Abstract
A taxonomic review of the North American band-winged grasshopper genus Encoptolophus Scudder (Orthoptera: Acrididae: Oedipodinae) was conducted. This genus is hypothesized to be non-monophyletic following a cladistic analysis of the genera in the Chortophaga genus group. We examined all species currently classified in this genus group for morphological characters and one behavioral character. The phenotypic character data were combined with three mitochondrial genes: cytochrome c oxidase subunit II, 16S rRNA and 12S rRNA. A parsimony analysis was performed on the combined data resulting in two equally parsimonious trees. Encoptolophus, as historically defined, is resolved in three separate clades. The results support erection of a new genus, Nebulatettix Gómez, Lightfoot & Miller gen.n. to comprise one of the groups historically classified in Encoptolophus. In addition, we transfer the species Encoptolophus californicus Bruner to Chimarocephala Scudder, comb.n., a combination used historically. The evolution of certain characters in the Chortophaga group is discussed, and a key to the genera is provided.
Keywords: Systematics, taxonomy, character evolution, morphology, Chimarocephala, Chortophaga, Shotwellia
Introduction
Band-winged grasshoppers (Acrididae: Oedipodinae) occur worldwide, particularly in temperate zones and most commonly in semi-arid regions. They are the most widely distributed Acrididae, and the diversity in North America is high with 181 described species (Eades et al. 2011). Oedipodinae are often conspicuous in their mating displays, which include crepitation (sounds produced in flight), stridulation, “femur-shaking”, and flight displays with variously patterned wings. Crepitation patterns vary between genera and species as species-specific mating signals used in potential mate identification, which can contribute to reproductive isolation (Otte 1970; Weissman & Rentz 1980). Oedipodinae, much like other subfamilies of Acrididae with more complex sexual behavior such as Gomphocerinae and Acridinae, are challenging taxonomically. Recent molecular analyses of many Oedipodinae genera worldwide has shown several genus groups and tribes to be non-monophyletic, suggesting that the characters used to define this rank may be largely the result of convergence (Fries et al. 2007; Chapco & Contreras 2011).
The genus Encoptolophus Scudder is currently placed within the Chortophaga genus group (Otte 1984), one of seven North American genus groups within Oedipodinae (Otte 1984). The Chortophaga group includes three additional genera: Chortophaga Saussure, Shotwellia Gurney and Chimarocephala Scudder. Otte (1984) presented the first hypothetical phylogenetic tree of all members of the group (Fig. 1). The group has members throughout much of North America and is considered sister to the rest of the Oedipodinae (Fries et al. 2007; Chapco & Contreras 2011). Chapco & Contreras (2011) hypothesized that the Chortophaga group, along with the monotypic genera Machaerocera Saussure and Melanotettix Bruner (currently classified within the Oedipodineae and Gomphocerinae, respectively), evolved in North America about 90 million years ago when Laurasia was intact, while all other North American Oedipodinae lineages originated in Africa and subsequently radiated throughout North America beginning approximately 50 million years ago. Fries et al. (2007) and Chapco & Contreras (2011) hypothesized the monophyly of the group based on molecular analysis of four mitochondrial genes from two species of the 15 currently in the Chortophaga group. These studies, along with an additional one by Chapco et al. (1997) represent the few molecular phylogenies of North American Oedipodinae. DNA analyses have resolved challenging problems, like placement of Machaerocera, an oedipodine, and Melanotettix, a questionably classified gomphocerine, sister to the Chortophaga group (Fries et al. 2007; Chapco & Contreras 2011), when morphological data has been lacking or difficult to homologize.
Fig. 1.

Tree adapted from Otte (1984) depicting phylogenetic hypotheses for the Chortophaga group. Mapped characters are not reproduced here.
Another recent molecular study of this group conducted by Edelman et al. (2010) supported the phylogenetic position of Shotwellia isleta Gurney as sister to the remaining Chortophaga group. That investigation also revealed that Encoptolophus, as currently defined, is seemingly not monophyletic with some species more closely related to Chimarocephala (Edelman et al. 2010), but that study included only three species, did not include the type of the genus, and no changes to the classification were made.
Each of these previous analyses did not have sufficient taxon sampling to address a more comprehensive examination of the naturalness of the classification. The goal of this research is to more thoroughly test the monophyly of Encoptolophus and phylogenetic relationships among the Chortophaga group genera using combined morphological and DNA sequence data and to revise the classification as needed.
Materials and Methods
Taxon sampling
Twenty-four species (including 11 non-Chortophaga group outgroup species) were included in the analysis with specimens of 22 of these successfully DNA sequenced (Table 1). We were unable to procure DNA from E. californicus Bruner, despite several failed attempts to collect fresh specimens and to sequence old dried museum specimens. We were also unable to include sequence data for Chortophaga cubensis (Scudder). Only morphological data were included for these two species (Tables 2 and 3). Specimens were collected by DCL, RAG, D.B. Weissman (Department of Entomology, California Academy of Sciences) and A. Joern (Department of Ecology, University of Kansas), and identifications were verified by D.C.L. and R.A.G. Voucher specimens and additional DNA tissue samples are deposited in the Division of Arthropods, Museum of Southwestern Biology at the University of New Mexico (MSBA, K.B.M.). Additional specimens were borrowed from the Essig Museum of Entomology at UC Berkeley (R. Gillespie), the California Academy of Sciences (D.H. Kavanaugh), the Cornell University Insect Collection (J.K. Leibherr), the Albert J. Cook Arthropod Research Collection at Michigan State University (A. Cognato), the Academy of Natural Sciences of Philadelphia (D. Otte), and the Museum of Entomology of the Florida State Collection of Arthropods (M.C. Thomas). The lectotype of E. californicus (Figs 8, 15, 21 and 22) and lectotype of E. robustus Rehn & Hebard (Figs 17 and 19) were also examined and borrowed from the Academy of Natural Sciences of Philadelphia.
Table 1.
Exemplars and locality information for the respective taxa used for phylogenetic analysis.
