept for a minute that the tree of life consists of three domains and that the root lies in the branch leading to Bacteria (Gogarten et al. 1989; Iwabe et al. 1989; Brown and Doolittle 1995). If we plot biological characteristics that are common to two of the three domains on that tree, there are three possible patterns (Fig. 1). We use parsimony to interpret how each of these three patterns evolved, but our logic is not applied consistently, in a fascinating way.
Figure 1.
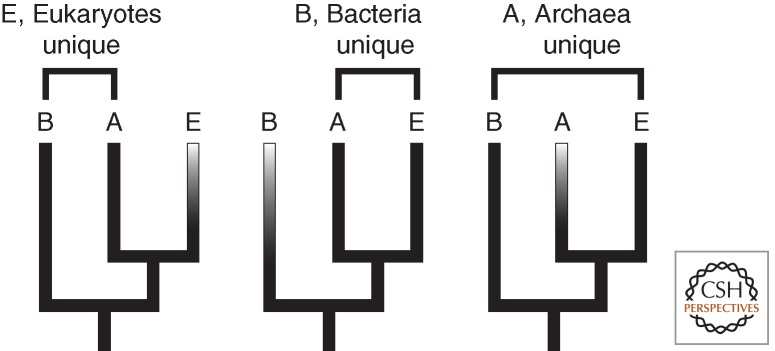
Distribution of character states between three domains on the tree of life and how we interpret them. When Bacteria and Archaea share a state and eukaryotes are different (type E), we typically conclude that the shared state is ancestral to the LUCA and eukaryotes changed. When eukaryotes and Archaea share a state and Bacteria are unique (type B), we similarly conclude that the shared state was ancestral to the LECA, and either Bacteria or the lineage leading to the LECA changed. But when Bacteria and eukaryotes share a state and Archaea are unique (type A), we seem to have trouble applying the same logic, which would at face value simply be to conclude that the shared state was ancestral to the LUCA and that Archaea had changed.
In the case in which Bacteria and Archaea are the same and eukaryotes are different, we historically tend to conclude that the bacterial/archaeal state is ancestral and eukaryotes simply evolved a different state (type E in Fig. 1, e.g., having a cytoskeleton vs. not, or having a rotary motor driving flagella vs. dynein motors driving flagellar motion). In cases in which Archaea and eukaryotes are the same and Bacteria are different (type B in Fig. 1, e.g., TATA-binding protein vs. σ factor, or Shine-Delgarno vs. ternary complex-based translation initiation), we tend to conclude that the eukaryotic/archaeal state was ancestral to these two groups (and whether Bacteria, the archaeal/eukaryotic ancestor, or neither is ancestral is ambiguous). Both of these conclusions are based on the same logic and are consistent with one another. It is therefore interesting that the third pattern is handled differently. In cases in which Bacteria and eukaryotes are the same and Archaea differs (type A in Fig. 1, e.g., Embden-Meyerhof vs. Entner-Doudoroff glycolysis, or ester-linked fatty acid lipids vs. ether-linked isoprenyl lipids), applying the same logic would lead to the conclusion that the bacterial/eukaryotic state is ancestral and Archaea changed. But when faced with these characters (at a time when the tree structure described above was still widely accepted), we seemed to think they were somehow harder to explain. It is as though the question of how the Archaea can be more closely related to eukaryotes but differ in character states is uniquely problematic; but, in fact, the logic is no different from eukaryotes having a unique character state that they do not share with Archaea, which we readily fit into existing structures of thought.
