Abstract
SET domain-containing proteins belong to a group of enzymes named after a common domain that utilizes the cofactor S-adenosyl-L-methionine (SAM) to achieve methylation of its substrates. Many SET domain-containing proteins have been shown to display catalytic activity towards particular lysine residues on histones, but emerging evidence also indicates that various non-histone proteins are specifically targeted by this clade of enzymes. Here, we summarize the most recent findings on the biological functions of the major families of SET domain-containing proteins catalyzing the methylation of histones 3 on lysines 4, 9, 27, and 36 (H3K4, H3K9, H3K27, and H3K36) and histone 4 on lysine 20 (H4K20) as well as candidates that have been reported to regulate non-histone substrates.
Keywords: SET domain-containing proteins, histone lysine methylation, non-histone substrates
SET domain-containing proteins
SET domain-containing proteins exist in all eukaryotes studied to date. This protein family is characterized by an approximately 130 amino-acid-long domain called the SET domain, which was named after the Drosophila proteins Suppressor of variegation 3-9 (Su(var)3-9), Enhancer of zeste (E(z)), and Trithorax (Trx). The SET domain possesses catalytic activity towards the ε-amino group of lysine residues. Depending on the context and their biochemical properties, SET domain-containing proteins are able to mono-, di-, or trimethylate their lysine substrates by utilizing the cofactor S-adenosyl-L-methionine (SAM). In vivo, lysine methylation is often dynamically regulated by the opposing actions of lysine methyltransferases and lysine demethylases. Initially reported to catalyze the methylation of histones, it has now become increasingly clear that SET domain-containing proteins also target many non-histone substrates, some of which constitute regulators of signaling pathways, transcription factors, and tumor suppressors (Tables 1-3, Boxes 1-2). The grouping of SET domain-containing proteins based on sequence similarity of their SET domains often closely reflects an already reported specificity for certain substrates (Figure 1, showing human SET domain-containing proteins). Here, we provide an overview on the biological functions of the major groups of SET domain-containing histone lysine methyltransferases (KMTs) based on their substrate specificity towards histones. Apart from affecting chromatin states either by directly methylating histones (Figures 1-2) and thus altering the chromatin environment to enhance or suppress the binding of co-factors, KMTs have also been reported to target non-histone proteins (Table 1, Box 1). Importantly, many other SET domain-containing proteins are only known to methylate non-histone substrates and do not appear to target histones directly (Tables 2-3, Box 2). Furthermore, the SET domain does not generally exist as an independent entity, as in many proteins it co-occurs with multiple other protein domains (Figure 1). Some SET domain-containing proteins are found in complexes or interact with proteins that regulate their target specificity and catalysis (Figure 3).
Table 1. Non-histone targets of H3K4, H3K9, H3K27, H3K36 and H4K20 KMTs.
| KMT family | SET domain-containing protein | Substrate | Lysine(s) | Function | Reference |
|---|---|---|---|---|---|
| H3K4 KMT | Set1 (yeast) | Dam1 (yeast) | 233 (me2) | Proper chromosome segregation | [243, 244] |
| H3K9 KMT | G9A | ACIN1 | 654 (me2, me3) | Unknown | [248] |
| H3K9 KMT | G9A | CDYL | 135 (me1, me2, me3) | Decreased interaction with H3K9me3 | [248] |
| H3K9 KMT | G9A | DNMT1 | 70 (me2) | Unknown | [248] |
| H3K9 KMT | G9A/G9a GLP/Glp | DNMT3/Dnmt3a | 47 (me1, me2) (human), 44 (me1, me2) (mouse), | Binding of MPHOSPH8 to DNMT3A | [288] |
| H3K9 KMT | G9A | G9A | 165 (me2, me3) | Required for binding of HP1α and HP1γ | [109] |
| H3K9 KMT | G9A | G9A | 185 (me2, me3) | Unknown | [248] |
| H3K9 KMT | G9A GLP | P53 | 373 (me2) | Unknown | [247] |
| H3K9 KMT | G9A | RUVBL2/REPTIN | 67 (me1) | Negative regulation of hypoxia-inducible genes | [246] |
| H3K9 KMT | G9A | WIZ | 305 (me2, me3) | Unknown | [248] |
| H3K9 KMT | GLP | GLP | 205 | Binding of MPHOSPH8 to GLP | [288] |
| H3K9 KMT | SETDB1 | Tat (HIV protein) | 50, 51 | Inhibition of HIV transcription | [250] |
| H3K9 KMT | SUV39H1 | CBX4/PC2 | 191 (me2) | TUG1 ncRNA-dependent recruitment to Polycomb bodies | [245] |
| H3K27 KMT | EZH2 | GATA4 | 299 (me1) | Decreased GATA4-dependent transcriptional activation | [251] |
| H3K27 KMT | EZH2 | RORA | 38 (me1) | Enhanced proteasomal degradation | [252] |
| H3K27 KMT | EZH2 | STAT3 | 180 | Increased STAT3 phosphorylation, enhanced STAT3 activity | [253] |
| H3K36 KMT | NSD1 | RELA/P65 | 218 (me1), 221 (me2) | Activation of NF-κB target genes | [255] |
| H3K36 KMT | SETMAR | SETMAR (automethylation) | 485 (me1) | Decreased Topoisomerase IIα-mediated decatenation | [256] |
| H4K20 KMT | SETD8/PR-SET7/SET8 | NUMB | 158 (me2), 163 (me2) | Decreased interaction with P53, ubiquitination-dependent P53 degradation | [258] |
| H4K20 KMT | SETD8/PR-SET7/SET8 | P53 | 382 (me1) | Promotes interaction with L3MBTL1 to represses P53-dependent transcription | [226, 227] |
| H4K20 KMT | SETD8/PR-SET7/SET8 | PCNA | 248 (me1) | Stabilization of PCNA, proper DNA replication | [259] |
The methylation state of the respective lysine (if known) is indicated in brackets with me1 for monomethylation, me2 for dimethylation and me3 for trimethylation. However, many reports do not necessarily exclude the possibility that other methylation states exist on the same lysine.
Table 3. Non-histone substrates of other SET domain-containing proteins.
| SET domain-containing protein | Substrate | Lysine(s) | Function | Reference |
|---|---|---|---|---|
| Rubisco LSMT (tobacco) | Rubisco LSMT (tobacco) | 14 (me3) | Not known | [315, 316] |
| Rkm1 (yeast) | Rpl23ab (yeast) | 105 (me2), 109 (me2) | Not known | [317, 318] |
| Rkm2 (yeast) | Rpl12ab (yeast) | 3 (me2), 10 (me3) | Not known | [319] |
| Rkm3 (yeast) | Rpl42ab (yeast) | 40 (me1) | Not known | [320] |
| Rkm4 (Set7) (yeast) | Rpl42ab (yeast) | 55 (me1) | Not known | [320] |
| SETD6 | RELA/P65 | 310 (me1) | Repression of NF-κB target genes via the H3K9 methyltransferase GLP | [321, 322] |
| SMYD2 | P53 | 370 (me1) | Represses P53-dependent transcription | [323-326] |
| SMYD2 | HSP90 | 209, 615 (me1), 616 (me1) | Myofilament organization | [327, 328] |
| SMYD2 | RB1 | 810 (me1) | Stimulates E2F1-mediated transcription and cell cycle progression | [329] |
| SMYD2 | RB1 | 860 (me1) | Enhances L3MBTL1 binding to RB1 | [330] |
| SMYD3 | FLT1/VEGFR1 | 831 (me2) | Enhanced VEGFR1 kinase activity | [27] |
The methylation state of the respective lysine (if known) is indicated in brackets with me1 for monomethylation, me2 for dimethylation and me3 for trimethylation. However, many reports do not necessarily exclude the possibility that other methylation states exist on the same lysine.
Box 1. Non-histone substrates of H3K4, H3K9, H3K27, H3K36 and H4K20 methyltransferases (KMTs).
H3K4 KMTs (Table 1)
In Set1, Rad6, and Bre1 mutants, Dam1 methylation on lysine 233 is strongly reduced affecting proper chromosome segregation in yeast. Dam1 methylation negatively affects its phosphorylation by Ipl kinase on neighboring serines. Thus, Rad6/Bre1-mediated H2B ubiquitination appears to provide a platform on kinetochores for Set1-mediated methylation of Dam1 [243, 244].
H3K9 KMTs (Table 1)
SUV39H1 methylates CBX4 (PC2) on lysine 191. This methylation event is required for the binding of CBX4 to the ncRNA TUG1, which then targets methylated CBX4 to Polycomb bodies to repress transcription [245]. Under hypoxic conditions, G9A methylates the chromatin remodeler RUVBL2 (REPTIN) on lysine 67. Methylated RUVBL2 in turn binds to the promoters of hypoxia-responsive genes and negatively affects their expression allowing a balanced response to HIF1α-induced gene expression [246]. G9A/GLP also methylate P53 on lysine 373 [247]; WIZ on lysine 305; CDYL on lysine 135; ACIN1 on lysine 654 [248]; and C/EBPβ [249] and SETDB1 have been reported to target the HIV-1 protein Tat [250].
H3K27 KMTs (Table 1)
EZH2 directly methylates the transcription factor GATA4, the orphan nuclear receptor RORA and STAT3. GATA4 methylation on lysine 299 disrupts its interaction with the acetyltransferase p300 causing attenuated transcriptional activity of GATA4 targets [251]. Methylation of RORA on lysine 38 provides a binding platform for the DCAF1/DDB1/CUL4 ubiquitin ligase complex to dynamically control protein stability [252] and EZH2-mediated methylation of STAT3 on lysine 180 results in increased STAT3 phosphorylation and enhanced STAT3 activity in glioblastoma stem-like cells [253]. A catalytically active cytoplasmic version of PRC2 has also been reported to be involved in actin polymerization via VAV1 [254]. Currently, it is unknown whether this role of PRC2 involves its catalytic activity and the methylation of non-histone substrates in the cytoplasm.
H3K36 KMTs (Table 1)
NSD1 methylates lysines 218 and 221 of P65, a component of the NF-κB complex [255] and SETMAR automethylation mediates decreased Topoisomerase IIα chromosome decatenation [256, 257].
H4K20 KMTs (Table 1)
SETD8 directly methylates P53 on lysine 382 which promotes L3MBTl1-P53 interaction to repress P53-dependent transcription [226, 227]. Methylation of NUMB on lysines 158 and 163 by SETD8 decreases the interaction of NUMB with P53 resulting in increased P53 degradation and reduced NUMB -mediated apoptosis [258]. SETD8 plays an important role in DNA replication. In this context, methylation of PCNA by SETD8 on lysine 248 has been shown to stabilize PCNA to allow for proper DNA replication [259].
Box 2. Other SET domain-containing proteins.
Apart from the H3K4, H3K9, H3K27, H3K36 and H4K20 KMTs discussed here, many other SET domain-containing proteins exist which often are involved in important developmental processes (Figure 1). Whether their catalytic function as methyltransferases is important in this context is not always clear, often because of a lack of a reported substrate or independent experimental confirmation. Particularly, the PRDM family and also other SET domain-containing proteins have been reported to be crucial developmental regulators, but either their substrates are unknown, or in many cases as for histones, questionable [68, 69]. In Tables 2 and 3, we have summarized the known biological functions of these SET domain-containing proteins, if a methylated substrate has been described. The enzyme with the most non-histone substrates to date is SETD7. Among others, SETD7methylates the tumor suppressors P53 and RB1, nuclear hormone receptors, but also components of major signaling pathways and transcription factors (Table 2). Other SET domain-containing proteins with known substrates are yeast Rkm1-4, SETD6, SETMAR, SMYD2, and SMYD3 (Table 3).