| Genus group or tribe | Genus and species | Locality | GenBank accession number (12S,16S,COII) | UNM MSBA voucher number (MSBA, KBM lab) |
|---|---|---|---|---|
| Chortophaga | Chimarocephala elongata | USA. California, San Benito Co., 3.2 km N New Idria, 36°32′13.2″ N 120°50′0.6″ W, 9 Jun 2011, D.C. Lightfoot, coll. | MSBA-22887 KBMC-OR86 |
|
| Chortophaga | Chimarocephala pacifica | USA. California, San Mateo Co., Golden Gate National Recreation Area, Milagra Ridge, S05-44, 37°22′52.1″ N 122°17′4.3″ W, 31 Mar 2005, D.B. Weissman, coll. | MSBA-22890, KBMC-OR89 | |
| Chortophaga | Chortophaga australior | USA. Florida, Alachua Co., Archer Road, 5 May 1997, J.M. Squitier, coll. | MSBA-22894 KBMC-OR68 |
|
| Chortophaga | Chortophaga mendocino | USA. California, Sonoma Co., Salt Point State Park, along U.S. highway 2.2 km W Plantation at mile marker 42.1, 38°35′50.8″ N 123°20′26.8″ W, 22 April 2011, D.B. Weissman, coll. | MSBA-22895 KBMC-OR80 |
|
| Chortophaga | Chortophaga virdifasciata | USA. New Mexico, Socorro Co., nr. Bernardo, Hwy. 60, 34°25′01″ N 106°47′49″ W, 11 May 2005, D.C. Lightfoot, coll. | GU476952, GU476966, GU476988 | MSBA-22899 KBMC-OR43 |
| Chortophaga | Encoptolophus costalis | USA. New Mexico, Torrance Co., 12 km E Estancia, 34°46′7.0″ N 105°55′52.2″ W, 29 August 2009, D.C. Lightfoot, coll. | MSBA-22896 KBMC-OR65 |
|
| Chortophaga | Encoptolophus fuliginosus | MEXICO. Michoacan, km 291 on Hwy. 15, 19°55′56″ N 101°36′43″ W, 01 Jun 2008, D.C. Lightfoot, D.B. Weissman, coll. | GU476950, GU476965, GU476985 | MSBA-22904 KBMC-OR48 |
| Chortophaga | Encoptolophus otomitus | MEXICO. Puebla, nr. Oriental, Hwy. 129, 20°43′29″ N 49°22′03″ W, 22 Jun 2006, D.C. Lightfoot, D.B. Weissman, coll. | GU476932, GU476956, GU476977 | MSBA-22895 KBMC-OR39 |
| Chortophaga | Encoptolophus pallidus | USA. Nevada, Nye Co., Ash Meadows National Wildlife Refuge, 2.2 km N State Line Rd., along road to Visitor Center, 36°22′0.1″ N 116°18′6.4″ W, 5 June 2011, D.C. Lightfoot, coll. | MSBA-24689 KBMC-OR85 |
|
| Chortophaga | Encoptolophus robustus | MEXICO. Baja California Sur, 17.7 km N fork of Highways 1 and 22, near Ciudad Constitución, 25°5′53.9″ N 111°42′16.4″ W, 31 Jan 2011. R.A. Gómez, D.C. Lightfoot, D.B. Weissman, coll. | MSBA-22899 KBMC-OR79 |
|
| Chortophaga | Encoptolophus sordidus | USA. Kansas, Riley Co., Konza Prairie Biological Station, Watershed N4A, CGRO sample, 07 Aug 2007, sweep, A. Joern, coll. | MSBA-24898 KBMC-OR90 |
|
| Chortophaga | Encoptolophus subgracilis | MEXICO. San Luis Potosi, 6 km N. Palmira, 21°42′25″ N 98°56′12″ W, 24 Jun 2006, D.C. Lightfoot, D.B. Weissman, coll. | GU476949, GU476955, GU476990 | MSBA-22894 KBMC-OR38 |
| Arphia | Lactista azteca | USA. Texas, Brewster Co., nr. Alpine, 30°20′44″ N 103°41′50″ W, 14 Oct 2004, D.C. Lightfoot, coll. | GU476944, GU476959, GU476987 | MSBA-22890 KBMC-OR34 |
| Arphia | Lactista elota | MEXICO. Jalisco, 43 km W, Guadalajara, km 151 on Hwy. 15, 20°53′19″ N 103°56′12″ W, 31 May 2008, D.C. Lightfoot, D.B. Weissman, coll. | GU476935, GU476953, GU476982 | MSBA-22906 KBMC-OR50 |
| Arphia | Lactista gibbosus (osalare) | USA. California, Los Angeles Co., Topanga Canyon Blvd, 34°00′50″N 118°36′04″ W, 23 Aug 2006, D.C. Lightfoot, D.B. Weissman, coll. | GU476946, 16S*, GU476994 | MSBA-22897 KBMC-OR41 |
| Arphia | Lactista punctatus | MEXICO. Veracruz, 12 km SE Tuzamapan, 19°21′50″ N 96°49′24″ W, 21 Jun 2006, D.C. Lightfoot, D.B. Weissman, coll. | GU476934, GU476963, GU476996 | MSBA-22896 KBMC-OR40 |
| Machaerocera | Machaerocera mexicana | MEXICO. Veracruz, Cofre de Perote, North side, 6 km S Highway 140, 19°36′33″ N 97°7.2′35″ W, June 2006, D.C. Lightfoot, D.B. Weissman, coll. | MSBA-24690 KBMC-OR72 |
|
| Eritettix | Opeia obscura | USA. New Mexico, Socorro Co., Sevilleta National Wildlife Refuge, 15 km SE Bernardo, 34°19′58.8″ N 106°43′15.6″ W, 2 September 2005, D.C. Lightfoot, coll. | MSBA-24691 KBMC-OR83 |
|
| Scyllinini | Rhammatocerus viatorialus | MEXICO. Oaxaca, 3 km S Gaelepo along Highway 175, 17°18′23″ N 96°30′30″ W, 19 Jun 2006, D.C. Lightfoot, D.B. Weissman, coll. | MSBA-24692 KBMC-OR82 |
|
| Chortophaga | Shotwellia isleta | USA. New Mexico, Dona Ana Co., Lake Issac, nr. Las Cruces, 32°27′ 38″ N 106°43′ 12″ W, 04 Oct 2004, D.C. Lightfoot, coll. | GU476931, GU476968, GU476976 | MSBA-22887 KBMC-OR31 |
| Amblytropidiini | Syrbula montezuma | USA. New Mexico, Socorro Co., Sevilleta National Wildlife Refuge, 15 km SE Bernardo 34°19′58.8″ N 106°43′15.6″ W, 28 Sep 2004, D.C. Lightfoot, coll. | MSBA-24693 KBMC-OR76 |
|
| Parapleurini | Stethophyma lineatum | USA. New Mexico, Sandoval Co., Valles Caldera National Preserve, Alamo Bog-wetland area, 5 Aug 2009, R.R. Parmenter, coll. | MSBA-24899 KBMC-OR73 |
|
| Sphingonotini | Trimerotropis pallidipennis | USA. Arizona, Chiricahua Mts., Cave Creek Canyon, 31°52′43″ N 109°13′21″ W, 21 Jun 2008, W.C. Edelman, D.C. Lightfoot, coll. | GU476938, GU476964, GU476998 | MSBA-22908 KBMC-OR52 |
| Hippiscus | Xanthippus corallipes | USA. New, Mexico, Socorro Co., Sevilleta National Wildlife Refuge, 34°20′02″ N 106°37′53″ W, 04 Jun 2005, D.C. Lightfoot, coll. | GU476940, GU476957, GU476984 | MSBA-22902 KBMC-OR46 |
GenBank accession numbers and UNM MSBA voucher information are also included. Genes that failed to amplify are noted with an asterisk for the specified taxon and gene(s).
Table 2.