This inconsistency is particularly interesting because the unique characters of Archaea have been featured (often post hoc) in several chimeric hypotheses, even if the root evidence of the theory is generally molecular trees. For example, eukaryotes are sometimes said to have information systems from Archaea and metabolism from Bacteria, which is derived from molecular phylogenies but linked to the unusual metabolism of Archaea (Rivera et al. 1998). This is a result of looking at the shared similarities of eukaryotic and bacteria metabolism as indicating direct transmission (via chimerism, HGT, EGT, etc.); but another way to look at it is that archaeal metabolism has simply changed a great deal and thus no longer shares many genes with eukaryotes, and some of those that they do share are very divergent. Moreover, this pattern of character-state changes observed for the three domains is not unique—if you select any three lineages within the tree, you can likely assemble sets of characters representing all three possible patterns, and likely they would all be explained by differential descent with modification (i.e., one lineage simply changed). Which raises the question, why are character-state patterns that distinguish the Archaea not interpreted as simply reflecting change in that lineage? One possible factor that may influence our thinking is the way in which Archaea were perceived at the time their significance was discovered. Many of the unique features of eukaryotes are cytological and relatively obvious, and these were accordingly recognized very early, and form the foundations of the “prokaryotic–eukaryotic dichotomy” that so profoundly influenced our thinking historically (Stanier and Van Niel 1962; Stanier 1970). Only subsequently did the distinctions between Archaea and Bacteria become clear, and only when molecular phylogenies showed that they were a distinct domain and then suggested a close relationship between Archaea and eukaryotes did the importance of archaeal characteristics begin to have an impact on larger evolutionary schemes. Our concept of prokaryote was based primarily on Bacteria, and because many of the characteristics of Archaea that differ from Bacteria are at the molecular level and were discovered concurrently with the discovery that they are distinct and closer to eukaryotes, they became the exceptions. At the same time, the powerful notion that Archaea were an ancient lineage took hold (Balch et al. 1977; Woese and Fox 1977a,b). Although there is no evidence to support this idea, it nevertheless strongly affected how Archaea were and still are interpreted. Indeed, it is entirely plausible that the notion that Archaea are ancient or primitive continues to undermine the interpretation of unique features of the group as being due to relatively recent modifications.
Ideally, how we evaluate evidence to arrive at a likelihood through logical thinking like parsimony should be a commutative process (which is to say, the order of operations will not affect the outcome, e.g., 1 + 2 = 2 + 1), but it is not. The order in which we discover or learn things contributes to how we interpret their meaning because we do not interpret information in a vacuum but, rather, in the context of established structures. It is interesting to consider how our thinking might have differed had Archaea been the mainstream microbes of study that formed our beliefs about prokaryotes. Bacteria are both more distantly related to eukaryotes and more different in their molecular character states; thus, it is not unreasonable to think that they might now be considered the more strange and unusual prokaryotes. A similar case may be seen in the symbiogenic hypotheses for the simultaneous origin of mitochondria and eukaryotes. These arose from our recognition that mitochondria did not demonstrably postdate other eukaryotic features (like the nucleus, endomembrane, and cytoskeleton), but all this really says is that mitochondria may be as old or older than these features, but it does not suggest that mitochondria must be older, because the absence of evidence for an archezoan intermediate is not evidence for its absence, any more than that logic can be applied to any other feature (e.g., we cannot say the absence of a cytoskeleton-lacking eukaryote means that its evolution must have coincided with the origin of mitochondria). Indeed, the idea that mitochondria arose after the diversification of extant eukaryotes is itself a relatively recent idea (Stewart and Mattox 1980; Cavalier-Smith 1983). If our understanding of mitochondrial diversity had been established 10 years earlier, the Archezoa hypothesis would never have been proposed, and would never have been disproven. Without all this context, the simultaneous symbiogenic hypotheses might never have been proposed.
BIASED BY DATA: WHAT COULD COUNTERFACTUAL HISTORIES TELL US?
In his seminal review on the origin of eukaryotes, Roger Stanier (1970) made his now-famous remark that evolutionary speculation was “… a relatively harmless habit, like eating peanuts, unless it assumes the form of an obsession; then it becomes a vice.” If excessive evolutionary speculation can be a vice, then excessively speculating about how we think about evolutionary speculation surely is much worse. Indeed, taking into account how we might think about new information can only really tell us how the history of ideas might affect what we think, but does it contribute anything to suggest what version of the evolutionary origin of eukaryotes might actually reflect what really happened? The answer is, maybe, but in any event it is worthwhile to return to the question really being asked, and the data itself.