Figure 1. Relationship and structure of human SET domain-containing proteins.
51 human SET domain-containing proteins were aligned according to their annotated SET domain by using ClustalO v. 1.1.0. The length of each tree branch to the next branch point constitutes a readout for the unit change per amino acid as displayed on the axis at the bottom of the tree. For each SET domain-containing protein the name(s) and corresponding domain structure are provided. The allocation and annotation of each domain structure follows SMART or NCBI (for PRDM10, SETD3, SETD4 and SETD9) as accessed on April 9, 2013. Symbols and names for depicted domains are displayed in the box labeled “Domains”. Approximately 2000 amino acids (∼2K aa) from the domain structure of MLL3 and MLL4 were removed as indicated by two parallel slashes. SET domain-containing proteins that show specificity towards the same histone residue (see also Figure 2) are highlighted in the same color. Colors are: green: histone H3K4 lysine methyltransferases (KMTs); red: H3K9 KMTs; orange: H3K27 KMTs; blue: H3K36 KMTs; purple: H4K20 KMTs.
Figure 2. Histone lysine methyltransferase target specificities of mammalian SET domain-containing proteins.
Green: H3K4 histone lysine methyltransferases (KMTs); red: H3K9 KMTs; orange: H3K27 KMTs; blue: H3K36 KMTs; purple: H4K20 KMTs. Non-histone substrates have been described for members of all five KMT families, SETD7 and other SET domain-containing proteins. A summary of all non-histone targets described to date can be found in Tables 1-3.
Table 2. Non-histone substrates of SETD7/SET7/SET9.
| SET domain-containing protein | Substrate | Lysine(s) | Function | Reference |
|---|---|---|---|---|
| SETD7/SET7/SET9 | Androgen receptor (AR) | 630 and/or 632 (me1) | Stimulation of AR-dependent transcription | [289, 290] |
| SETD7/SET7/SET9 Setd7/Set7/Set9 | DNMT1/Dnmt1 | 142 (me1), 1094 1096 (mouse) | Enhanced proteasomal degradation | [291-293] |
| SETD7/SET7/SET9 | E2F1 | 185 (me1) | Enhanced proteasomal degradation, prevention of apoptosis | [294] |
| SETD7/SET7/SET9 | Estrogen receptor α (ERα) | 302 (me1) | Stabilization of and increased transactivation by ERα | [295] |
| SETD7/SET7/SET9 | FOXO3 | 271 (me1) | Decreased protein stability | [296] |
| SETD7/SET7/SET9 | NR1H4 (FXR) | 206 | Enhanced binding to response elements and transactivation | [297] |
| SETD7/SET7/SET9 Setd7/Set7/Set9 | P53 | 372 (me1) 369 (me1) (mouse) | Increased half-life by preventing proteasomal degradation and transactivation potential (function challenged by two new reports) | [298-302] |
| SETD7/SET7/SET9 | PCAF | 78 (me1), 89 (me1), 638 (me1) | Not known | [303] |
| SETD7/SET7/SET9 | RB1 | 810 (me1) | Cell cycle arrest | [304] |
| SETD7/SET7/SET9 | RB1 | 873 | Interaction with HP1, transcriptional repression | [305] |
| SETD7/SET7/SET9 | RELA/P65 | 37 (me1) | Enhanced promoter | [306, 307] |
| binding, activation of NF-κB target genes | ||||
| SETD7/SET7/SET9 | RELA/P65 | 314 (me1), 315 (me1) | Enhanced proteasomal degradation, repression of NF-κB target genes | [308, 309] |
| SETD7/SET7/SET9 | SIRT1 | Multiple lysines | Unknown | [310] |
| SETD7/SET7/SET9 | STAT3 | 140 (me2) | Downregulation of STAT3-dependent transcription | [311] |
| SETD7/SET7/SET9 | TAF10 | 189 (me1) | Stimulation of TAF10-dependent transcription | [312] |
| SETD7/SET7/SET9 | Tat (HIV protein) | 51 (me1) | Stimulation of HIV transcription | [313] |
| SETD7/SET7/SET9 | YAP1 | 494 (me1) | Cytoplasmic retention of YAP1 | [314] |
The methylation state of the respective lysine (if known) is indicated in brackets with me1 for monomethylation, me2 for dimethylation and me3 for trimethylation. However, many reports do not necessarily exclude the possibility that other methylation states exist on the same lysine. For a more detailed review on the function of SETD7/SET7/SET9 methylated non-histone substrates we refer to [16].
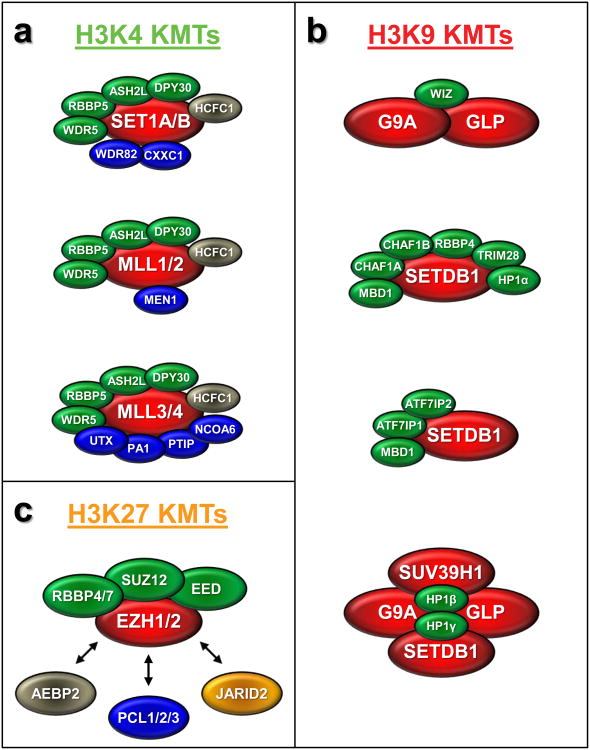
Figure 3. Mammalian protein complexes of SET domain-containing proteins described to date.
All SET domain-containing proteins are highlighted in red. (a) Six COMPASS-like complexes for mammalian H3K4 KMTs have been described consisting of three “subbranches” (SET1A/B, MLL1/2 and MLL3/4). All complexes share core subunits which are indicated in green. Complex-specific subunits are highlighted in blue. All mammalian COMPASS-like complexes additionally contain HCFC1 highlighted in gray which is not conserved in yeast. (b) Four complexes for mammalian H3K9 KMTs have been reported: a heterodimeric G9A/GLP complex together with the multi-zinc finger protein WIZ; an S-phase specific SETDB1 complex with the subunits MBD1, the chromatin assembly factor subunits CHAF1 A, CHAF1B, RBBP4 and the subunits TRIM28 and HP1α; a SETDB1 complex with the subunits MBD1, ATF7IP1 and ATF7IP2; a quaternary mega complex consisting of the H3K9 KMTs G9A, GLP, SUV30H1, SETDB1 and HP1β, HP1γ (c) H3K27 KMT complexes consist of either EZH1 or EZH2, the core subunits RBBP4 or RBBP7, SUZ12, EED (highlighted in green) and accessory factors such as AEBP2 (gray), PCL1 or PCL2 or PCL3 (all in blue) and JARID2 (orange) that either modulate PRC2 activity and/or are required for the context-specific recruitment of PRC2 to chromatin.
Histone H3K4 methyltransferases
Substrate specificity
H3K4 KMTs are conserved in plants, yeast, Drosophila, and mammals. Yeast Set1 is the sole enzyme that catalyzes the mono-, di-, and trimethylation of H3K4 [1, 2]. In Drosophila, yeast Set1 is represented by three homologous proteins: Set1, Trithorax (Trx), and Trithorax-related (Trr). Mammals contain six yeast Set1-related proteins with SET1A and SET1B (related to Drosophila Set1); MLL1 and MLL2 (related to Drosophila Trx); and MLL3 and MLL4 (related to Drosophila Trr) [1] (Figure 1). Drosophila Set1 constitutes a major di- and trimethyltransferase and this function is redundantly shared by mammalian SET1A/B [3-6]. Although the loss of Trx/MLL1/MLL2 does not result in bulk changes of any H3K4 methyl mark in most contexts, Mll2 is globally required for H3K4 trimethylation in oocytes [3, 7, 8]. Trr (and based on homology we predict mammalian MLL3/4 in a redundant fashion) is a major H3K4 monomethyltransferase [9]. Besides Trr, which is only homologous to the C-terminal portion of mammalian MLL3/4, Drosophila also contains another protein, LPT, with homology to the N-terminus of MLL3/4, and like Trr, LPT is also required for bulk H3K4 monomethylation [3, 9, 10]. Thus in Drosophila, two proteins, Trr (bearing the catalytic SET domain) and LPT, constitute the functional homolog of mammalian MLL3/4. All three Drosophila H3K4 KMTs are essential for viability [5, 11, 12], and Mll1 and Mll2 knockout mice show embryonic lethality confirming the importance of these genes in development [13, 14]. However, selective deletion of the Mll1 SET domain yields viable offspring albeit with various defects [15]. This suggests that the function of MLL1 and possibly most SET domain-containing proteins cannot simply be reduced to their methyltransferase activity as many of these enzymes contain important domains that might function independently of the SET domain.
With the yeast Set1 target Dam1, only one non-histone substrate among the H3K4 KMTs has been reported to date (Table1, Box 1). Additionally, SETD7 (SET7/SET9) has been demonstrated to be an H3K4 monomethyltransferase. However, SETD7 only displays limited activity on nucleosomal substrates, making it an unlikely candidate for H3K4 methylation in vivo. In fact, the identification of many non-histone substrates of SETD7 implies that they represent the functionally relevant SETD7 targets in vivo (Table 2, Box 2) [16]. Although Drosophila Ash1/mammalian ASH1L and MLL5 have been described as H3K4 KMTs [17-19], these initial reports could not be confirmed by others [20-23]. Instead, Ash1/ASH1L constitute an H3K36 dimethyltransferase; and UpSET, the Drosophila homolog of MLL5, restricts chromatin accessibility by binding to a Rpd3/Sin3-containing histone deacetylase complex [22]. Finally, SMYD3 can methylate H3K4, H4K5, and H4K20, but also targets non-histone substrates such as FLT1 (Table 3) [24-27]. Considering that SMYD3 does not display distinct substrate specificity on histones and also targets non-histone substrates, it is unlikely that it represents an H3K4 KMT in vivo.