Characters states per species as coded in the matrix.
| Species | 0000000001 | 1111111112 | 2222222223 | 3333333334 |
|---|---|---|---|---|
| 1234567890 | 1234567890 | 1234567890 | 1234567890 | |
| Chimarocephala elongata | 0004301100 | 1???001110 | 000?010011 | 000010110? |
| Chimarocephala pacifica | 0004301100 | 11010*1200 | **00010011 | 000010110? |
| Chortophaga australior | 0003101000 | 000101?100 | 0100010111 | 000010110? |
| Chortophaga cubensis | 0003101000 | 000101?100 | 0100010100 | ????10110? |
| Chortophaga mendocino | 0003$01000 | 000101?200 | 0000010011 | 000010110? |
| Chortophaga virdifasciata | 0003101000 | 000101?200 | 0000010011 | 0000101101 |
| Encoptolophus californicus | 0004#01100 | *111001110 | 0000010111 | 000010110? |
| Encoptolophus costalis | 0002200000 | 011101?110 | 0100010111 | 0000201101 |
| Encoptolophus fuliginosus | 0001300000 | 0???002110 | 010?010111 | 0000100101 |
| Encoptolophus otomitus | 0004300000 | 0111002110 | 0*00010100 | ????200101 |
| Encoptolophus pallidus | 0001$00000 | 011111?011 | 0*00010110 | ????101100 |
| Encoptolophus robustus | 0001200000 | 011101?110 | 0000010111 | 0000101100 |
| Encoptolophus sordidus | 0002200000 | 011101?$10 | 0100010111 | 000020110? |
| Encoptolophus subgracilis | 0001200000 | 011111?*11 | 0000010111 | 0000101100 |
| Lactista azteca | 002?$10011 | 012001?010 | 1001011121 | 1111011101 |
| Lactista elota | 002?210011 | 012001?210 | 1011011121 | 1111011101 |
| Lactista gibbosus | 002?210011 | 012001?210 | 1001011121 | 1111011101 |
| Lactista punctatus | 002?210011 | 012001?110 | 1011011121 | 111101110? |
| Machaerocera mexicana | 00042000?0 | 012101?110 | 000100?021 | 2?00011100 |
| Opeia obscura | 113?000010 | 0102004020 | 000010?000 | ????011010 |
| Rhammatocerus viatorius | 013?100010 | 01100030$* | 000210?000 | ????011010 |
| Shotwellia isleta | 0000100000 | 01110??020 | 0000011111 | 0000011100 |
| Stethophyma lineatum | 003?100010 | 0120000020 | 0000110?00 | ????011000 |
| Syrbula montezuma | 113?000010 | 0102003020 | 000?10?020 | ????011010 |
| Trimerotropis pallidipennis | 002?200010 | 012001?010 | 0001011121 | 1110011101 |
| Xanthippus corallipes | 001?100011 | 012001?110 | 1001011021 | 1110011000 |
? refers to a missing or unknown character state. Polymorphisms are referred to by the following symbols: * for (0 + 1), $ for (1 + 2), # for (2 + 3).
Table 3.
Number of specimens per species and their geographic origin examined and scored for characters in the phylogenetic analysis.
| Species | Number of specimens examined (n) | Country: State |
|---|---|---|
| Chimarocephala pacifica | 186 | USA: California |
| Chimarocephala elongata | 11 | USA: California |
| Chortophaga australior | 38 | USA: Florida |
| Chortophaga australior | 14 | USA: Georgia |
| Chortophaga cubensis | 66 | Cuba: Havana Province |
| Chortophaga mendocino | 16 | USA: California |
| Chortophaga virdifasciata | 3 | USA: Alabama |
| Chortophaga virdifasciata | 1 | USA: Colorado |
| Chortophaga virdifasciata | 7 | USA: Missouri |
| Chortophaga virdifasciata | 1 | USA: New Mexico |
| Chortophaga virdifasciata | 37 | USA: New York |
| Chortophaga virdifasciata | 2 | USA: South Carolina |
| Chortophaga virdifasciata | 6 | USA: Texas |
| Encoptolophus californicus | 26 | USA: California |
| Encoptolophus costalis | 1 | USA: Kansas |
| Encoptolophus costalis | 29 | USA: New Mexico |
| Encoptolophus costalis | 14 | USA: North Dakota |
| Encoptolophus costalis | 1 | USA: South Dakota |
| Encoptolophus costalis | 1 | USA: Texas |
| Encoptolophus fuliginosus | 2 | México: Michoacán |
| Encoptolophus otomitus | 2 | México: Puebla |
| Encoptolophus pallidus | 15 | USA: California |
| Encoptolophus pallidus | 7 | USA: Nevada |
| Encoptolophus robustus | 20 | USA: California |
| Encoptolophus robustus | 29 | México: Baja California Norte |
| Encoptolophus robusuts | 3 | México: Baja California Sur |
| Encoptolophus sordidus | 1 | USA: Connecticut |
| Encoptolophus sordidus | 2 | USA: Indiana |
| Encoptolophus sordidus | 1 | USA: Kansas |
| Encoptolophus sordidus | 2 | USA: Massachusetts |
| Encoptolophus sordidus | 21 | USA: Michigan |
| Encoptolophus sordidus | 1 | USA: Montana |
| Encoptolophus sordidus | 63 | USA: New York |
| Encoptolophus sordidus | 3 | USA: North Dakota |
| Encoptolophus sordidus | 1 | USA: Ohio |
| Encoptolophus subgracilis | 20 | USA: Arizona |
| Encoptolophus subgracilis | 20 | USA: California |
| Encoptolophus subgracilis | 5 | USA: New Mexico |
| Encoptolophus subgracilis | 5 | USA: Texas |
| Encoptolophus subgracilis | 20 | México: Baja California Norte |
| Encoptolophus subgracilis | 6 | México: Baja California Sur |
| Encoptolophus subgracilis | 2 | México: Jalisco |
| Encoptolophus subgracilis | 2 | México: Michoacán |
| Encoptolophus subgracilis | 2 | |
| Encoptolophus subgracilis | 12 | |
| Lactista azteca | 2 | México: Baja California Norte |
| Lactista azteca | 3 | USA: Arizona |
| Lactista azteca | 3 | USA: New Mexico |
| Lactista azteca | 2 | USA: Texas |
| Lactista elota | 3 | México: Jalisco |
| Lactista gibbosus | 8 | México: Baja California Sur |
| Lactista gibbosus | 1 | México: San Luis Potosí |
| Lactista punctatus | 2 | México: Veracruz |
| Machaerocera mexicana | 5 | México: Veracruz |
| Opeia obscura | 15 | USA: New Mexico |
| Rhammatocerus viatorius | 13 | México: Baja California Sur |
| Shotwellia isleta | 14 | USA: New Mexico |
| Stethophyma lineatum | 2 | USA: New Mexico |
| Stethophyma lineatum | 3 | USA: Oregon |
| Syrbula Montezuma | 11 | USA: New Mexico |
| Trimerotropis pallidipennis | 23 | USA: New Mexico |
| Xanthippus corallipes | 12 | USA: Arizona |
Figs 2–9.
Chortophaga group species, fastigium and vertical ridge, dorsal aspect. (2) Shotwellia isleta, (3) Encoptolophus sordidus, (4) Encoptolophus otomitus, (5) Encoptolophus fuliginosus, (6) Chimarocephala virdifasciata, (7) Encoptolophus subgracilis, (8) Encoptolophus californicus LT, (9) Chortophaga virdifasciata. Scale bars = 1 cm. This figure is published in colour in the online edition of this journal, which can be accessed via http://booksandjournals.brillonline.com/content/1876312x
Figs 10–18.
Chortophaga group species, entire pronotum, dorsal aspect. (10) Shotwellia isleta, (11) Encoptolophus sordidus, (12) Encoptolophus fuliginosus, (13) Chortophaga virdifasciata, (14) Chimarocephala elongata, (15) Encoptolophus californicus LT, (16) Encoptolophus pallidus, (17) Encoptolophus robustus LT, (18) Encoptolophus subgracilis. Scale bars = 2.5 cm. This figure is published in colour in the online edition of this journal, which can be accessed via http://booksandjournals.brillonline.com/content/1876312x
Figs 19–24.