The question of how eukaryotes originated was initially a cytological one. Early theories focused on explaining the significant differences between prokaryotic and eukaryotic cells and how the many unique characteristics of eukaryotes arose. Currently, however, we are obsessed as a field with genomic data. Evolutionary biology and molecular biology have rapidly advanced together, as have cell biology and molecular biology. In contrast, however, a strongly integrated evolutionary cell biology has lagged behind; thus, molecular and genomic data have come to dominate recent thinking about the origin of eukaryotes. This is also aided by the relative ease of molecular methods and, in particular, their scalability (-omics), and perhaps a certain degree of genomic arrogance that arose from the transformational changes that molecular biology genomics have legitimately wrought on other biological disciplines, giving this kind of data an air of objectivity and superiority. Consequently, modern chimeric theories that arose from molecular and genomic data were typically not formulated to explain the characteristics of eukaryotic cells but, rather, to explain inconsistencies in the molecular and genomic data itself. In Doolittle’s prescient and referential review at the 25th anniversary of Stanier’s (Doolittle 1996), he concluded, “Peanuts are often just snacks before supper. Such speculations based on limited quantifiable data whet our appetites for the multi-course meal of genome sequences about to be served, but by the time we get to the dessert of comparative genome analysis, we will have forgotten many of them.” This has certainly proved true, and, going further, it seems that we have forgotten why we applied genomics to this question in the first place. Even if the genome of the nucleocytoplasmic lineage is ultimately proven to be derived from a fusion of two cells to produce a genomic chimera, this only adds one step in the origin of eukaryotes, but by itself does not explain the cellular innovations like the cytoskeleton, endomembrane system, or potentially even the differences in information storage and processing systems, which represent the hallmark eukaryotic features initially in need of explaining. We have called upon genomic data to help explain these changes, but having generated the data, it is proving in some ways to be a distraction, and now we are like children mesmerized by a new toy.
It is interesting, therefore, to consider what kinds of conclusions we might draw if we learned what we know today in a different order, or learned it all at once. Counterfactual histories refer to the intellectual exercise of “what if,” where some historical facts are altered, and then we consider what effects those changes would have on other facts. This kind of thinking is mostly applied to political histories (e.g., what if Hitler was born a woman), where there is no set outcome and an infinite number of possibilities that might play out. These ideas have been applied to several fields of science (Pessoa 2000, 2010; Radick 2008), but there are two ways to do this. Science, as opposed to the history of science, is typically seeking to discover something that exists whether we discover it or not. Changing some of these facts would be implausible, even silly, and do not really tell us much about reality (like, what if ice was heavier than water). But, in other cases, reality is sufficiently removed from our direct knowledge and involves a sufficient degree of chance to allow informative changes to be imposed, like replaying evolutionary history to see if humans are likely (Gould 1990; Dawkins 2005). Thus, counterfactual histories of science are constrained by the facts that exist to be discovered, but if we focus on alternative histories of the human activity of science, then these constraints limit the universe of possible histories in a way that may allow us to consider whether our interpretation of the facts stands up to scrutiny. If, for example, we were to develop a series of counterfactual histories in which a certain set of now-accepted scientific observations was made in different chronological order, would we always interpret them in the same way? The answer is probably no.
If we go back to the original question we set out to address, the list of eukaryotic innovations that need explanation has not changed much. The similarities in information systems of Archaea and Eukarya push back some characteristics to before the origin of eukaryotes (Ouzonis and Sander 1992; Creti et al. 1993; Langer and Zillig 1993; Zillig et al. 1993; Kaine et al. 1994; Marsh et al. 1994; Keeling and Doolittle 1995a,b; Langer et al. 1995; Ouzonis et al. 1995), and we have identified distant relatives of many other uniquely eukaryotic proteins (Sanchez et al. 1994; Erickson 1995; Erickson et al. 1996), but cytologically much of what Roger Stanier suggested in 1970 still makes sense, regardless of whether some chimerical hypotheses are correct or not. Stanier’s intuition was to propose characteristics that most likely preceded other characteristics because they made subsequent changes more plausible. For example, he identified the cytoskeleton as a key innovation because it would logically precede the endomembrane system, which itself had to be in place to make the nucleus (which is a part of the endomembrane system) and for endocytosis to take place, which is itself a logical predecessor of endosymbiosis, which gave rise to mitochondria. Stanier was wrong on some details because our understanding of the endosymbiotic origin of mitochondria and plastids was in its infancy, and he did not elaborate on the details of why some of these events could happen. Now, however, many molecular-based hypotheses fail not only to explain why these changes took place, but do not even explain how they might have anymore. The noteworthy exceptions serve to prove the rule. Martin has approached the problem from an energetics point of view that also takes into account the actual cellular transition from prokaryote to eukaryote (Martin and Muller 1998; Sousa et al. 2013), whereas Cavalier-Smith has for many years been developing variations of the neomuran hypothesis that, while based on cytological principles, incorporate molecular data (phylogenies in particular) and are about as close in form to the original cytological explanations as we now have (e.g., see Cavalier-Smith 1987a, 2014).