Protein complexes (Figure 3)
In order to exert its catalytic activity as an H3K4 KMT, yeast Set1 requires the presence of additional subunits, which together with the catalytic SET domain-containing enzyme form a complex named COMPASS (Complex of Proteins Associated with Set1). In yeast, these subunits are Cps60, Cps50, Cps40, Cps35, Cps30, Cps25, and Cps15 [1]. The Drosophila and mammalian H3K4 KMTs exist in COMPASS-like complexes and all share the core subunits Ash2 in Drosophila/ASH2L in mammals (related to Cps60); RBBP5 (related to Cps50); Wds in Drosophila/WDR5 in mammals (related to Cps30); Dpy-30L1 in Drosophila/DPY30 in mammals (related to Cps25); and Hcf in Drosophila/HCFC1 in mammals (no yeast homolog). Besides these common subunits, each of the three “subbranches” contains complex-specific subunits. The Drosophila Set1 and the mammalian SET1A/B complexes share Cxxc1/CXXC1 (related to Cps40); Wdr82/WDR82 (related to Cps35); the Trx and MLL1/2 complexes Mnn1 in Drosophila/MEN1 in mammals; and the Trr and MLL3/4 complexes Ncoa6/NCOA6, Pa1/PA1, Ptip/PTIP, and Utx/UTX [1, 3].
Biochemical reconstitution followed by three dimensional cryo-electron microscopy of the yeast SET domain along with the core components Cps60, Cps50, Cps30, and Cps25 revealed that the yeast COMPASS “core” exists in a Y-shaped configuration with Cps50/Cps30 forming the top two adjacent lobes of the “Y” and Cps60/Cps25 constituting the stem. The localization of the SET domain at the juncture of Cps50/Cps30 and Cps60/Cps25 results in a channel that can only be accessed by a flexible peptide suggesting that COMPASS can only add methyl groups to amino acid residues that are located very close to the start or end of a protein [28]. Similarly, WDR5, RBBP5, and ASH2L pose fundamental components for the mammalian COMPASS-like complexes [28, 29]. WDR5 is essential for the assembly of all mammalian COMPASS-like complexes through its interaction with the WDR5 interaction (Win) motif located just N-terminally of the SET domain; and basic and acidic patches in Cps50/RBBP5 are required for its interaction with Set1/SET1A/SET1B [30-33]. Additionally, interaction between WDR5 and RBBP5 is crucial for complex assembly and activity [34, 35]. Both ASH2L and RBBP5 together with the catalytic SET domain are able to directly contact the substrate SAM to form a joint catalytic center [36]. The DNA-binding properties of the ASH2L N-terminus are necessary for optimal H3K4 KMT activity and transcriptional output of target genes [37, 38].
Transcription and recruitment of H3K4 KMTs
Set1/SET1A/SET1B are mainly involved in regulating H3K4 tri- and dimethylation on promoters and bodies of actively transcribed genes [1, 6]. An E2/E3 ubiquitin ligase module consisting of RAD6/BRE1 directly mediates H2B monoubiquitination and is required for proper implementation of H3K4 di- and trimethylation by Set1/SET1A/SET1B [1]. The Cps35/WDR82 subunit of COMPASS interacts with chromatin and COMPASS in an H2B monoubiquitination dependent manner and is required for H3K4 di- and trimethylation similar to both RAD6 and BRE1 [1, 39-42]. In yeast, asymmetric H3 arginine 2 (H3R2) dimethylation prevents the recruitment of Cps40, and thus, globally affects H3K4 trimethylation via Set1 [43]. Similar results were observed in mammals but have been attributed to MLL1 recruitment on Homeobox (HOX) genes [44, 45]. Therefore, it is possible that asymmetric H3R2 dimethylation might interfere with the recruitment of various COMPASS-like complexes in mammals. In mammals, the Set1a/b complex-specific subunit Cxxc1 (Cps40 in yeast) is also recruited to unmethylated CpG islands to implement H3K4 trimethylation; and artificial CpG sequences in the absence of promoters are able to recruit Cxxc1 and induce H3K4 trimethylation [46]. However, depending on the context, Cxxc1 is not only required to implement H3K4 trimethylation on promoter-proximal sequences, but also prevents introduction of H3K4 trimethylation on promoter-distal elements [47]. How this is achieved is currently unknown.
Drosophila studies have described Trx as a positive regulator of Hox genes and important developmental genes. This was also subsequently confirmed in mammals [1]. Indeed, while Mll1 is only required for the H3K4 trimethylation of a subset of Hox genes, Men1 knockout results in a dramatic loss of H3K4 trimethylation and gene expression on all Hox gene clusters in mouse embryonic fibroblasts providing evidence that Mll1/2 function redundantly on Hox genes and specifically target this gene class [8]. In yeast, the RNA Polymerase II (Pol II) interacting Paf1 complex has a dual function by controlling H2B monoubiquitination through Rad6/Bre1 and by providing a landing pad for Pol II to promote Set1-dependent H3K4 di- and trimethylation. This role has been confirmed for MLL1 on HOX genes in mammals as well [1, 48-51]. Long noncoding (nc) RNAs could also play a potential role in targeting the Trx/MLL1/MLL2 COMPASS-like complexes. For example, the ncRNA HOTTIP is transcribed from the 5′ tip of the human HOXA locus and binds WDR5 to recruit MLL1 across the HOXA cluster and the ncRNA Mistral activates transcription of Hoxa6 and Hoxa7 by recruiting Mll1 in mouse ES cells [52, 53].
Trr/MLL3/MLL4 interact with hormone receptors and are recruited to the promoters of target genes where they induce H3K4 trimethylation and transcription in Drosophila and mammals [1, 54-57]. The process of nuclear receptor signaling via Trr in Drosophila is highly regulated. In the absence of hormone (ecdysone), the ecdysone receptor resides in the cytoplasm, but upon binding to ecdysone, forms a heterodimer with the hormone receptor Ultraspiracle. The heterodimeric complex then translocates together with Trr into the nucleus to activate ecdysone-inducible genes [58]. Mammalian MLL3/4 are also involved in the P53-mediated DNA damage response. By binding to P53 the MLL3/4 complex-specific subunit NCOA6 recruits MLL3/4 COMPASS-like complexes to the promoters of the P53 target genes to catalyze H3K4 trimethylation and induce transcription [59].
Functions of MLL1 during the cell cycle
The protein levels of MLL1 are tightly controlled during the cell cycle and peak at the G1/S and G2/M transition. SCF and APC E3 ubiquitin ligase complexes ensure that MLL1 is degraded by the proteasome during the S and M phase, respectively [60]. Apart from controlling cell cycle regulators in G1, MLL1 also plays roles in origin of replication firing during S phase. Upon DNA damage, the checkpoint kinase ATR phosphorylates MLL1 on serine 516 preventing its degradation by SCF. MLL1 accumulation on chromatin prevents loading of the essential pre-replication complex component CDC45 which results in replication blockage [61]. In contrast to most transcription factors, MLL1 remains associated with mitotic chromosomes during M phase. Genes that are exclusively bound by MLL1 in mitosis are particularly highly expressed during interphase. Thus, MLL1 is thought to provide a “priming” platform for highly expressed S phase genes [60, 62].
Trr/MLL3/MLL4 and enhancer-associated H3K4 monomethylation
H3K4 monomethylation is a mark that is broadly distributed across the genome of Drosophila and mammals and enriched on the bodies of actively transcribed genes and cis-regulatory elements called enhancers [63, 64]. Although Drosophila Trr and its mammalian homologs MLL3/4 have been described as promoter-proximal H3K4 trimethyltransferases in hormone receptor-mediated transcription, studies based on a global H3K4 monomethylation reduction in Utx and trr mutant tissue show that in fact Trr constitutes a major H3K4 monomethyltransferase. Trr localizes to promoters but is also enriched on promoter-distal elements together with the Trr complex-specific subunits LPT and Utx, the histone acetyltransferase CBP, and broad domains of H3K4 monomethylation. Trr is able to convey enhancer-mediated activation of the cut locus in vivo and knockdown of Trr results in reduced H3K4 monomethylation, H3K27 acetylation, and a concomitant increase of H3K27 trimethylation on enhancers. This suggests that Trr/MLL3/MLL4 COMPASS-like complexes also function in enhancer-mediated processes by providing a hub for H3K27 demethylation of inactive/”poised” enhancers via Utx/UTX, followed by H3K4 monomethylation by Trr/MLL3/MLL4, and subsequent acetylation by CBP/p300 to create an activated enhancer state [9, 65].
Histone H3K9 methyltransferases
Substrate specificity
SUV39H1 was the first SET domain-containing protein reported to contain methyltransferase specificity in general and towards H3K9 in particular [66, 67]. Subsequently, the list of H3K9 KMTs has been expanded considerably, and in mammals, now includes SUV39H1, SUV39H2, G9A, GLP, SETDB1, SETDB2, PRDM3, and PRDM16 [66-68] (Figures 1-2). PRDM3 and PRDM16 are members of the PRDM family and contain an N-terminal PR domain, which is closely related to the SET domain [68, 69]. With a panoply of possible candidates in this group, it has been difficult to clearly separate state-specific functions in H3K9 methylation for any particular enzyme. It appears that the mono-, di-, and trimethylated states of H3K9 are in many cases either regulated redundantly or context-specifically based on the catalytic function, recruitment mechanisms to chromatin, and potentially varying expression patterns of the respective enzymes. SUV39H1/2 mediate the bulk of H3K9 trimethylation [70]. They preferentially localize to pericentric heterochromatin and other regions that contain repetitive DNA elements such as telomeres. On telomeres SUV39H1/2 also mediate dimethylation of H3K9 [67, 71]. In contrast, G9A and GLP are euchromatic H3K9 methyltransferases and act in a heterodimeric complex together with WIZ, a multi-zinc finger protein, to globally achieve mono-and dimethylation of H3K9 [66, 70, 72-74]. SETDB1 has also been ascribed bulk di- and trimethyltransferase activity and directs repression of euchromatic genes with the chromatin-associated factor ATF7IP (MCAF, AM), aiding in the conversion from the H3K9 dimethylated state to the trimethylated state [75]; whereas SETDB2 affects H3K9 trimethylation at pericentric heterochromatin [76]. How SUV39H1, SUV39H2, G9A, GLP, and SETDB1 can display overlapping state-specific, bulk H3K9 methylation changes is currently not clear. One explanation might be the co-occurrence of SUV39H1, G9A, GLP, and SETDB1 in a mega-complex [77]. Therefore, removing the function of one H3K9 KMT might also result in the loss of activity of the other candidates on common chromatin targets. In contrast, Prdm3 and Prdm16 function redundantly as cytoplasmic H3K9 monomethyltransferases [78]. Other histone substrates for H3K9 KMTs such as linker histone H1 mono- and dimethylation on lysine 26 and H3K27 and H3K56 methylation by G9A and GLP have been described [79-82] and several non-histone targets for H3K9 KMTs have been reported (Box 1).
Drosophila orthologs exist for all of the mammalian H3K9 KMTs including Su(var)3-9 for SUV39H1/2 [83], G9a for G9A/GLP [84], Eggless for SETDB1/2 [85, 86], and Hamlet for PRDM3/16. H3K9 KMT activities have been reported for Drosophila Su(var)3-9, G9a, and Eggless, but not for Hamlet. Generally, the Drosophila studies confirm the mammalian findings with occasional discrepancies as to the state-specific effects of the respective enzymes on H3K9 methylation [83, 85, 87, 88]. In Drosophila, Eggless and Hamlet are the only H3K9 KMT members that are essential for viability [85, 89], which is in contrast to the mammalian system where Suv39h1/2, G9a, Glp, Setdb1, Prdm3, and Prdm16 are all required for proper embryonic development. However, some Suv39h1/2 double-knockout mice survive into adulthood [72, 73, 90-93].