Chortophaga group species. (19–21) occiput, dorsal aspect. (19) Encoptolophus robustus LT, (20) Chimarocephala elongata, (21) Encoptolophus californicus LT. (22–24) left metafemur, dorsal aspect. (22) Encoptolophus californicus LT, (23) Encoptolophus subgracilis, (24) Encoptolophus fuliginosus. Scale bars adjacent to Figs 19–21 = 1 cm. Scale bars adjacent to Figs 22–24 = 5 cm. This figure is published in colour in the online edition of this journal, which can be accessed via http://booksandjournals.brillonline.com/content/1876312x
Outgroups included species in the Oedipodinae genera Lactista Saussure of the Arphia genus group, Xanthippus Saussure of the Hippiscus genus group, and Trimerotropis Stål of the Sphingonotini. Additional outgroups were species of Machaerocera and Stethophyma Fischer based on the former’s hypothesized sister group relationship to the Chortophaga group and the latter’s classification having recently shifted from Gomphocerinae to Oedipodinae (Fries et al. 2007; Chapco & Contreras 2011). Otte (1981) initially placed Stethophyma with the Gomphocerinae because of similarity in behavior and appearance, but noted that Stethophyma could have also been placed with Oedipodinae based on stridulatory apparatus (Otte 1981). Recently, Stethophyma has been informally placed in the Oedipodinae tribe Parapleurini (Eades et al. 2011). The cladogram was rooted between three gomphocerine representatives from three different tribes and Oedipodinae. Taxa included in the analysis along with their collection and voucher data are in Tables 1 and 2.
We note that the various “tribes” increasingly referenced in the literature and attributed to Orthoptera Species File (OSF2) (Eades et al. 2011) are not properly published according to the International Code of Zoological Nomenclature and are, therefore, unavailable, and we do not use them here. We use the term “genus group” following Otte (1984), referring to a group of related genera, rather than a group of related species within one genus. There are, however, a small number of valid tribe names, which we do use (e.g., Parapleurini Brunner von Wattenwyl, Sphingonotini Johnston).
DNA
Immediately after collection, whole mesothoracic or metathoracic legs were removed and placed in 95% ethanol. These legs were then kept frozen at −20°C until DNA extraction. DNA was extracted from large legs from dissected muscle tissue. For smaller legs, an entire leg was crushed and extracted. DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA) using the animal tissue protocol.
Three mitochondrial genes were sequenced: 16S rRNA (16S), 12S rRNA (12S) and cytochrome c oxidase subunit II (COII). Nuclear genes were not sequenced because even distantly related taxa were previously shown to have highly conserved gene sequences (e.g., histone III and 28S rRNA, see Edelman et al. 2010). Nearly all specimens were sequenced for each gene, but some failed to amplify or sequence (Table 1). Primers used for amplification and sequencing are listed in Table 4.
Table 4.
Primers used for amplification and sequencing.
| Gene | Primer | Direction | Sequence (5′→3′) |
|---|---|---|---|
| COII | F-luea | For | TCT AAT ATG GCA GAT TAG TGC |
| COII | R-lysa | Rev | GAG ACC AGT ACT TGC TTT CAG TCA TC |
| 16S | 16SAb | For | CGC CTG TTT ATC AAA AAC AT |
| 16S | 16SBb | Rev | CTC CGG TTT GAA CTC AGA TCA |
| 12S | 12Saic | For | AAA CTA CGA TTA GAT ACC CTA TTA T |
| 12S | 12Sbic | Rev | AAG AGC GAC GGG CGA TGT GT |
Gene amplification and PCR protocols closely followed Miller et al. (2005), Miller et al. (2007), and Edelman et al. (2010). DNA fragments were amplified using PCR with TaKaRa Ex Taq (Takara Bio, Otsu, Shiga, Japan) on an Eppendorf Mastercycler ep gradient S Thermal Cycler (Eppendorf, Hamburg, Germany) and visualized using gel electrophoresis with 1% TAE gel. Gels were stained using SYBR® Safe DNA gel stain (Invitrogen, Carlsbad, CA, USA). PCR products were then purified using ExoSAP-IT (USB-Affymetrix, Cleveland, OH, USA) and cycle sequenced following an ABI Prism Big Dye Terminator Reaction (version 3.1, Fairfax, VA, USA) using the same primers involved in amplification. Sequencing reaction products were purified using Sephadex G-50 Fine (GE Healthcare, Uppsala, Sweden) and sequenced with an ABI 3130xl Genetic analyzer (Molecular Biology Facility, UNM). All amplicons were sequenced in both directions and were edited using Sequencher 4.10.1 (Genecodes 1991–2010).
Before any sequence was used in the analysis, we applied several recommended regulatory techniques prescribed by Song et al. (2008) to reduce the potential for inclusion of pseudogenes or numts. These paralogous nuclear mitochondrial pseudogenes are a concern for systematists because of the potential for incorrect homology and are a particular problem in Orthoptera (Song et al. 2008). In order to prevent this, sequences were aligned against confirmed final sequences, investigated using basic alignment BLAST (ncbi.gov), and, in COII, examined for an open reading frame.
Data acquired and used by Edelman et al. (2010) were also used in this analysis (Table 1) with one exception that requires explanation. Their specimen identified as Chimarocephala elongata Rentz appears to be a highly rugose specimen of Chimarocephala pacifica (Thomas) based on reassessment by RAG and DCL. Because of this, we extracted DNA from a different individual more convincingly identified as C. elongata (by R.A.G. and D.C.L.) and amended the entries in GenBank associated with the misidentified specimen (KBMC-OR32) to reflect this new determination (see KBMC-OR89, Table 1).
Morphology and non-molecular characters
High-resolution images of key morphological characters across the Chortophaga group were acquired using a Visionary Digital BKplus Lab Imaging system (http://www.visionarydigital.com). In addition, illustrations of additional characters used in this analysis can be found in Otte (1981, 1984). In order to understand the phylogenetic relationships of the Chortophaga group, we examined multiple phenotypic characters for variation across taxa. Internal and external male genitalic morphology were examined across all taxa, but no variation useful for a cladistic analysis was discovered. Near uniformity in cerci shape and male genitalic morphology is characteristic of Oedipodinae, in contrast with the large majority of Acrididae (Roberts 1941; Dirsh 1956) and, therefore, this character system is not represented among the phenotypic data (Table 2). The relative positions of the intercalary and ulnar veins of the tegmen have been historically used in Oedipodinae for diagnosis at the genus rank (Bruner 1905; Rehn & Hebard 1912; Hebard 1934; Strohecker et al. 1968). However, we found that character to be unreliable in separating Chortophaga from Chimarocephala and Encoptolophus, an observation also made by Rentz (1977), and that character was not included. A total of 39 morphological characters and one behavioral character were included, most of which are characters traditionally used in the identification of Oedipodinae (see Table 2). Some of these characters were used by Stroehecker et al. (1968), Otte (1981, 1984), and Edelman et al. (2010). However, all relevant characters from these works were reassessed for homology, which in some cases, differed from those analyses.