This can all be summed up rather simply. Scientific hypotheses are the products of several factors, the data being only one of them. As scientists, we focus on the data and are largely unconscious of the many other factors influencing how we interpret it, but the influence of history should not be underestimated. If we can occasionally slough off history and rethink the problem and what information we have that might address it, without being quite so bound to prevailing ideas, the problem can sometimes be simplified or at least seen more clearly. The origin of eukaryotes began as a cytological problem, and if we return to thinking about the cells as a whole, I suspect it will lead us back to autogenous mechanisms to explain the most important transitions. Molecular biology and genomics are powerful tools to test hypotheses that explain cytological changes, but it is a mistake to let these kinds of data dominate the formulation of hypotheses with no reference to their cytological implications. This leads us away from the transformation of cellular architecture, which, as Stanier also pointed out (Stanier et al. 1957), remains “… the greatest single evolutionary discontinuity to be found in the present day world.”
ACKNOWLEDGMENTS
Nearly all of the reading and thinking behind this work was done during the tenure of a fellowship from the John Simon Guggenheim Memorial Foundation, from 2012 to 2013. I also thank the Canadian Institute for Advanced Research for long-term support. This review is the direct result of a rash letter sent to my co-editor, Eugene Koonin, and I thank him for his, probably misguided, encouragement. I also thank Ford Doolittle, Michael Gray, Fabien Burki, Todd Handy, and Maureen O’Malley for valuable discussions on this topic and extremely helpful guidance and criticism.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Balch WE, Magrum LJ, Fox GE, Wolfe RS, Woese CR 1977. An ancient divergence among the bacteria. J Mol Evol 9: 305–311 [DOI] [PubMed] [Google Scholar]
- Bell PJ 2006. Sex and the eukaryotic cell cycle is consistent with a viral ancestry for the eukaryotic nucleus. J Theor Biol 243: 54–63 [DOI] [PubMed] [Google Scholar]
- Bonen L, Doolittle WF 1975. On the prokaryotic nature of red algal chloroplasts. Proc Natl Acad Sci 72: 2310–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L, Doolittle WF 1976. Partial sequences of 16S RRNA and the phylogeny of blue-green algae and chloroplasts. Nature 261: 669–673 [DOI] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF 1995. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci 92: 2441–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF 1997. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev 61: 456–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1978. The evolutionary origin and phylogeny of microtubules, mitotic spindles and eukaryote flagella. Biosystems 10: 93–114 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1983. A 6-kingdom classification and a unified phylogeny. In Endocytobiology II: Intracellular space as oligogenetic ecosystem (ed. Schenk HEA, Schwemmler WS), pp. 1027–1034 Walter De Gruyter, New York [Google Scholar]
- Cavalier-Smith T 1987a. The origin of eukaryote and archaebacterial cells. Ann NY Acad Sci 503: 17–54 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1987b. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann NY Acad Sci 503: 55–71 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1993. Kingdom protozoa and its 18 phyla. Microbiol Rev 57: 953–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 1997. Cell and genome coevolution: Facultative anaerobiosis, glycosomes and kinetoplastan RNA editing. Trends Genet 13: 6–9 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2010. Kingdoms protozoa and chromista and the eozoan root of the eukaryotic tree. Biol Lett 6: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cavalier-Smith T 2014. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton E 1925. Pannsporella perplexa. Réflexions sur la biologie et la phylogénie des protozoaires. Ann Sci Nat Zool 10e: 1–84 [Google Scholar]