Protein complexes (Figure 3)
In contrast to H3K4 and H3K27 KMTs which form stereotypic stable complexes, the H3K9 KMTs appear to form more transient interactions or have the ability to change based on the context and local environment and requirements. For example, SETDB1 is recruited to euchromatic regions by autosumoylated TRIM28 (KAP1) to silence transcription in an HP1α-dependent mechanism [94-96]. SETDB1 has been described to form an S phase-specific protein complex with the methyl-CpG-binding protein MBD1 and the chromatin assembly factors CHAF1A (CAF1P150), CHAF1B (CAF1P60), and RBBP4 to achieve H3K9 methylation during replication [97, 98]. Cell-type specific recruitment of this TRIM28/CHAF1A/CHAF1B/RBBP4-containing SETDB1 complex is mediated by zinc finger transcription factors such as ZNF274 [99]. MBD1 also forms a SETDB1-containing complex with ATF7IP1 and ATF7IP2 to induce heterochromatin formation via HP1α, -β, and -γ [100]. More recently, a subset of SUV39H1, SETDB1, G9A, and GLP has been shown to exist in a multi-meric mega-complex that targets satellite repeats and regulates G9A target genes [77].
Repressive domains of H3K9 tri- and dimethylation
Heterochromatic regions such as pericentric chromatin and other regions contain repetitive DNA elements that are characterized by strong enrichment of H3K9 trimethylation, which is important in maintaining genome stability. SUV39H1/2-directed pericentric chromatin formation affects chromosomal stability and is involved in regulation of telomere length [71, 90]. The H3K9 di-and trimethyl binder HP1α interacts with SUV39H1 and has been implicated in a self-enforcing heterochromatin-spreading mechanism in which an initial H3K9 methylation event by SUV39H1 is enforced by the recruitment of HP1α and the subsequent spreading of the H3K9 trimethyl mark through continued SUV39H1 activity [66]. Likewise, loss of the Drosophila SUV39H1/2 ortholog Su(var)3-9 results in spontaneous heterochromatic DNA damage which is accompanied by translocation defects [101]. Interestingly, H3K9 monomethylation by PRDM3/16 may be a prerequisite for further H3K9 trimethylation by SUV39H1/2 as co-depletion of the murine H3K9 monomethyltransferases Prdm3/16 results in heterochromatic defects that resemble those of Suv39h1/2 loss including a bulk reduction in H3K9 trimethylation and derepressed satellite transcription [78]. However, because PRDM3 interacts with SUV39H1, it is possible that the role of PRDM3 and/or PRDM16 in H3K9 trimethylation and heterochromatic silencing might be mediated by this physical interaction and not their H3K9 monomethylation activity per se [102-104]. A recent study in C. elegans supports the importance of H3K9 methylation in repeat-rich heterochromatin to maintain interaction with the nuclear lamina as depletion of MET-2, the C. elegans homolog of SETDB1 and SET-25 another H3K9 trimethyl-specific KMT, results in chromosome detachment from the nuclear periphery [105].
The mammalian genome also contains large megabase-sized domains that are enriched for H3K9 di- and trimethylation and nuclear lamins, but are distinct from the previously described constitutive heterochromatin. They emerge during differentiation, localize to the nuclear periphery, and are thought to prevent the transcription of gene blocks that need to be silenced [67, 106, 107]. G9a represses genes within these domains of facultative heterochromatin, but is not required for their localization to the nuclear periphery [66, 108]. HP1α interacts with the G9A and GLP complexes, which requires the automethylation of G9A on a histone-like sequence [109, 110]. Both the chromodomain of HP 1α and the ankyrin repeats in G9A/GLP can bind H3K9 dimethyl marks [111]. Analogous to the spreading mechanism of HP1α and SUV39H1 in constitutive heterochromatin, this allows spreading of H3K9 dimethylation over large domains [66].
Transcriptional repression (and activation)
Various proteins, including mainly transcription factors and corepressors, have been proposed to interact and/or recruit H3K9 KMTs, and thus, provide a possible rational for target specificity. Generally, most studies suggest a function in transcriptional repression for the respective H3K9 KMTs. Gene repression by H3K9 KMTs usually correlates with increased H3K9 methylation on promoters and in some cases has been demonstrated to depend directly on the catalytic activity of the enzyme. Transcriptional repression by H3K9 KMTs is required in many developmental and physiological contexts including neurogenesis and the hypoxia and stress response (Box 3). However, evidence also exists for a direct involvement of some H3K9 KMTs in transcriptional activation by either RNA Polymerase I or II [112-114].
Box 3. H3K9 KMTs, neurogenesis and the hypoxia and stress response.
Derepression of neuronal genes can be observed in brains of G9a or Glp knockout mice and is accompanied by various behavioral phenotypes such as cognitive disabilities and autistic-like features. This role in neurogenesis also appears to be conserved for Drosophila G9a [260-262]. Furthermore, cocaine-induced vulnerability to stress has been linked to a loss of G9a function in the nucleus accumbens (NAc), an important brain reward region. Cocaine exposure results in decreased G9a/Glp levels and H3K9 dimethylation in NAc neurons by the upregulation of the transcription factor ΔFosB. Overexpression of G9a is able to reverse the depressive-like symptoms from cocaine exposure implicating G9a in this stress response pathway [263, 264]. However, G9A also represses neuronal genes in nonneuronal cells in cooperation with the Mediator and REST complexes [265, 266]. Hamlet, the Drosophila homolog of PRDM3/16, specifies the neuronal fate in the peripheral nervous system and plays a role in nascent olfactory receptor neurons where it opposes Notch signaling-induced transcriptional responses [89, 267]. Defects in the peripheral nervous system have also been reported for Prdm3 mutant mice and Prdm16 expression appears to be sensitive to Notch pathway activity in the telencephalon [92, 268]. Many reports have implicated PRDM3/16 in transcriptional repression and activation via interaction with other co-repressors and co-activators, but currently it is unclear whether these also depend on the H3K9 monomethyltransferase function of PRDM3/16 [68, 69]. As PRDM3/16 have been reported to monomethylate H3K9 in the cytoplasm, the nuclear role in transcription activation and repression might be independent of the KMT activities of PRDM3/16.
Some H3K9 KMTs are also involved in cellular response mechanisms to hypoxia. For instance, transcriptional repression of the tumor suppressor RUNX3 under hypoxic conditions depends on the catalytic activity of G9A [269]. Upon hypoxic stress in the fetal lung, Suv39h1/2 localize to the promoter of the SP-A gene resulting in increased H3K9 di- and trimethylation counterbalancing its expression through hypoxic transcription factors [270]. Sirtuins are NAD+-dependent deacetylases that are sensors of oxidative stress. SIRT1 recruits SUV39H1 to chromatin and deacetylates the SET domain of SUV39H1 positively regulating its activity [271]. The activation of SUV39H1 can be reversed by DBC1, an inhibitor that can bind to the SET domain of SUV39H1 [272]. Additionally, SIRT1 controls the half-life of SUV39H1 by preventing its polyubiquitination through MDM2 on lysine 87. This stabilizes SUV39H1 and implies a major role for SUV39H1 in the control of oxidative and metabolic stress [273].
Transcriptional repression of cell cycle components
BCL1 1B (CTIP2) affects cell cycle progression by recruiting SUV39H1 to the cell cycle inhibitor gene P21[115]. Similarly, G9A has been reported to be required for repression of P21, which is also a known P53 target [116]. Therefore, not surprisingly SUV39H1 and GLP have been shown to regulate the P53 pathway. The E3 ubiquitin ligase MDM2 interacts with SUV39H1 and GLP to drive formation of P53/SUV39H1 and P53/GLP “complexes” that are able to repress P21 transcription during P53-mediated stress response [117]. Conversely, inhibition of P53 target genes via SUV39H1 and GLP can be relieved by binding of the P53 activator CDKN2A (ARF) to MDM2 [117]. Furthermore, GLP also methylates P53 directly on lysine 373 resulting in the inhibition of P53 activity (Table 1). This inhibition can be released by the P53 activator CDKN2A [117]. Evidence for silencing of another cell cycle inhibitor, P16, by SUV39H2 functionally connects the Polycomb machinery with H3K9 KMTs. Silencing of the P16 gene is thought to be initiated by the Polycomb homolog CBX7. CBX7-mediated recruitment of SUV39H2 and the subsequent trimethylation of H3K9 then results in further compaction of the P16 locus [118].
Histone H3K27 methyltransferases
Substrate specificity
Drosophila Enhancer of zeste (E(z)) is a member of the Polycomb group of proteins functioning in the negative regulation of genes that are crucial for the developmental patterning of organisms – the so called homeotic genes [119-121]. Later, it was shown that E(z) and its mammalian homologs, EZH1 and EZH2 (Figures 1-2), are the catalytically active components of a protein complex named Polycomb repressive complex 2 (PRC2), which is able to methylate H3K27 [119] (Figure 2). E(z) null mutant larvae in Drosophila lose all H3K27 mono-, di-, and trimethylation [122]. EZH2 is responsible for the bulk di- and trimethylation of H3K27 in mammals but has also been described to mono- and dimethylate linker histone H1 on lysine 26 [123] and to methylate non-histone targets (Table 1, Box 1). EZH1 is a weaker H3K27 di- and trimethyltransferase and is more highly expressed in nonproliferative cells [124]. Similar to Drosophila E(z), EZH1/2 function redundantly to implement all three H3K27 methylation states [125]. Both Drosophila E(z) and mouse Ezh2 are essential genes (Ezh1 knockout mice have not been reported to date) highlighting the importance of H3K27 KMTs in development [126, 127].
Protein complexes (Figure 3)
Like the COMPASS family of H3K4 KMTs E(z)/EZH1/EZH2 require the presence of other proteins to exert catalytic activity towards H3K27. The core subunits are comprised of E(z), Su(z), Esc/Escl, and Nurf55 in Drosophila and EZH1/2, SUZ12, EED, and RBBP4/7 in mammals [119]. EED exists in four isoforms of which the largest assembles in a particular PRC2 complex that is able to mono- or dimethylate histone H1 on lysine 26 resulting in transcriptional repression [123]. EED in the core PRC2 complex appears to be required for H3K27 di- and trimethylation spreading as binding of the EED/Esc C-terminus to trimethylated H3K27 is required for the allosteric activation of PRC2 [128, 129]. In contrast, H3K4 trimethylation and H3K36 di- and trimethylation allosterically inhibit PRC2 via the SUZ12/Su(z)12 C-terminus [130, 131]. SUZ12 is also capable of “sensing” its chromatin environment and binds to an amino acid stretch (aa 31-42) on H3 of neighboring nucleosomes. Optimal H3K27 methyltransferase activity is only achieved if the neighboring nucleosome is close enough to be contacted by SUZ12. Local chromatin compaction preceding H3K27 methylation was illustrated for the gene Cyp26a1 [132]. The recent publication of the molecular architecture of human PRC2 supports these roles of EED/Esc and SUZ12/Su(z)12, and localizes the points of interaction of the PRC2 core subunits with each other in more detail [133].
Modulation of PRC2 activity and recruitment
Other accessory factors have been described that either modulate PRC2 activity and/or are required for the context-specific recruitment of PRC2 to chromatin [119]. These include Jing, Pcl, and Jarid2 in Drosophila[134-136] and AEBP2, PCL1-3 (PHF1, MTF2, PHF19), and JARID2 in mammals [137-141].