Four morphometric characters found to be useful in diagnosing Chortophaga group taxa and included in this analysis were: (1) the ratio of the greatest width to greatest length of the pronotum, (2) the ratio of the dorsal length of the head capsule to the length of the pronotum, (3) the angle of the posterior margin of the pronotal disc and (4) the ratio of the length of the prozona to the length of the metazona. Quantitative characters are typically not useful in a cladistic analysis because of relatively continuous variation and difficulty in coding (see below), and ratios in measurement characters were used to standardize individual specimens of variable body size. Measurements were made using a Zeiss SteREO Discovery V8 microscope with an Achromat S FWD 63 mm lens at a 1 unit to 1 mm ratio (Zeiss, Berlin, Germany), and Adobe Photoshop CS5 (Adobe, San Jose, CA, USA) in conjunction with high-resolution photographs to accurately measure the angle of the hind margin of the pronotum. The characters coded for the analysis are described below, and coded states per species are presented in Table 2.
Head
-
1
Shape of head in lateral aspect: (0) nearly vertical or weakly slanted; (1) strongly slanted. State 1 has historically defined numerous gomphocerine taxa.
-
2
Shape of compound eyes: (0) ovoid and directed downward; (1) ovoid, anterodorsally narrowed. State 1 is characteristic of numerous gomphocerine taxa, including those examined for this analysis.
-
3
General shape of fastigium: (0) triangulate (Figs 2–9); (1) hexagonal; (2) ovoid; (3) domed. Members of the Chortophaga group have a triangulate fastigium with variation in width, length and depth across the group.
-
4
Specific shape of fastigium when triangulate: (0) parallel-sided basally, pointed apically (Fig. 2); (1) moderately narrow and weakly arcuate basally, pointed apically (Figs 5 and 7); (2) distinctly wider than long basally (Fig. 3); (3) strongly triangulate (Fig. 9); (4) elongate, narrow and bullet-shaped (Figs 4, 6 and 8). This character has states similar to those evaluated by Otte (1984).
-
5
Fastigium depth: (0) convex; (1) weakly depressed/shallow (Figs 2 and 9); (2) depressed (Figs 3, 5 and 7); (3) markedly depressed (Figs 4, 6 and 8). This particular continuous character has historically been used in diagnosing species of Oedipodinae and was challenging to code. It was coded as additive since state 1 exhibits intermediate similarity between states 0 and 2.
-
6
Apex of fastigium: (0) not directly leading into frontal ridge (Figs 2–9); (1) directly leading into frontal ridge. Members of Lactista have the fastigium leading directly into the frontal ridge, whereas all other taxa examined for this analysis were coded with state 0.
-
7
Frontal ridge at level of vertex in lateral aspect: (0) more rounded; (1) angulate. This character is similar to a Chortophaga group character presented by Otte (1984) who described this character as grouping Chortophaga with Chimarocephala.
-
8
Projection of frontal ridge: (0) not distinctly projecting beyond compound eyes; (1) distinctly projecting beyond compound eyes. Otte (1984) coded Chortophaga as having a moderately projecting frontal ridge with Chimarocephala having a markedly projecting frontal ridge. The intermediate state for this character presented by Otte (1984) was excluded from this analysis because it appeared to be inconstant across the specimens observed.
-
9
Median carinula of fastigium: (0) does not extend to the apex of the fastigium (Figs 2–9); (1) extends to the apex of the fastigium. Machaerocera mexicana Saussure was coded as ambiguous for this character as the specimens observed did not appear to have a median carinula.
-
10
Fastigium internal sculpturing: (0) with 4 rounded depressions; (1) without 4 rounded depressions (Figs 2–9). State 0 is characteristic of outgroup members Lactista and Xanthippus.
-
11
Median carinula of occiput: (0) indistinct (Fig. 19); (1) distinct (Figs 20 and 21). This character is adapted from Strohecker et al. (1968). The character was coded as polymorphic for E. californicus as not all specimens were observed to have a distinct occipital carinula.
-
12
Head length to pronotum length (males): (0) less than or equal to 0.52; (1) greater than or equal to 0.52. Character state 1 is characteristic of members of Chortophaga, which have a distinctly elongate pronotum, and this character appears to not have been used in an analysis previous to this. The description of this character and other similar morphometric characters presented here are based on males because of the greater number of male specimens observed, in general.
Pronotum
-
13
Width to length ratio of pronotum (males): (0) ranging from 0.5–0.6, with averages ranging from 0.46–0.5; (1) ranging from 0.5–0.8, with averages ranging from 0.6–0.72; (2) ranging from 0.64–0.89, with averages ranging from 0.74–0.85 (2). Additive, as this is another continuous character, which we attempted to separate into distinct bins.
-
14
Prozona length to metazona length ratio (males): (0) 0.4–0.65; (1) 0.67–0.9; (2) > 1. Additive (see explanation above). State 1 is characteristic of members of the Chortophaga group and Machaerocera mexicana, which have a slightly longer metazona than prozona but not to the extent of state 0.
-
15
Elevation of lateral carinae: (0) more elevated and/or more distinct on prozona (Figs 11–15 and 17); (1) less elevated and less distinct on prozona (Figs 16 and 18). This character was included in the key to the species of Encoptolophus presented by Strohecker et al. (1968) for separating E. subgracilis Caudell (Fig. 18) and E. pallidus Bruner (Fig. 16), which were coded with state 1, from E. robustus (Fig. 17), which was coded with state 0.
-
16
Lateral carinae: (0) continuous (Figs 14 and 15) or nearly continuous post-sulcus (Fig. 12M); (1) discontinuous and clearly separated post-sulcus (Figs 11, 13 and 16–18). State 0 is characteristic of several taxa coded for in this analysis, including members of the gomphocerine outgroup taxa, E. otomitus (Saussure), E. fuliginosus Bruner, E. californicus, Ch. elongata, and Ch. pacifica. However, this character alone does not adequately capture the variation in this feature. Shotwellia isleta was coded as ambiguous for this character because the pronotum of S. isleta does not have lateral carinae.
-
17
Shape of lateral carinae when continuous: (0) arcuate; (1) weakly divergent across pronotum (Figs 14 and 15); (2) weakly divergent on prozona and moderately divergent on metazona (Fig. 12); (3) distinctly constricted; (4) parallel-sided. This variation was coded using two characters in order to capture the different shapes of the lateral carinae in order to prevent coding all continuous lateral carinae as the same. Otte (1984) treats this character in his key to the species of Encoptolophus but did not assess it in species of Chimarocephala, which were both coded with state 1.
-
18
Height of pronotal median carina: (0) low (Fig. 10); (1) medium (Figs 12 and 14–18); (2) high (Figs 11 and 13). The height of the median carina or median keel of the pronotum has historically been used to identify species of Oedipodinae and continues to be useful for this purpose, but the feature has continuous variation that was difficult to separate into distinct bins. This character is a good example of the difficulty of using traditional morphological characters associated with the Chortophaga group in inferring phylogeny. It was coded as additive since the low median carina of Shotwellia isleta (Fig. 10), for example, appears more similar to the moderately high median carina of E. fuliginosus (Fig. 12) and others (Figs 14–18) than the high median carina of species such as E. sordidus (Burmeister) (Fig. 11).
-
19
Posterior angle of metazonal disc: (0) less than 95 degrees (Figs 13 and 14); (1) greater than 95 but less than 120 degrees (Figs 11, 12 and 15–18); (2) greater than 120 degrees (Fig. 10). This character is similar to a character presented by Otte (1984) for distinguishing the genus Chortophaga and its members from other Chortophaga group members. However, we measured specimens for more careful assessment and added state 2 for certain outgroup taxa and Shotwellia.