- Chatton E 1938. Titre et travaux scientifique (1906–1937) De Edouard Chatton. Sottano, Sette, Italy [Google Scholar]
- Creti R, Londei P, Cammarano P 1993. Complete nucleotide sequence of an archaeal (Pyrococcus woesei) gene encoding a homolog of eukaryotic transcription factor IIB (TFIIB). Nucleic Acids Res 21: 2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R 2005. Ancestor’s tale: A pilgrimage to the dawn of evolution. Houghton Mifflin Harcourt, New York [Google Scholar]
- Dodge JD 1965. Chromosome structure in the dinoflagellates and the problem of the mesokaryotic cell. Excerpta Med Int Congr Ser 91: 339–345 [Google Scholar]
- Doolittle WF 1996. Some aspects of the biology of cells and their possible evolutionary significance. In Evolution of microbial life (ed. Roberts DM, et al. ), pp. 1–22 University of Cambridge Press, Cambridge, UK [Google Scholar]
- Dougherty EC, Allen MB 1960. Is pigmentation a clue to protistan phylogeny? In Comparative biochemistry of photoreactive systems (ed. Allen MB), pp. 129–144 Academic, New York [Google Scholar]
- Erickson HP 1995. FtsZ, a prokaryotic homolog of tubulin? Cell 80: 367–370 [DOI] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci 93: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P 2006. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res 117: 5–16 [DOI] [PubMed] [Google Scholar]
- Forterre P 2011. A new fusion hypothesis for the origin of eukarya: Better than previous ones, but probably also wrong. Res Microbiol 162: 77–91 [DOI] [PubMed] [Google Scholar]
- Gibor A, Granick S 1964. Plastids and mitochondria: Inheritable systems. Science 145: 890–897 [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Kiblak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson NF, Poole RJ, Date T, Oshima T, et al. 1989. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc Natl Acad Sci 86: 6661–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksoyr J 1966. Evolution of eucaryotic cells. Nature 214: 1161. [DOI] [PubMed] [Google Scholar]
- Golding GB, Gupta RS 1995. Protein-based phylogenies support a chimeric origin for the eukaryotic genome. Mol Biol Evol 12: 1–6 [DOI] [PubMed] [Google Scholar]
- Gornik SG, Ford KL, Mulhern TD, Bacic A, McFadden GI, Waller RF 2012. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr Biol 22: 2303–2312 [DOI] [PubMed] [Google Scholar]
- Gould SJ 1990. Wonderful life: The Burgess Shale and the nature of history. Norton, New York [Google Scholar]
- Gourguechon S, Holt LJ, Cande WZ 2013. The Giardia cell cycle progresses independently of the anaphase-promoting complex. J Cell Sci 126: 2246–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Doolittle WF 1982. Has the endosymbiont hypothesis been proven? Microbiol Rev 46: 1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS 1995. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol Microbiol 15: 1–11 [DOI] [PubMed] [Google Scholar]
- Gupta RS 1999. Origin of eukaryotic cells: Was metabolic symbiosis based on hydrogen the driving force? Trends Biochem Sci 24: 423–424 [DOI] [PubMed] [Google Scholar]
- Gupta RS, Golding GB 1993. Evolution of Hsp70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol 37: 573–582 [DOI] [PubMed] [Google Scholar]
- Gupta RS, Singh B 1994. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol 4: 1104–1114 [DOI] [PubMed] [Google Scholar]
- Hamkalo BA, Rattner JB 1977. The structure of mesokaryote chromosome. Chromosoma 60: 39–47 [DOI] [PubMed] [Google Scholar]
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.” Proc Natl Acad Sci 106: 3859–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H 1984. The origin of the eukaryotic cell. Speculations Sci Technol 7: 77–81 [PubMed] [Google Scholar]