Beside the histone modifications mentioned above, H3K27 acetylation is implemented by CBP/p300, and by nature of modifying the same residue, is mutually exclusive with H3K27 trimethylation on PRC2 target genes [142-144]. H3 serine 28 phosphorylation (H3S28) constitutes another opposing histone mark to H3K27 trimethylation. MSK1 and MSK2 have been identified as the responsible kinases that phosphorylate H3S28 resulting in a transitional H3K27 trimethylation-S28 phosphorylation double modification, which subsequently is resolved to H3K27 acetylation-S28 phosphorylation. H3K27 trimethylation-S28 phosphorylation is thought to displace PRC2 to recruit transcriptional activators resulting in derepression of PRC2 target genes [145, 146].
In Drosophila, specific sequences called Polycomb response elements (PREs) are sufficient to recruit PRC2 via its interaction with PRE-binding transcription factors, supporting evidence for this mechanism in the mammalian system is rather sparse [147-150]. However, in mammals, PRC2 interacts with many ncRNAs, some of which directly recruit PRC2 in cis or trans to establish H3K27 trimethylation and transcriptional silencing [151, 152]. For example, the ncRNA Xist directly recruits PRC2 to the female X-chromosome during X-chromosome inactivation [153, 154], and the ncRNA HOTAIR which is expressed from a region of the human HOXC locus silences a portion of the HOXD cluster in trans via recruitment of PRC2 [155, 156]. Interestingly, the phosphorylation of EZH2 on threonine 350 (threonine 345 in mice) by the cell cycle-regulated Cyclin-dependent kinase 1 (CDK1) increases its binding to HOTAIR [157]. Another study reports that phosphorylation of human EZH2 on threonine 350 results in the increased recruitment of PRC2 and the hypersilencing of many PRC2 target loci [158]. However, matters are further complicated by the finding that phosphorylation of EZH2 on threonine 492 (threonine 487 in mice) appears to destabilize the PRC2 complex and that the phosphorylation of mouse Ezh2 on threonine 345 and 487 promotes Ezh2 ubiquitylation and subsequent degradation via the proteasome [159, 160]. In summary, this implies that phosphorylation of EZH2 affects recruitment of PRC2 either through ncRNAs or other factors to regulate PRC2 target gene expression in a cell cycle-dependent manner, but that at the same time, mechanisms exist that allow for the timely degradation of phosphorylated EZH2 as cells transition from one phase of the cell cycle to the next. Other kinases such as AKT and mitogen-activated protein kinase P38 have also been reported to phosphorylate EZH2 on serine 21 and threonine 372, respectively [161, 162]. For example, phosphorylation of EZH2 on serine 21 by AKT negatively regulates EZH2 activity and p38 activation in satellite cells promotes the interaction of Ezh2 with the transcription factor Yy1 via phosphorylation on threonine 372 to repress Pax7 transcription [161, 162].
miRNAs provide yet another mechanism by which EZH2 protein levels are controlled. They bind to the UTRs of the EZH2 transcripts to limit the transcriptional output of EZH2. A negative feedback loop between PRC2 and miRNA loci ensures that this process remains tightly controlled [163-165]. As miRNAs are often expressed tissue specifically, they might provide one explanation of how EZH2 levels can be regulated in specific cell types.
Transcriptional regulation
H3K27 monomethylation tends to be associated with constitutive heterochromatin (along with H3K9 trimethylation) and inactive/poised enhancers, whereas H3K27 di- and trimethylation occur largely on facultative heterochromatin, but can also be found on intergenic and subtelomeric chromatin regions [119, 166]. H3K27 trimethylation is often enriched over large domains that can comprise up to 10 kb in Drosophila and 100 kb in mammals. Mammalian studies have shown that these domains increase in size upon differentiation resulting in even stronger transcriptional silencing [167]. However, trimethylated H3K27 also localizes more distinctly on transcriptionally inactive genes outside these domains [168]. This is in agreement with the finding that PRC2 has been reported to maintain gene repression of HOX genes and many other developmentally regulated genes [119]. Nonetheless, Pol II phosphorylated on serine 5 (a mark for transcription initiation) can be detected on a significant number of PRC2-targeted promoters. Additionally, a Drosophila study in esc mutant embryos reports augmented levels of Pol II on many PRC2 target genes suggesting a role for PRC2 in Pol II pausing [119, 169]. Indeed, some PRC2-repressed genes that are bivalently marked by H3K4 and H3K27 trimethylation in primary T and ES cells are characterized by Pol II recruitment and the transcription of 50-200 nucleotide long short RNAs. These short RNAs form stem-loop structures which interact with SUZ12 and are thought to result in gene repression in cis [170].
However, increasing evidence suggests that PRC2 can also function in active transcriptional processes. For example, H3K27 mono- and trimethylation can be enriched on actively transcribed genes as well [166, 171], and PRC2 (Suz12) is recruited to differentiation-activated genes in mouse ES cells [172]. Ezh1 overlaps with the H3K4 trimethyl mark on promoters of actively transcribed genes in mouse myoblasts [173]. Furthermore, Ezh1 depletion resulted in reduced Pol II occupancy and delayed transcriptional activation supporting a role for PRC2 in transcriptional elongation [173]. Drosophila studies support this idea as E(z) and Jarid2 loss have been described to result in repression of some PRC2-bound genes [134].
EZH2 can also activate genes independently of the PRC2 complex which potentially occurs under conditions where increased levels of EZH2 cannot be bound by other PRC2 core components such as in cancer cells. For example, EZH2 functions as a transactivator of the NF-κB pathway by interacting with RELA and RELB and has been shown to positively integrate the estrogen receptor α (ERα) and Wnt signaling pathways by binding to ERα and the Wnt pathway components TCF and β-Catenin. In both cases, this occurs independently of its methyltransferase activity [174, 175]. Furthermore, in a SET domain-dependent process EZH2 was found to be phosphorylated on lysine 21 by AKT leading to Androgen receptor (AR) binding and activation of AR target genes [176].
Histone H3K36 methyltransferases
Substrate specificity
In mammalian cells, H3K36 KMTs include SET2 (SETD2), NSD1, NSD2 (WHSC1, MMSET), NSD3 (WHSC1L), ASH1L [177] and SETMAR [178] (Figure 1). Set2/SET2 is conserved from yeast to mammals whereas NSD1-3 and ASH1L are homologous to Maternal-effect sterile 4 (Mes-4)/MES-4 and Ash1/LIN-59 in Drosophila and C. elegans [177, 179-182]. In yeast, Set2 is required for all of the mono-, di-, and trimethylation of H3K36 [183, 184]. C. elegans, Drosophila and mammalian Set2/SET2 constitute a major H3K36 trimethyltransferase [185-187] whereas NSD1-3 and their Drosophila ortholog Mes-4 preferentially mono- and dimethylate H3K36 [179, 188-191]. A reported loss of H3K36 trimethylation in the absence of NSD1 and Nsd2 could be explained by a requirement of mono- or dimethylated H3K36 for SET2/Set2 function [189, 192]. Among the family of SET domain-containing proteins, ASH1L is most closely related to SET2 and NSD1-3, and therefore, not surprisingly, also catalyzes H3K36 dimethylation [21, 23, 193]. Interestingly, both ASH1L and NSD1 contain an autoinhibitory loop between their SET and Post-SET domains, and at least in the case of NSD1 autoinhibition, can allosterically be relieved by interaction of the enzyme with nucleosomal DNA. Similarly, SETMAR (METNASE) regulates H3K36 dimethylation on histone octamers in vitro and DNA double-strand breaks in vivo [178, 194]. Non-histone substrates have been described for NSD1 and SETMAR (Table 1, Box 1) and different histone substrate specificities besides H3K36 have been reported for SET2, NSD1-3, and ASH1L. Other SET domain-containing proteins such as SMYD2 and SETD3 were also shown to regulate H3K36 methylation [177]. It appears that at least some of these findings are either the result of utilizing non-physiological substrates or can be attributed to indirect effects [177].
Transcriptional properties
Initial experiments showed that yeast Set2 is able to act as a transcriptional repressor of a lacZ reporter construct and the endogenous GAL4 gene [177, 195]. Set2 interacts with the hyperphosphorylated C-terminus of Pol II and establishes H3K36 trimethylation along gene bodies during transcriptional elongation [177]. At the same time, recruitment of the Rpd3S histone deacetylase complex via Set2 preserves a deacetylated state to prevent spurious intragenic transcription [177]. In yeast, H3K36 methylation-dependent recruitment of the Rpd3S complex is achieved by the combinatorial requirement for two Rpd3S complex-specific subunits –Eaf3 (MRG15 in mammals) and Rco1- and their respective chromo- and PHD-domains [196, 197]. Apart from its role in keeping the coding regions of genes in a hypoacetylated state, Set2 also suppresses chaperone-mediated histone exchange to prevent the incorporation of acetylated histones on actively transcribed genes [198].
Association of MES-4 in C. elegans early embryos of previously active maternal genes and persistence of Pol II after loss of MES-4 suggests a role for the NSD family in transcriptional repression [199, 200]. Likewise, human NSD1 localizes to the promoter region of the HOX gene MEIS1 and is required for its repression [201]. Nsd2 in conjunction with various transcription factors represses target genes such as the Nkx2-5 target Pdgfra[192]. However, NSD1-3 bind to the promoters of certain genes and appear to be required for transcription initiation [189, 202, 203]. They also control the H3K36 methylation state within the body and/or promoters of these genes. Furthermore, NSD3 forms a complex with the H3K4 demethylase LSD2 (AOF1) which is enriched genome-wide within the body of actively transcribed genes [113]. Similarly, NSD2 controls the H3K36 dimethylation pattern over the body of genes [191]. Thus, taken together, the NSD family seems to be involved in transcriptional repression, initiation, and elongation events.
Drosophila Ash1 and its mammalian homolog, ASH1L, are required for Hox gene expression [60, 204]. Based on the finding that ASH1L constitutes a H3K36 KMT and seems to associate with active genes, this suggests that it might be involved in the process of transcriptional elongation [60, 144]. Indeed, the bithoraxoid ncRNA in Drosophila recruits Ash1 in cis to induce transcription of Ubx, and a recent mammalian study implicated ASH1L in a similar event [60, 205]. Here, the ncRNA DBE-T drives derepression of a repeat element in muscular dystrophy patients by recruiting ASH1L. ASH1L in turn catalyzes the dimethylation of H3K36 at this locus resulting in transcriptional induction [205].
Histone H3K36 methylation and splicing
Lately, correlative evidence points towards a possible role for H3K36 methylation in splicing events. Several studies reported that nucleosomes and H3K36 trimethylation are generally more highly enriched on exons than introns [206-208]. However, confirmatory experimental evidence has largely been lagging behind. For example, SET2 has been implicated in controlling splice site switching. Exons, which are being excluded, are thought to be more highly enriched for H3K36 trimethylation, allowing recruitment of MRG15 via its chromodomain. MRG15 in turn is able to bind polypyrimidine tract-binding protein (PTB), which is known to antagonize exon inclusion, thus providing a possible explanation for exon definition in tissue-specific splicing [209]. On the other hand, the splicing mechanism itself is required for appropriate SET2 recruitment and proper H3K36 methylation suggesting a bidirectional reinforcement between SET2-dependent H3K36 methylation and the splicing machinery [210, 211].