-
20
Posterolateral margins of metazonal disc: (0) not as in state 1 (Figs 10–15 and 17); (1) weakly arcuate forming a broadly triangular point basally (Figs 16 and 18). State 1 is characteristic of Encoptolophus pallidus and E. subgracilis, but not E. robustus (Fig. 17). This character does not appear to not have been described prior to this analysis. It is treated separately from character 19 because that character addresses the shape mathematically but does not capture qualitative homology assessments like this.
-
21
Pronotum sculpturing: (0) not distinctly rugose; (1) distinctly rugose, particularly in the prozona. Members of Lactista and Xanthippus have a distinctly rugose pronotum, and Ch. pacifica was coded with both states as numerous specimens were observed to have a distinctly rugose pronotum. Machaerocera mexicana was coded with state 0 despite having significant sculpturing because its pronotum is heavily tuberculate.
-
22
Markings of pronotal metazonal disc: (0) without 2–3 conspicuous maculations extending from the hind margin inward (Figs 10 and 13–18); (1) with 2–3 conspicuous maculations extending from the hind margin inward (Figs 11 and 12). 2–3 conspicuous maculations on the dorsum of the pronotum are present in several members of the Chortophaga genus group, and this historic character (Otte 1984) has often been used to distinguish members of Encoptolophus, in particular.
-
23
Tooth of lateral lobe: (0) absent; (1) present. L. elota Otte and L. punctatus Stål have a small tooth-like projection at the posterodistal margin of the lateral lobe. All of the remaining taxa examined for the analysis do not have this projection.
Venter
-
24
Shape of metasternal interspace: (0) of male linear, of female quadrate; of male quadrate; (1) of female at least 1.5 times wider than long; (2) of male and female linear. This was adapted directly from the key to the genera of Oedipodinae presented by Strohecker et al. (1968). The metasternal interspace of males is linear and of females quadrate in species of the Chortophaga group, Stethophyma, and Opeia McNeill.
Tegmin
-
25
White streak of tegmin: (0) without white streak above metafemur; (1) with white streak above metafemur. This was adapted from Otte (1981) for identifying the species of Stethophyma, and species of the outgroup gomphocerine taxa and Stethophyma lineatum (Scudder) were coded with state 1.
-
26
Intercalary vein of tegmin: (0) absent; (1) present. This was also adapted from Otte (1981).
-
27
Location of intercalary vein: (0) parallel to medial and cubital veins; (1) angled toward medial vein distally. The states for this character are different from previous work on the group as addressed earlier in the methods.
-
28
Patterning of tegmin: (0) without 1–4 distinct bands; (1) with 1–4 distinct bands. Banding in the Chortophaga group is variable with respect to intensity and number of bands, and these character data have long been used to diagnose species in the group, which may be problematic as this variation appears to be rather plastic and correlated with seasonality (see discussion on character evolution).
Hind wing
-
29
Disc of hind wing: (0) transparent (Fig. 28); (1) faintly pigmented (Figs 25–27); (2) distinctly pigmented. Most Chortophaga group species have a faintly pigmented disc of hind wing, unlike the outgroup species examined. Certain species in the Chortophaga group have a completely transparent wing (e.g., E. otomitus (Fig. 28) and C. cubensis), and were coded as ambiguous for characters 31–34 (Table 2).
-
30
Hind wing banding: (0) without band (Fig. 28); (1) with band (Figs 25–27). This character was included in addition to the previous character because Syrbula montezuma (Saussure), for example, has a distinctly pigmented hind wing but lacks a band.
-
31
Band of hind wing: (0) faint/smoky (Figs 25–27); (1) distinct black band that does not occupy more than 1/2 of hind wing; (2) large band that occupies more than 1/2 of hind wing. Character state 0, despite different terminology, is similar to the synapomorphy for the Chortophaga group as revealed by the parsimony analysis performed by Edelman et al. (2010).
-
32
Location of band of hind wing: (0) distal (Figs 25–27); (1) medial-distal. Machaerocera mexicana was coded as ambiguous for this character because the band does not appear homologous to either state 0 or 1 in character 31 and takes up a significantly larger amount of space, which made it challenging to code as either of the two listed described here. A distal band of the hind wing is present in all members of the Chortophaga group that have a pigmented hind wing.
-
33
Band of hind wing: (0) without spur (Figs 25–27); (1) with spur. Outgroup species included in the analysis of Lactista, Trimerotropis and Xanthippus have a spur along the band of the hind wing.
-
34
Hind wing: (0) without dark apex (Figs 25–27); (1) with dark apex. Members of Lactista have a dark apex of the hind wing.
-
35
Cells in anterior region of hind wing, costal-subcostal sector: (0) unenlarged (Fig. 25); (1) enlarged (Figs 26 and 27); (2) dramatically enlarged (Fig. 28). This character was significant in the phylogeny presented by Otte (1984, Fig. 1) who applied it only to certain species within Encoptolophus. It was the basis for the previous placement of E. otomitus, E. fuliginosus, E. sordidus and E. costalis Scudder in a clade separate from other members that lacked enlarged cells in the first fold of the hind wing (Otte 1984). When we closely examined this character, it was determined that several taxa have enlarged cells but not so dramatically modified as in E. otomitus (Fig. 28), E. sordidus and E. costalis including E. fuliginosus (Fig. 27). Encoptolophus fuliginosus was coded as intermediate. Additive.
-
36
Hind wing, anterior media Ma and posterior media Mp sector: (0) without vein (Fig. 28) or with very short vein arising near distal edge (Figs 26 and 27); (1) with vein, typically arising posterior to apex of hind wing (Fig. 25). Otte (1984) mapped this character on a hypothetical phylogenetic tree of the genus groups of Oedipodinae but excluded it from an ingroup analysis of the group, which would have provided support for grouping Chortophaga and Chimarocephala as nested within the Chortophaga group.
Figs 25–28.
Chortophaga group species, left hind wing, dorsal aspect. (25) Shotwellia isleta, with arrow indicating vein between anterior medial and posterior medial veins, (26) Encoptolophus subgracilis, (27) Encoptolophus fuliginosus, (28) Encoptolophus otomitus. Scale bars = 5 cm. This figure is published in colour in the online edition of this journal, which can be accessed via http://booksandjournals.brillonline.com/content/1876312x
Metalegs
-
37
Dorsal surface of metafemur: (0) with narrow, oblique dark band (Figs 24 and 28); (1) with other coloration (e.g., Figs 22 and 23). E. fuliginosus and E. otomitus have a narrow, oblique dark band on the dorsal surface of the metafemur. Banding on the metafemur is variable across the group, but state 0 appears unique and has historically been used to identify E. otomitus (Otte 1984).
-
38
Patterning of medial surface of metafemur: (0) without banding; (1) with banding. Character state 1 is characteristic of Oedipodinae but is difficult to homologize except as absent or present. Machaerocera mexicana does have banding along the medial surface of the metafemur.
-
39
Stridulatory file of medial face of metafemur (males): (0) absent; (1) present. This character has historically been used as a method of separating Gomphocerinae from Acridinae and Oedipodinae (Otte 1981).
Sound production
-
40
Crepitation: (0) do not crepitate; (1) crepitate. This is the only behavioral character included in this study. Despite the number of taxa for which character information was missing, this character was still included because crepitation patterns do appear to be informative with respect to phylogeny (Weissman & Rentz 1980). We also included this character because crepitation is among the more compelling features of Oedipodinae, and perhaps, character descriptions such as this will promote further studies into this behavioral feature. Observations by DCL suggest that S. isleta, E. subgracils, E. robustus and E. pallidus do not crepitate, whereas crepitation has been observed in C. virdifasciata (DeGeer), E. otomitus, E. fuliginosus and E. costalis.