- Hashimoto T, Hasegawa M 1996. Origin and early evolution of eukaryotes inferred from the amino acid sequences of translation elongation factors 1α/Tu and 2/G. Adv Biophys 32: 73–120 [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Nakamura Y, Nakamura F, Shirakura T, Adachi J, Goto N, Okamoto K, Hasegawa M 1994. Protein phylogeny gives a robust estimate for early divergences of eukaryotes: Phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol Biol Evol 11: 65–71 [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Nakamura Y, Kamaishi T, Nakamura F, Adachi J, Okamoto K, Hasegawa M 1995. Phylogenetic place of mitochondrion-lacking protozoan, Giardia lamblia, inferred from amino acid sequences of elongation factor 2. Mol Biol Evol 12: 782–793 [DOI] [PubMed] [Google Scholar]
- Hinkle G, Sogin ML 1993. The evolution of the Vahlkampfiidae as deduced from 16S-like ribosomal RNA analysis. J Eukaryot Microbiol 40: 599–603 [DOI] [PubMed] [Google Scholar]
- Iwabe N, Kuma K-I, Hasegawa M, Osawa S, Miyata T 1989. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci 86: 9355–9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Dutcher SK 1991. Molecular studies of linkage group XIX of Chlamydomonas reinhardtii: Evidence against a basal body location. J Cell Biol 113: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaine BP, Mehr IJ, Woese CR 1994. The sequence, and its evolutionary implications, of a Thermococcus celer protein associated with transcription. Proc Natl Acad Sci 91: 3854–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaishi T, Hashimoto T, Nakamura Y, Nakamura F, Murata S, Okada N, Okamoto K-I, Shimzu M, Hasegawa M 1996. Protein phylogeny of translation elongation factor EF-1α suggests microsporidians are extremely ancient eukaryotes. J Mol Evol 42: 257–263 [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L, Mrazek J, Campbell AM, Spormann AM 1999. A chimeric prokaryotic ancestry of mitochondria and primitive eukaryotes. Proc Natl Acad Sci 96: 9190–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA 1998. Changing perspectives on the origin of eukaryotes. Trends Ecol Evol 13: 493–497 [DOI] [PubMed] [Google Scholar]
- Katz LA 2002. Lateral gene transfers and the evolution of eukaryotes: Theories and data. Int J Syst Evol Microbiol 52: 1893–1900 [DOI] [PubMed] [Google Scholar]
- Katz LA 2012. Origin and diversification of eukaryotes. Annu Rev Microbiol 66: 411–427 [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Doolittle WF 1995a. Archaea: Narrowing the gap between prokaryotes and eukaryotes. Proc Natl Acad Sci 92: 5761–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Doolittle WF 1995b. An archaebacterial elF-1A: New grist for the mill. Mol Microbiol 17: 399–400 [DOI] [PubMed] [Google Scholar]
- Klein RM, Cronquist A 1967. A consideration of the evolutionary and taxonomic significance of some biochemical, micromorphology, and physiological characters in the thallophytes. Q Rev Biol 42: 105–296 [PubMed] [Google Scholar]
- Koonin EV 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Rivera MC 1994. Was the nucleus the first endosymbiont? Proc Natl Acad Sci 91: 2880–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer D, Zillig W 1993. Putative tflls gene of Sulfolobus acidocaldarius encoding an archaeal transcription elongation factor is situated directly downstream of the gene for a small subunit of DNA-dependant RNA polymerase. Nucleic Acids Res 21: 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer D, Hain J, Thuriaux P, Zillig W 1995. Transcription in Archaea: Similarity to that in Eucarya. Proc Natl Acad Sci 92: 5768–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbeater B, Dodge JD 1967. An electron microscope study of nuclear and cell division in a dinoflagellate. Arch Mikrobiol 57: 239–254 [DOI] [PubMed] [Google Scholar]
- Li JY 1983. Studies on dinoflagellate chromosomal basic protein. Biosystems 16: 217–225 [PubMed] [Google Scholar]