Histone H4K20 methyltransferases
Substrate specificity
X-ray crystallographic studies of SETD8 (PR-SET7, SET8) in combination with nuclear magnetic resonance and in vitro assays on recombinant nucleosomes indicate that SETD8 is a major H4K20 monomethyltransferase [212, 213]. In mice Suv420h1 and Suv420h2 are redundantly required to implement bulk H4K20 di- and trimethylation [214, 215] (Figures 1-2). Implementation of H4K20 monomethylation by Setd8 is essential for proper H4K20 di- and trimethylation via Suv420h1/2 [216]. Therefore, not surprisingly, loss of Setd8 results in a more severe developmental phenotype than either loss of Suv420h1 or Suv420h2 alone or a combined loss of Suv420h1/2 function. Setd8 knockout leads to lethality before the embryonic eight-cell stage, and this developmental arrest depends on the catalytic activity of Setd8 [216]. Suv420h1 knockout and Suv420h1/2 double-knockout mice either die perinatally or develop normally as in the case of Suv420h2 knockouts [214]. In agreement with the mammalian findings, the Drosophila homologs of SETD8 and SUV420H1/2 –Pr-Set7 and Hmt4-20- are H4K20 mono-and H4K20 di- and trimethyltransferases, respectively [215, 217-219]. Usually, Drosophila pr-set7 null mutants show lethality during the late third instar larval stage [217] while Hmt4-20 null mutants are viable into adulthood [219]. Different non-histone substrates are also direct H4K20 KMT targets (Table 1, Box 1).
Transcriptional properties
The function of SETD8 in transcription is mechanistically not very well understood. Initially thought to be involved in transcriptional repression, accumulating evidence suggests that SETD8 is also required for the activation of gene expression. A role for Setd8 in transcriptional repression is supported by the finding that SETD8/Setd8 functions in chromatin compaction [216, 220, 221] and Pr-Set7 has been shown to be a suppressor of variegation in Drosophila[217]. Furthermore, some categories of repressed genes are enriched for H4K20 monomethylation and derepressed upon depletion of SETD8 [222]. For example, SETD8 controls gene repression through L3MBTL1 which is recruited to chromatin by H4K20 monomethylation [223, 224]; and another example is SET-1, the C. elegans ortholog of mammalian SETD8, which controls the repression of X-linked genes during dosage compensation [225]. SETD8 also regulates gene repression indirectly by monomethylating the tumor suppressor P53 on lysine 382 [226] (Table 1). L3MBTL1 can bind to monomethylated P53, and thus keep highly responsive P53 target genes repressed [227]. Repressive roles in transcription regulation for both SUV420H1 and SUV420H2 have been reported [228-230]. SUV420H1 is a suppressor of glucocorticoid receptor (GR)-mediated induction of gene expression via its interaction with GRIP1, a coregulator of the GR pathway [229], and is involved in repression of the fetal γ globin gene [230]. However, it has been proposed that the implementation of H4K20 trimethylation through SUV420H2 represses the promoter escape of Pol II by preventing recruitment of the histone acetyltransferase MOF [228]. Interestingly, SUV420H1 exists in at least two isoforms which significantly differ in their localization patterns. Isoform1 of SUV420H1 and SUV420H2 are confined to pericentric chromatin, while isoform 2 displays a more widespread distribution pattern [231]. Therefore, it is conceivable that isoform 2 could represent a more general regulator of gene repression than isoform 1. On the other hand, H4K20 monomethylation has been reported to be highly enriched over actively transcribed genes [168, 232]; and SETD8 is involved in the activation of Wnt target genes through its interaction with lymphoid enhancing factor-1 (LEF1)/TCF4 [233]. SETD8 is also required for the regulation of TWIST1 target genes during the epithelial-mesenchymal transition (EMT) [234]. However, in this context, a dual role for SETD8 in transcriptional repression and activation for different EMT genes has been proposed confirming the above-mentioned dichotomy in SETD8 function [234].
Regulation of H4K20 KMTs
SETD8 (and with it H4K20 monomethylation) is strictly controlled during the cell cycle with the highest amounts of SETD8 in G2/M and early G1, while there are undetectable levels in the S phase [216, 235, 236]. Various enzymes regulate Setd8 stability posttranslationally at different points of the cell cycle. Degradation of SETD8 by the E3 ubiquitin ligases SCF/SKP2 [237] and CRL4Cdt2[235, 236, 238] might provide a rationale for the low levels of SETD8 during the late G1 and S phase, although only CRL4Cdt2 was shown to directly ubiquitylate SETD8. Despite its high levels during mitosis, SETD8appears to be turned over by APCCdh1, another E3 ubiquitin ligase, after being “primed” by a phosphorylation/dephosphorylation switch mediated by CDK1/CCND1 and CDC14 [239].
Concluding Remarks
The involvement of SET domain-containing proteins in many diverse mechanisms such as transcriptional regulation, enhancer function, mRNA splicing, DNA replication, and the DNA damage response (Box 4) either by means of methylating histones or targeting non-histone substrates (Tables 1-3, Boxes 1-2) highlights the importance of this protein family in maintaining proper tissue homeostasis. Therefore not surprisingly, the misregulation of certain histone methylation marks and the misregulation and mutation of various SET domain-containing proteins are increasingly correlated with various forms of cancer and other diseases [240, 241]. For example, the important role of MLL1 translocations in acute myeloid and lymphoid leukemia depends on the methyltransferase activity of an intact MLL1 on the sister chromosome [242]. However, for most other SET domain-containing proteins, it is currently unknown whether the SET domain plays a disease-relevant role, and if yes, whether the pathological effects are mediated via the methylation of histones or non-histone substrates. Future research will need to address some of these questions in more detail.
Box 4. KMTs, DNA replication and the DNA damage response.
H3K4 KMTs
In yeast, H3K4 trimethylation marks meiotic recombination sites, and in Set1 mutants double strand break formation (DSB) formation is strongly impaired [274]. For efficient meiotic recombination to occur, in-loop sequences in promoters must be connected with DSB proteins on chromosome axes. Set1/COMPASS achieves this by means of its Cps40 subunit. Cps40 binds to promoters via a PHD finger domain and promotes DSB formation by tethering in-loop sequences to chromosome axes [275, 276]. Interestingly, DSB repair by non-homologous end joining in yeast also depends on Set1; and Set1 and H3K4 trimethylation increase upon DNA damage on newly created DSB sites, possibly implying a more general role for Set1 and H3K4 trimethylation in DSB-mediated processes [277].
H3K36 KMTs
Set2 is vital for the process of DNA replication as loss of Set2 causes delayed loading of Cdc45 at origins of replication as supported by studies in budding and fission yeast [278-280]. Furthermore, NSD2 coordinates the DSB response through a module that includes the γH2AX-MDC1-dependent recruitment of NSD2 to H4K20-methylated sites of DSBs, which is followed by 53BP1 binding [281, 282]. Whether the effects on H4K20 methylation during DSB repair are the direct result of NSD2 function is not clear. Based on what is known about the substrate specificity of H4K20 KMTs, we propose that either SETD8 and/or SUV420H1/2 may be affected by NSD2 in this context. Likewise, SETMAR controls DSB repair by nonhomologous end-joining by catalyzing H3K36 dimethylation on DSBs to enhance recruitment of early DSB factors such as NBS1 and KU70 [178, 194].
H4K20 KMTs
SETD8 promotes the assembly of pre-replication complex components during late M and early G1 phase to allow for proper replication origin licensing, which also depends on SUV420H1/2 and H4K20 trimethylation. Interaction of SETD8 with PCNA at replication origins through a conserved motif results in degradation of SETD8 providing an explanation for the low SETD8 levels from the late G1 to S phase [283-287] (Table 1). Furthermore, SETD8 and SUV420H1/2 also constitute major regulators of the DNA damage response by providing an H4K20 monomethyl/dimethyl platform for 53BP1, and at least in the case of SETD8, recruitment upon DNA damage is achieved by PCNA [214, 216, 218, 220, 235, 236, 283].
Highlights.
An overview on the substrates of SET domain-containing proteins is provided.
Protein complexes of SET domain-containing proteins are described.
Biological functions of different classes of SET domain-containing proteins are discussed.