Analysis
COII was not length variable across the included taxa and was aligned manually. 16S and 12S are length variable and were aligned using Muscle (Edgar 2004) and the default settings.
Combined morphological and molecular character data were analyzed using equal-weights parsimony with NONA (Goloboff 1995) as implemented by WinClada (Nixon 2002). The commands used in NONA were: hold 10000, hold/60, mult 60 and max*. Characters 5, 13, 14, 18 and 35 were treated as additive (see character descriptions above for reasoning). All remaining multistate characters were treated as non-additive. This analysis included 1613 aligned base pairs, (395 parsimony-informative). Gaps were treated as missing data. Trees and characters were examined under various optimizations using WinClada (Nixon 2002).
Branch support (bs) was evaluated using bootstrap values in NONA as implemented by WinClada (Nixon 2002) based on 1000 replications sampling about 10% of characters, and saving the consensus for each replication. Partitioned and total Bremer support values (Baker and DeSalle 1997) were calculated in PAUP* 4.0 (Swofford 2002) using a batch file generated by TreeRot v2 (Sorensen 1999).
Results
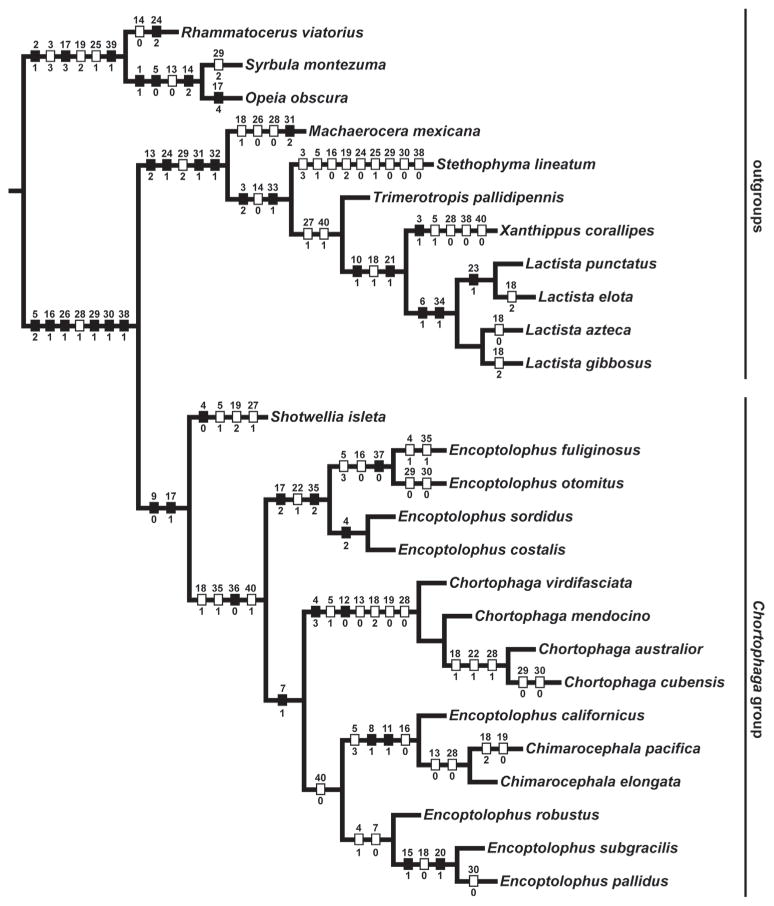
Parsimony analysis of the combined data resulted in two most parsimonious cladograms (Figs 1 and 2) with length 1367 (1171 with uninformative characters deactivated) and CI = 0.48, RI = 0.64. In both trees the Chortophaga group is resolved as monophyletic with high support (bs = 87%). The main conflict between the two trees is the relative placement of the clade comprising Chortophaga species and one of the Encoptolophus clades. The placement of Machaerocera mexicana is weakly resolved (bs < 50%, Bremer = 2). Stethophyma lineatum groups with Oedipodinae, similar to findings by Fries et al. (2007) and Chapco & Contreras (2011) (bs > 50%, Bremer = 3). Shotwellia isleta is resolved as sister to all other Chortophaga group members, similar to findings by Edelman et al. (2010), bs > 85%, Bremer support = 7. The clade containing Encoptolophus, Chortophaga and Chimarocephala is monophyletic with high support (bs > 95%, Bremer = 22). Members of Encoptolophus are not monophyletic in either solution and are resolved in three different places (Fig. 3). The first is a clade that includes the type species of Encoptolophus, E. sordidus and three other species (Encoptolophus sensu stricto), with high support (bs > 80%, Bremer = 6). The second is a clade with three species, E. robustus, E. subgracilis and E. pallidus, with weak support (bs < 50%, Bremer support = 2). The third is the single species E. californicus, which is resolved as sister to Chimerocephala (bs > 80%, Bremer = 3).
Discussion
These results confirm the difficulty of diagnosing groups of Oedipodinae taxa using morphology because of considerable homoplasy in traditionally-used features (Figs 1 and 2), some of which were revealed to be of little phylogenetic value and/or difficult to homologize. A notable example of considerable convergent evolution within this group is Stethophyma lineatum, which is strongly grouped with Oedipodinae taxa despite sharing some superficially similar morphological character states with Gomphocerinae outgroup taxa, a conclusion also reached by Fries et al. (2007) and Chapco & Contreras (2011). Nevertheless, results from this analysis (Figs 1 and 2) have important implications for the classification of the Chortophaga group of genera.
The Chortophaga group is well-supported as monophyletic, which corroborates previous analyses (Fries et al. 2007; Edelman et al. 2010), and the group is characterized by (1) an incomplete, distal, faint or smoky band on the flight wing (Edelman et al. 2010), 92) absence of distinct pigmentation in the basal half of the hind wing and (3) fastigium with a short median carinula that does not extend to the apex of the fastigium (character 9), which is a previously undiscovered synapomorphy for the group. The internal phylogenetic structure in the Chortophaga group indicates a need for reclassification (Figs 1 and 2).
Machaerocera mexicana has never been formally (or validly) placed within a genus group or tribe (Otte 1984). Evidence from our analysis indicates that the species should best be classified in its own Machaerocera genus group (Fig. 3). The species does exhibit distinct differences from Chortophaga group members such as the strongly tuberculate sculpturing of the pronotum, the tegmen lacking an intercalary vein, the apical margin of the tegmen slanted and not rounded like many Acridinae, the metathoracic space of the male quadrate and of the female longer than the mesothoracic space, and the hind wing distinctly pigmented with a large band that occupies a large area of the hind wing disc. Additionally, a recent molecular phylogenetic analysis of Oedipodinae placed Machaerocera and Melanotettix as sister to the Chortophaga group (Chapco & Contreras 2011). Machaerocera is also ecologically different from taxa of the Chortophaga group, and is often found directly on vegetation in subtropical woodlands (Otte 1984), instead of on open bare soil in relatively arid environments (D.C.L., unpublished).