- Li JY, Wu CF 2005. New symbiotic hypothesis on the origin of eukaryotic flagella. Naturwissenschaften 92: 305–309 [DOI] [PubMed] [Google Scholar]
- Mahler HR, Raff RA 1975. The evolutionary origin of the mitochondrion: A nonsymbiotic model. Int Rev Cytol 43: 1–124 [DOI] [PubMed] [Google Scholar]
- Margulis L 1970. Origin of eukaryotic cells. Yale University Press, New Haven, CT [Google Scholar]
- Margulis L 1981. Symbiosis in cell evolution. Freeman, San Francisco [Google Scholar]
- Margulis L 1990. Words as battle cries—Symbiogenesis and the new field of endocytobiology. Bioscience 40: 673–677 [PubMed] [Google Scholar]
- Margulis L, Dolan MF, Guerrero R 2000. The chimeric eukaryote: Origin of the nucleus from the karyomastigont in amitochondriate protists. Proc Natl Acad Sci 97: 6954–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh TL, Reich CI, Whitlock RB, Olsen GJ 1994. Transcription factor IID in the Archaea: Sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci 91: 4180–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W 1999. A briefly argued case that mitochondria and plastids are descendants of endosymbionts, but that the nuclear compartment is not. Proc R Soc Lond Ser B-Biol Sci 266: 1387–1395 [Google Scholar]
- Martin W, Muller M 1998. The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- Martin W, Rotte C, Hoffmeister M, Theissen U, Gelius-Dietrich G, Ahr S, Henze K 2003. Early cell evolution, eukaryotes, anoxia, sulfide, oxygen, fungi first (?), and a tree of genomes revisited. IUBMB Life 55: 193–204 [DOI] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P 1998. Symbiosis between methanogenic Archaea and δ-proteobacteria as the origin of eukaryotes: The syntrophic hypothesis. J Mol Evol 47: 517–530 [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317: 1921–1926 [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR, Overbeek R 1994. The winds of (evolutionary) change: Breathing new life into microbiology. J Bacteriol 1765: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzonis C, Sander C 1992. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell 71: 189–190 [DOI] [PubMed] [Google Scholar]
- Ouzonis C, Kyrpides N, Sander C 1995. Novel protein families in Archaean genomes. Nucleic Acids Res 23: 565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa O Jr 2000. Counterfactual histories: The beginning of quantum physics. Philos Sci 68: S519–S530 [Google Scholar]
- Pessoa O Jr 2010. Modeling the causal structure of the history of science. In Model-based reasoning in science & technology (ed. Magnani L, et al. ), pp. 643–654 Springer, Berlin [Google Scholar]
- Piaget J 1953. The origin of intelligence in the child. Routledge and Kegan Paul, New York [Google Scholar]
- Radick G 2008. Conterfactuals and the historian of science. Isis 99: 547–551 [Google Scholar]
- Raff RA, Mahler HR 1972. The non symbiotic origin of mitochondria. Science 177: 575–582 [DOI] [PubMed] [Google Scholar]
- Raff RA, Mahler HR 1975. The symbiont that never was: An inquiry into the evolutionary origin of the mitochondrion. Symp Soc Exp Biol 1975: 41–92 [PMC free article] [PubMed] [Google Scholar]
- Rivera MC, Lake JA 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431: 152–155 [DOI] [PubMed] [Google Scholar]
- Rivera MC, Jain R, Moore JE, Lake JA 1998. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci 95: 6239–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo PJ 1991. The enigma of the dinoflagellate chromosome. J Protozool 38: 246–252 [Google Scholar]
- Rizzo PJ, Morris RL 1983. Some properties of the histone-like protein from Crypthecodinium cohnii (Hcc). Biosystems 16: 211–216 [DOI] [PubMed] [Google Scholar]
- Rizzo PJ, Nooden LD 1972. Chromosomal proteins in the dinoflagellate alga Gyrodinium cohnii. Science 176: 796–797 [DOI] [PubMed] [Google Scholar]
- Robinow CF 1956. The chromatin bodies of bacteria. Bacteriol Rev 20: 207–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow CF 1962. Morphology of the bacterial nucleus. Br Med Bull 18: 31–35 [DOI] [PubMed] [Google Scholar]