Acknowledgments
We thank Drs. Edwin Smith and Marc Morgan for insightful discussions and for the critical reading of this manuscript, and Lisa Kennedy for editorial assistance. Studies in Shilatifard's laboratory regarding the subject of this review are supported in part by funding through National Institute of Health grants R01CA150265, R01CA89455, and R01GM069905 to AS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shilatifard A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annual review of biochemistry. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider J, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Molecular cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Mohan M, et al. The COMPASS family of H3K4 methylases in Drosophila. Molecular and cellular biology. 2011;31:4310–4318. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardehali MB, et al. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. The EMBO journal. 2011 doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallson G, et al. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190:91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Molecular and cellular biology. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreu-Vieyra CV, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Molecular and cellular biology. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herz HM, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes & development. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chauhan C, et al. Histone recognition and nuclear receptor co-activator functions of Drosophila Cara Mitad, a homolog of the N-terminal portion of mammalian MLL2 and MLL3. Development. 2012;139:1997–2008. doi: 10.1242/dev.076687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sedkov Y, et al. Molecular genetic analysis of the Drosophila trithorax-related gene which encodes a novel SET domain protein. Mechanisms of development. 1999;82:171–179. doi: 10.1016/s0925-4773(98)00246-9. [DOI] [PubMed] [Google Scholar]
- 12.Mazo AM, et al. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu BD, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 14.Glaser S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 15.Terranova R, et al. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics : official journal of the DNA Methylation Society. 2011;6:1059–1067. doi: 10.4161/epi.6.9.16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 18.Beisel C, et al. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- 19.Byrd KN, Shearn A. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11535–11540. doi: 10.1073/pnas.1933593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sebastian S, et al. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka Y, et al. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397:161–168. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Rincon-Arano H, et al. UpSET Recruits HDAC Complexes and Restricts Chromatin Accessibility and Acetylation at Promoter Regions. Cell. 2012;151:1214–1228. doi: 10.1016/j.cell.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An S, et al. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. The Journal of biological chemistry. 2011;286:8369–8374. doi: 10.1074/jbc.M110.203380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamamoto R, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nature cell biology. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 25.Foreman KW, et al. Structural and functional profiling of the human histone methyltransferase SMYD3. PloS one. 2011;6:e22290. doi: 10.1371/journal.pone.0022290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Aller GS, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics : official journal of the DNA Methylation Society. 2012;7:340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunizaki M, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer research. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi YH, et al. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20526–20531. doi: 10.1073/pnas.1109360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southall SM, et al. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Molecular cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 30.Patel A, et al. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. The Journal of biological chemistry. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 31.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. The Journal of biological chemistry. 2008;283:35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P, et al. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic acids research. 2012;40:4237–4246. doi: 10.1093/nar/gkr1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mersman DP, et al. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. The Journal of biological chemistry. 2012;287:2652–2665. doi: 10.1074/jbc.M111.280867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odho Z, et al. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. The Journal of biological chemistry. 2010;285:32967–32976. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avdic V, et al. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19:101–108. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 36.Cao F, et al. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PloS one. 2010;5:e14102. doi: 10.1371/journal.pone.0014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, et al. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO reports. 2011;12:797–803. doi: 10.1038/embor.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarvan S, et al. Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain. Nature structural & molecular biology. 2011;18:857–859. doi: 10.1038/nsmb.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JS, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 40.Vitaliano-Prunier A, et al. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nature cell biology. 2008;10:1365–1371. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- 41.Zheng S, et al. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Molecular and cellular biology. 2010;30:3635–3645. doi: 10.1128/MCB.00324-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soares LM, Buratowski S. Yeast Swd2 is essential because of antagonism between Set1 histone methyltransferase complex and APT (associated with Pta1) termination factor. The Journal of biological chemistry. 2012;287:15219–15231. doi: 10.1074/jbc.M112.341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirmizis A, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guccione E, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 45.Hyllus D, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes & development. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clouaire T, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes & development. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood A, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Molecular cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 49.Wood A, et al. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. The Journal of biological chemistry. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 50.Milne TA, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Molecular cell. 2010;38:853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muntean AG, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertani S, et al. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Molecular cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Vicent GP, et al. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes & development. 2011;25:845–862. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim DH, et al. ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyltransferase activity. Molecular endocrinology. 2009;23:1556–1562. doi: 10.1210/me.2009-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S, et al. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Molecular endocrinology. 2009;23:610–619. doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo C, et al. Global identification of MLL2-targeted loci reveals MLL2's role in diverse signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17603–17608. doi: 10.1073/pnas.1208807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnston DM, et al. Ecdysone- and NO-mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Molecular cell. 2011;44:51–61. doi: 10.1016/j.molcel.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee J, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuettengruber B, et al. Trithorax group proteins: switching genes on and keeping them active. Nature reviews Molecular cell biology. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 61.Liu H, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blobel GA, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Molecular cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 64.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herz HM, et al. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Molecular and cellular biology. 2010;30:2485–2497. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes & development. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Black JC, et al. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- 69.Fog CK, et al. PRDM proteins: important players in differentiation and disease. BioEssays : news and reviews in molecular. cellular and developmental biology. 2012;34:50–60. doi: 10.1002/bies.201100107. [DOI] [PubMed] [Google Scholar]
- 70.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Molecular cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 71.Garcia-Cao M, et al. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nature genetics. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 72.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes & development. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes & development. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ueda J, et al. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. The Journal of biological chemistry. 2006;281:20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- 75.Wang H, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Molecular cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 76.Falandry C, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. The Journal of biological chemistry. 2010;285:20234–20241. doi: 10.1074/jbc.M109.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fritsch L, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Molecular cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 78.Pinheiro I, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150:948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 79.Trojer P, et al. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. The Journal of biological chemistry. 2009;284:8395–8405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiss T, et al. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics & chromatin. 2010;3:7. doi: 10.1186/1756-8935-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu H, et al. Histone methyltransferase G9a contributes to H3K27 methylation in vivo. Cell research. 2011;21:365–367. doi: 10.1038/cr.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu Y, et al. Histone H3 lysine 56 methylation regulates DNA replication through its interaction with PCNA. Molecular cell. 2012;46:7–17. doi: 10.1016/j.molcel.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schotta G, et al. Central role of Drosophila SU (VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. The EMBO journal. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seum C, et al. Drosophila G9a is a nonessential gene. Genetics. 2007;177:1955–1957. doi: 10.1534/genetics.107.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seum C, et al. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS genetics. 2007;3:e76. doi: 10.1371/journal.pgen.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tzeng TY, et al. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12691–12696. doi: 10.1073/pnas.0705534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mis J, et al. Identification of three histone methyltransferases in Drosophila: dG9a is a suppressor of PEV and is required for gene silencing. Molecular genetics and genomics : MGG. 2006;275:513–526. doi: 10.1007/s00438-006-0116-x. [DOI] [PubMed] [Google Scholar]
- 88.Brower-Toland B, et al. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics. 2009;181:1303–1319. doi: 10.1534/genetics.108.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moore AW, et al. hamlet, a binary genetic switch between single- and multiple- dendrite neuron morphology. Science. 2002;297:1355–1358. doi: 10.1126/science.1072387. [DOI] [PubMed] [Google Scholar]
- 90.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 91.Dodge JE, et al. Histone H3-K9 methyltransferase ESET is essential for early development. Molecular and cellular biology. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoyt PR, et al. The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mechanisms of development. 1997;65:55–70. doi: 10.1016/s0925-4773(97)00057-9. [DOI] [PubMed] [Google Scholar]
- 93.Aguilo F, et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood. 2011;117:5057–5066. doi: 10.1182/blood-2010-08-300145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schultz DC, et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & development. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ayyanathan K, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes & development. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Molecular cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Molecular cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 98.Loyola A, et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO reports. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frietze S, et al. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PloS one. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ichimura T, et al. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. The Journal of biological chemistry. 2005;280:13928–13935. doi: 10.1074/jbc.M413654200. [DOI] [PubMed] [Google Scholar]
- 101.Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS genetics. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spensberger D, Delwel R. A novel interaction between the proto-oncogene Evi1 and histone methyltransferases, SUV39H1 and G9a. FEBS letters. 2008;582:2761–2767. doi: 10.1016/j.febslet.2008.06.056. [DOI] [PubMed] [Google Scholar]
- 103.Cattaneo F, Nucifora G. EVI1 recruits the histone methyltransferase SUV39H1 for transcription repression. Journal of cellular biochemistry. 2008;105:344–352. doi: 10.1002/jcb.21869. [DOI] [PubMed] [Google Scholar]
- 104.Goyama S, et al. EVI-1 interacts with histone methyltransferases SUV39H1 and G9a for transcriptional repression and bone marrow immortalization. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U K. 2010;24:81–88. doi: 10.1038/leu.2009.202. [DOI] [PubMed] [Google Scholar]
- 105.Towbin BD, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 106.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 107.Wen B, et al. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nature genetics. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yokochi T, et al. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sampath SC, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Molecular cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 110.Chin HG, et al. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic acids research. 2007;35:7313–7323. doi: 10.1093/nar/gkm726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Collins RE, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono-and dimethyllysine binding modules. Nature structural & molecular biology. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yuan X, et al. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Molecular cell. 2007;27:585–595. doi: 10.1016/j.molcel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 113.Fang R, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Molecular cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Purcell DJ, et al. A distinct mechanism for coactivator versus corepressor function by histone methyltransferase G9a in transcriptional regulation. The Journal of biological chemistry. 2011;286:41963–41971. doi: 10.1074/jbc.M111.298463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cherrier T, et al. p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009;28:3380–3389. doi: 10.1038/onc.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim JK, et al. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic acids research. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen L, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. The EMBO journal. 2010;29:2538–2552. doi: 10.1038/emboj.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li Q, et al. Polycomb CBX7 directly controls trimethylation of histone H3 at lysine 9 at the p16 locus. PLoS genetics. 2010;5:e13732. doi: 10.1371/journal.pone.0013732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Simon J, et al. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development. 1992;114:493–505. doi: 10.1242/dev.114.2.493. [DOI] [PubMed] [Google Scholar]
- 121.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews Molecular cell biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 122.Ebert A, et al. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes & development. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kuzmichev A, et al. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Molecular cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 124.Margueron R, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Molecular cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O'Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and cellular biology. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Molecular cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 131.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yuan W, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 133.Ciferri C, et al. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Herz HM, et al. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Molecular and cellular biology. 2012;32:1683–1693. doi: 10.1128/MCB.06503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Savla U, et al. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- 136.Nekrasov M, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. The EMBO journal. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Herz HM, Shilatifard A. The JARID2-PRC2 duality. Genes & development. 2010;24:857–861. doi: 10.1101/gad.1921610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walker E, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell stem cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim H, et al. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic acids research. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sarma K, et al. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Molecular and cellular biology. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ballare C, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nature structural & molecular biology. 2012;19:1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pasini D, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic acids research. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schwartz YB, et al. Alternative epigenetic chromatin states of polycomb target genes. PLoS genetics. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gehani SS, et al. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Molecular cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 146.Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sing A, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 148.Woo CJ, et al. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Current opinion in genetics & development. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 150.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 151.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maenner S, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS biology. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaneko S, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes & development. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen S, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nature cell biology. 2010;12:1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wei Y, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nature cell biology. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. The Journal of biological chemistry. 2011;286:28511–28519. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Palacios D, et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell stem cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cha TL, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 163.Juan AH, et al. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Molecular cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kottakis F, et al. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Molecular cell. 2011;43:285–298. doi: 10.1016/j.molcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tzatsos A, et al. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. The Journal of biological chemistry. 2011;286:33061–33069. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell stem cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell stem cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 169.Chopra VS, et al. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Molecular cell. 2011;42:837–844. doi: 10.1016/j.molcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Molecular cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Young MD, et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic acids research. 2011 doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pasini D, et al. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Molecular and cellular biology. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mousavi K, et al. Polycomb Protein Ezh1 Promotes RNA Polymerase II Elongation. Molecular cell. 2012;45:255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 174.Lee ST, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Molecular cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 175.Shi B, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Molecular and cellular biology. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nature reviews Molecular cell biology. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee SH, et al. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bell O, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. The EMBO journal. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bender LB, et al. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chamberlin HM, Thomas JH. The bromodomain protein LIN-49 and trithorax-related protein LIN-59 affect development and gene expression in Caenorhabditis elegans. Development. 2000;127:713–723. doi: 10.1242/dev.127.4.713. [DOI] [PubMed] [Google Scholar]
- 182.Nakamura T, et al. huASH1 protein, a putative transcription factor encoded by a human homologue of the Drosophila ash1 gene, localizes to both nuclei and cell-cell tight junctions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7284–7289. doi: 10.1073/pnas.97.13.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Krogan NJ, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Molecular and cellular biology. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Strahl BD, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Molecular and cellular biology. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Edmunds JW, et al. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. The EMBO journal. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Larschan E, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Molecular cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 187.Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- 188.Li Y, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. The Journal of biological chemistry. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lucio-Eterovic AK, et al. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16952–1697. doi: 10.1073/pnas.1002653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Qiao Q, et al. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. The Journal of biological chemistry. 2011;286:8361–8368. doi: 10.1074/jbc.M110.204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kuo AJ, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Molecular cell. 2011;44:609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nimura K, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 193.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. The Journal of biological chemistry. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fnu S, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Landry J, et al. Set2-catalyzed methylation of histone H3 represses basal expression of GAL4 in Saccharomyces cerevisiae. Molecular and cellular biology. 2003;23:5972–5978. doi: 10.1128/MCB.23.17.5972-5978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li B, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 197.Li B, et al. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. The Journal of biological chemistry. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Venkatesh S, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 199.Furuhashi H, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics & chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rechtsteiner A, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Berdasco M, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Molecular and cellular biology. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yang P, et al. Histone methyltransferase NSD2/MMSET mediates constitutive NF-kappaB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Molecular and cellular biology. 2012;32:3121–3131. doi: 10.1128/MCB.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tanaka Y, et al. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression. PloS one. 2011;6:e28171. doi: 10.1371/journal.pone.0028171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature genetics. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Spies N, et al. Biased chromatin signatures around polyadenylation sites and exons. Molecular cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Schwartz S, et al. Chromatin organization marks exon-intron structure. 16. 2009:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 209.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.de Almeida SF, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nature structural & molecular biology. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 211.Kim S, et al. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xiao B, et al. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes & development. 2005;19:1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Couture JF, et al. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes & development. 2005;19:1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & development. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schotta G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & development. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Oda H, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Molecular and cellular biology. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Karachentsev D, et al. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes & development. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yang H, et al. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. The Journal of biological chemistry. 2008;283:12085–12092. doi: 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Sakaguchi A, et al. Functional characterization of the Drosophila Hmt4-20/Suv4-20 histone methyltransferase. Genetics. 2008;179:317–322. doi: 10.1534/genetics.108.087650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sakaguchi A, Steward R. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. The Journal of cell biology. 2007;176:155–162. doi: 10.1083/jcb.200607178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Houston SI, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. The Journal of biological chemistry. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Congdon LM, et al. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. Journal of cellular biochemistry. 2010;110:609–619. doi: 10.1002/jcb.22570. [DOI] [PubMed] [Google Scholar]
- 223.Kalakonda N, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sims JK, Rice JC. PR-Set7 establishes a repressive trans-tail histone code that regulates differentiation. Molecular and cellular biology. 2008;28:4459–4468. doi: 10.1128/MCB.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vielle A, et al. H4K20me1 Contributes to Downregulation of X-Linked Genes for C. elegans Dosage Compensation. PLoS genetics. 2012;8:e1002933. doi: 10.1371/journal.pgen.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Shi X, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Molecular cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.West LE, et al. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. The Journal of biological chemistry. 2010;285:37725–37732. doi: 10.1074/jbc.M110.139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kapoor-Vazirani P, et al. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Molecular and cellular biology. 2011;31:1594–1609. doi: 10.1128/MCB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Chinenov Y, et al. GRIP1-associated SET-domain methyltransferase in glucocorticoid receptor target gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20185–20190. doi: 10.1073/pnas.0810863105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rank G, et al. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood. 2010;116:1585–1592. doi: 10.1182/blood-2009-10-251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tsang LW, et al. Comparative analyses of SUV420H1 isoforms and SUV420H2 reveal differences in their cellular localization and effects on myogenic differentiation. PLoS genetics. 2010;5:e14447. doi: 10.1371/journal.pone.0014447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Li Z, et al. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3116–3123. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yang F, et al. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. The EMBO journal. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Oda H, et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Molecular cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Abbas T, et al. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Molecular cell. 2010;40:9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yin Y, et al. SET8 plays a role in controlling G1/S transition by blocking lysine acetylation in histone through binding to H4 N-terminal tail. Cell cycle. 2008;7:1423–1432. doi: 10.4161/cc.7.10.5867. [DOI] [PubMed] [Google Scholar]
- 238.Centore RC, et al. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Molecular cell. 2010;40:22–33. doi: 10.1016/j.molcel.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wu S, et al. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes & development. 2010;24:2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Albert M, Helin K. Histone methyltransferases in cancer. Seminars in cell & developmental biology. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 241.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature reviews Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Thiel AT, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang K, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Latham JA, et al. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146:709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yang L, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lee JS, et al. Negative regulation of hypoxic responses via induced Reptin methylation. Molecular cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Huang J, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. The Journal of biological chemistry. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rathert P, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nature chemical biology. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pless O, et al. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-beta. The Journal of biological chemistry. 2008;283:26357–26363. doi: 10.1074/jbc.M802132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Van Duyne R, et al. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology. 2008;5:40. doi: 10.1186/1742-4690-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.He A, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes & development. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lee JM, et al. EZH2 Generates a Methyl Degron that Is Recognized by the DCAF1/DDB1/CUL4 E3 Ubiquitin Ligase Complex. Molecular cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 253.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Su IH, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 255.Lu T, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Williamson EA, et al. The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic acids research. 2008;36:5822–5831. doi: 10.1093/nar/gkn560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Wray J, et al. Metnase mediates chromosome decatenation in acute leukemia cells. Blood. 2009;114:1852–1858. doi: 10.1182/blood-2008-08-175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Dhami GK, et al. Dynamic Methylation of Numb by Set8 Regulates Its Binding to p53 and Apoptosis. Molecular cell. 2013;50:565–576. doi: 10.1016/j.molcel.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 259.Takawa M, et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer research. 2012;72:3217–3227. doi: 10.1158/0008-5472.CAN-11-3701. [DOI] [PubMed] [Google Scholar]
- 260.Kramer JM, et al. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS biology. 2011;9:e1000569. doi: 10.1371/journal.pbio.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Balemans MC, et al. Reduced exploration, increased anxiety, and altered social behavior: Autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behavioural brain research. 2010;208:47–55. doi: 10.1016/j.bbr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 263.Covington HE, 3rd, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Molecular cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Mulligan P, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Molecular cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Endo K, et al. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nature neuroscience. 2012;15:224–233. doi: 10.1038/nn.2998. [DOI] [PubMed] [Google Scholar]
- 268.Kinameri E, et al. Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PloS one. 2008;3:e3859. doi: 10.1371/journal.pone.0003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lee SH, et al. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene. 2009;28:184–194. doi: 10.1038/onc.2008.377. [DOI] [PubMed] [Google Scholar]
- 270.Benlhabib H, Mendelson CR. Epigenetic regulation of surfactant protein A gene (SP-A) expression in fetal lung reveals a critical role for Suv39h methyltransferases during development and hypoxia. Molecular and cellular biology. 2011;31:1949–1958. doi: 10.1128/MCB.01063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 272.Li Z, et al. Inhibition of SUV39H1 methyltransferase activity by DBC1. The Journal of biological chemistry. 2009;284:10361–10366. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Bosch-Presegue L, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Molecular cell. 2011;42:210–223. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 274.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. The EMBO journal. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sommermeyer V, et al. Spp1, a Member of the Set1 Complex, Promotes Meiotic DSB Formation in Promoters by Tethering Histone H3K4 Methylation Sites to Chromosome Axes. Molecular cell. 2012 doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 276.Acquaviva L, et al. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339:215–218. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- 277.Faucher D, Wellinger RJ. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Biswas D, et al. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics. 2008;178:649–659. doi: 10.1534/genetics.107.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Kim HS, et al. Methylations of histone H3 lysine 9 and lysine 36 are functionally linked to DNA replication checkpoint control in fission yeast. Biochemical and biophysical research communications. 2008;368:419–425. doi: 10.1016/j.bbrc.2008.01.104. [DOI] [PubMed] [Google Scholar]
- 280.Pryde F, et al. H3 k36 methylation helps determine the timing of cdc45 association with replication origins. PloS one. 2009;4:e5882. doi: 10.1371/journal.pone.0005882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hajdu I, et al. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Pei H, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Jorgensen S, et al. The histone methyltransferase SET8 is required for S-phase progression. The Journal of cell biology. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Tardat M, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nature cell biology. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 285.Huen MS, et al. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. The Journal of biological chemistry. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Jorgensen S, et al. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. The Journal of cell biology. 2011;192:43–54. doi: 10.1083/jcb.201009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Beck DB, et al. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes & development. 2012 doi: 10.1101/gad.195636.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Chang Y, et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nature communications. 2011;2:533. doi: 10.1038/ncomms1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ko S, et al. Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Molecular endocrinology. 2011;25:433–444. doi: 10.1210/me.2010-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Gaughan L, et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic acids research. 2011;39:1266–1279. doi: 10.1093/nar/gkq861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nature genetics. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 292.Esteve PO, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Esteve PO, et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nature structural & molecular biology. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Molecular cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 295.Subramanian K, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Molecular cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Calnan DR, et al. Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging. 2012;4:462–479. doi: 10.18632/aging.100471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Balasubramaniyan N, et al. Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. American journal of physiology Gastrointestinal and liver physiology. 2012;302:G937–947. doi: 10.1152/ajpgi.00441.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 299.Ivanov GS, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Molecular and cellular biology. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kurash JK, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Molecular cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 301.Campaner S, et al. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Molecular cell. 2011;43:681–688. doi: 10.1016/j.molcel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 302.Lehnertz B, et al. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Molecular cell. 2011;43:673–680. doi: 10.1016/j.molcel.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 303.Masatsugu T, Yamamoto K. Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochemical and biophysical research communications. 2009;381:22–26. doi: 10.1016/j.bbrc.2009.01.185. [DOI] [PubMed] [Google Scholar]
- 304.Carr SM, et al. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. The EMBO journal. 2011;30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Munro S, et al. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene. 2010;29:2357–2367. doi: 10.1038/onc.2009.511. [DOI] [PubMed] [Google Scholar]
- 306.Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Li Y, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. The Journal of biological chemistry. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Yang XD, et al. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. The EMBO journal. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Yang XD, et al. Functional interplay between acetylation and methylation of the RelA subunit of NF-kappaB. Molecular and cellular biology. 2010;30:2170–2180. doi: 10.1128/MCB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Liu X, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Yang J, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kouskouti A, et al. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Molecular cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 313.Pagans S, et al. The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell host & microbe. 2010;7:234–244. doi: 10.1016/j.chom.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Oudhoff MJ, et al. Control of the hippo pathway by set7-dependent methylation of yap. Developmental cell. 2013;26:188–194. doi: 10.1016/j.devcel.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 315.Houtz RL, et al. Post-translational modifications in the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:1855–1859. doi: 10.1073/pnas.86.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ying Z, et al. Organization and characterization of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit epsilon N-methyltransferase gene in tobacco. Plant molecular biology. 1996;32:663–671. doi: 10.1007/BF00020207. [DOI] [PubMed] [Google Scholar]
- 317.Porras-Yakushi TR, et al. A novel SET domain methyltransferase modifies ribosomal protein Rpl23ab in yeast. The Journal of biological chemistry. 2005;280:34590–34598. doi: 10.1074/jbc.M507672200. [DOI] [PubMed] [Google Scholar]
- 318.Porras-Yakushi TR, et al. Yeast ribosomal/cytochrome c SET domain methyltransferase subfamily: identification of Rpl23ab methylation sites and recognition motifs. The Journal of biological chemistry. 2007;282:12368–12376. doi: 10.1074/jbc.M611896200. [DOI] [PubMed] [Google Scholar]
- 319.Porras-Yakushi TR, et al. A novel SET domain methyltransferase in yeast: Rkm2-dependent trimethylation of ribosomal protein L12ab at lysine 10. The Journal of biological chemistry. 2006;281:35835–35845. doi: 10.1074/jbc.M606578200. [DOI] [PubMed] [Google Scholar]
- 320.Webb KJ, et al. Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab. The Journal of biological chemistry. 2008;283:35561–35568. doi: 10.1074/jbc.M806006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Chang Y, et al. Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling. Nucleic acids research. 2011;39:6380–6389. doi: 10.1093/nar/gkr256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Levy D, et al. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nature immunology. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Xu S, et al. Structure of human lysine methyltransferase Smyd2 reveals insights into the substrate divergence in Smyd proteins. Journal of molecular cell biology. 2011;3:293–300. doi: 10.1093/jmcb/mjr015. [DOI] [PubMed] [Google Scholar]
- 324.Ferguson AD, et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19:1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 325.Wang L, et al. Structure of human SMYD2 protein reveals the basis of p53 tumor suppressor methylation. The Journal of biological chemistry. 2011;286:38725–38737. doi: 10.1074/jbc.M111.262410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 327.Abu-Farha M, et al. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. Journal of molecular cell biology. 2011;3:301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- 328.Donlin LT, et al. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes & development. 2012;26:114–119. doi: 10.1101/gad.177758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Cho HS, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14:476–486. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Saddic LA, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. The Journal of biological chemistry. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]