- Roger AJ 1999. Reconstructing early events in eukaryotic evolution. Am Nat 154: S146–S163 [DOI] [PubMed] [Google Scholar]
- Sagan L 1967. On the origin of mitosing cells. J Theoret Biol 14: 225–275 [DOI] [PubMed] [Google Scholar]
- Sanchez M, Valencia A, Ferrandiz MJ, Sander C, Vicente M 1994. Correlation between the structure and biochemical activities of Ftsa, an essential cell division protein of the actin family. EMBO J 1354: 4919–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp JL 1994. Evolution by association: A history of symbiosis. Oxford University Press, Oxford, UK [Google Scholar]
- Sapp JL 2005. The prokaryote–eukaryote dichotomy: Meanings and mythology. Microbiol Mol Biol Rev 69: 292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML 1991. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev 1: 457–463 [DOI] [PubMed] [Google Scholar]
- Sogin ML, Gunderson JH, Elwood HJ, Alonso RA, Peattie DA 1989. Phylogenetic meaning of the kingdom concept: An unusual ribosomal RNA from Giardia lamblia. Science 243: 75–77 [DOI] [PubMed] [Google Scholar]
- Sousa FL, Thiergart T, Landan G, Nelson-Sathi S, Pereira IA, Allen JF, Lane N, Martin WF 2013. Early bioenergetic evolution. Philos Trans R Soc Lond B Biol Sci 368: 20130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier RY 1970. Some aspects of the biology of cells and their possible evolutionary significance. In Organisation and control in prokaryotic and eukaryotic cells (ed. Charles HP, Knight BCJG), pp. 1–38 Cambridge University Press, Cambridge, UK [Google Scholar]
- Stanier RY, Van Niel CB 1962. The concept of a bacterium. Arch Mikrobiol 42: 17–35 [DOI] [PubMed] [Google Scholar]
- Stanier RY, Doudoroff M, Adelberg EA 1957. The microbial world. Prentice-Hall, Upper Saddle River, NJ [Google Scholar]
- Stewart KD, Mattox K 1980. Phylogeny of phytoflagellates. In Phytoflagellates, development in marine biology (ed. Cox ER), pp. 433–462 Elsevier, Amsterdam [Google Scholar]
- Takemura M 2001. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol 52: 419–425 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S 2012. Chromatin: Packaging without nucleosomes. Curr Biol 22: R1040–R1043 [DOI] [PubMed] [Google Scholar]
- Taylor FJ 1979. Symbionticism revisited: A discussion of the evolutionary impact of intracellular symbioses. Proc R Soc Lond B Biol Sci 204: 267–286 [DOI] [PubMed] [Google Scholar]
- Uzzell T, Spolsky C 1974. Mitochondria and plastids as endosymbionts: A revival of special creation? Am Sci 62: 334–343 [PubMed] [Google Scholar]
- Vossbrinck CR, Maddox JV, Friedman S, Debrunner-Vossbrinck BA, Woese CR 1987. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature 326: 411–414 [DOI] [PubMed] [Google Scholar]
- Williams BA, Keeling PJ 2003. Cryptic organelles in parasitic protists and fungi. Adv Parasitol 54: 9–68 [DOI] [PubMed] [Google Scholar]
- Woese CR 1987. Bacterial evolution. Microbiol Rev 51: 221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR 1994. Microbiology in transition. Proc Natl Acad Sci 9181: 1601–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE 1977a. The concept of cellular evolution. J Mol Evol 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE 1977b. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci 74: 5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci 87: 4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W 1991. Comparative biochemistry of Archaea and Bacteria. Curr Opin Genet Dev 1: 544–551 [DOI] [PubMed] [Google Scholar]
- Zillig W, Klenk HP, Palm P, Leffers H, Puhler G, Gropp F, Garrett RA 1989. Did eukaryotes originate by a fusion event? Endocytobiosis Cell Res 6: 1–25 [Google Scholar]
- Zillig W, Palm P, Klenk H-P, Langer D, Hüdepohl U, Hain J, Lanzendörfer M, Holz H 1993. Transcription in Archaea. In The biochemistry of Archaea (ed. Kates M, et al. ), pp. 544–551 Elsevier, Amsterdam [Google Scholar]