Encoptolophus californicus, described in Encoptolophus (Bruner 1905), was moved to the genus Chimarocephala by Strohecker et al. (1968) based on the distinct carina on the vertex and the median carina of the pronotum elevated. It was moved back to Encoptolophus by Otte (1984) without much discussion. Our results (based on morphological data alone since we were unable to obtain sequence data), provide convincing evidence that the species is, indeed, closely related to Chimarocephala, and not to the other species currently placed in Encoptolophus (Figs 1–3). Therefore we transfer this species from Encoptolophus and it becomes Chimarocephala californica (Bruner), comb.n.
Finally, results of this analysis indicate two larger clades historically recognized in a single genus, Encoptolophus. One of these includes the type of the genus, E. sordidus, and three other species, E. costalis, E. fuliginosus and E. otomitus. The other group is a clade with three species: E. robustus, E. subgracilis and E. pallidus.
Given the non-monophyly of Encoptolophus, there are several potential taxonomic actions based on these results. One option would be to sink Chimarocephala and Chortophaga into Encoptolophus, which has priority, thereby reclassifying the Chortophaga group with two genera: Shotwellia and Encoptolophus. This action would result in synonomizing two historically recognized, well-characterized, monophyletic genera. Another option would be to describe the clade composed of E. robustus, E. subgracilis and E. pallidus as a new monophyletic genus. Given a choice between sinking two historically recognized, well-characterized genera or erecting one new genus, in the interest of maintaining stability, we erect a new genus described below.
Nebulatettix Gómez, Lightfoot & Miller, gen.n
Type species: Encoptolophus subgracilis (Caudell)
Diagnosis and description
Nebulatettix is a member of the Chortophaga group based on: (1) presence of an incomplete, distal, faint or smoky band on flight wing (secondarily absent in nearly transparent wing of N. pallidus (Edelman et al. 2010)), (2) absence of distinct pigmentation in the basal half of the hind wing (Otte, 1984) and (3) the fastigium with a short median carinula that does not extend to the apex of the fastigium.
Adults of Nebulatettix can be distinguished from other genera in the Chortophaga group by the following combination of characters: (1) without a distinct projection of the frontal ridge beyond the compound eyes, (2) the fastigium triangulate apically, moderately narrow, and the length slightly greater than the width (Fig. 7), (3) the median carinula of the fastigium present and raised (Fig. 7), (4) without dark maculations on the pronotal shield (Figs 16–18) (present but polymorphic within N. pallidus), (5) the posterior angle of the pronotal shield broad, not acute, nor completely rounded basally (Figs 16), (6) the median carina of the pronotum not markedly elevated (Figs 16), (7) the width/length ratio of pronotum = 0.5/0.8 (average = 0.6/0.72), (8) the pronotal lateral carinae present and discontinuous (Figs 16), (9) the sector of the first fold between the anterior media and poster media veins without a vein, or with a very short vein arising near the distal margin of the wing (Fig. 26), (10) the pronotum typically without a pale X dorsally (Figs 16–18) (present in some dark individuals of N. subgracilis), (11) the dorsal surface of the metafemur typically with a large, dark triangular macula (Fig. 23) (faint in some pale individuals of N. pallidus) and (12) the tegmina with distinct banding.
Shotwellia, which is sister to the remaining members of the Chortophaga group, has a large number of autapomorphies (Edelman et al. 2010) that allow for easy identification. Shotwellia is superficially similar to N. pallidus, but differs in absence of lateral carinae (Fig. 10), greatly rounded pronotal disc (Fig. 10), costal-subcostal sector of hindwing with a vein (Fig. 25), and a more parallel-sided fastigium (Fig. 2). Nebulatettix differs from Chortophaga based on the shape of the fastigium not strongly triangulate as in Fig. 9 and the angle of the hind margin of the pronotal disc greater than 95° (Figs 16–18) instead of strongly acute (Fig. 13). Nebulatettix differs from Chimarocephala in the shape of the fastigium not strongly narrow, deep nor elongate as in Fig. 6, with conspicuously discontinuous, often indistinct, pronotal lateral carinae (Figs 16–18), absence of a distinct median occipital carinula (Fig. 19), and with a large, dark, triangular macula on the dorsal surface of the metafemur (Fig. 23) (faint in some pale individuals of N. pallidus).
Nebulatettix is superficially most similar to the genus Encoptolophus. Nebulatettix differs from Encoptolophus based on the following combination of characters: (1) cells in the costal-subcostal sector of apex of the hind wing not greatly enlarged in Nebulatettix (Fig. 26) instead of greatly enlarged cells (Fig. 28) (except E. fuliginosus which also has smaller cells (Fig. 27)), (2) Nebulatettix with the dorsal surface of the metafemur with a large triangular macula (Fig. 23), (3) the fastigium moderately narrow, angled, and pointed apically in Nebulatettix (Fig. 7) instead of wider than long (Fig. 3) or elongate, narrow and pointed (Fig. 4) (except E. fuliginosus (Fig. 5), which has a state similar to Nebulatettix)), (4) the fastigium less excavated in Nebulatettix (Fig. 7), (5) Nebulatettix without 2–3 conspicuous, dark maculations on the pronotal shield (Figs 16) (polymorphic in N. pallidus), which are present as distinct markings on the pronotum extending outward from the margin in Encoptolophus (Figs 11 and 12) (polymorphic in E. otomitus) and (6) Encoptolophus crepitate whereas Nebulatettix has not been observed to crepitate (Otte 1984; D.C.L., personal observation).
The figures presented herein in addition to a dichotomous key to the genera of the Chortophaga group presented below should aid in identification of Nebulatettix.
Etymology
This genus is named in honor of the dark markings present throughout the body of these dusky grasshoppers and is derived from the Latin nebula for “cloud” or “smoke” and from the Greek tettix for “grasshopper”.
Distribution and habitat
The genus is known from three species: N. robustus, which occurs in coastal southern California and Baja California of the United States and Mexico, N. pallidus, which is found in eastern California and western Nevada, and the most widespread species, N. subgracilis, which occurs in the Midwest and Southwest United States and throughout Mexico and Central America (Otte 1984). The species typically occupy relatively mesic and damp grasslands (Otte 1984), and N. robustus and N. pallidus have been collected on mesic patches of grassy habitat surrounded by widespread xeric environments (D.C.L., unpublished).
Chortophaga group character evolution
Many characters relevant to the relationships and classification of the Chortophaga group are associated with the aspects of the pronotum, particularly features of the lateral carinae and posterior margin, yet the height of the median carina has been shown to have little phylogenetic value and appears to be very plastic across the group, despite being useful in diagnosing species. Another character that has frequently been used historically but transforms frequently is patterning, in particular banding, of the tegmen (Otte 1984), which appears to be a plesiomorphy in the group that is lost in Chortophaga virdifasciata and C. mendocino Rentz and is regained in C. australior Rehn & Hebard and C. cubensis. Banding in Chortophaga appears to be indirectly influenced by seasonality and, therefore, may not be a fixed feature or phylogenetically informative (Brust et al. 2008), which appears to be the case in other genera in the group. The difficulty in determining naturalness of genera within the group is directly relevant to the use of more plastic characters in phylogenies, particularly continuous characters such as pronotal median carina height and fastigium depth, both of which were tested in this analysis.
Among the most notable derived losses in the group is the absence of flight displays and crepitation in Nebulatettix (D.C.L., personal observation). Despite missing data for some ingroup taxa (Table 2), crepitation appears to be a unique oedipodine behavior that has been lost and gained numerous times. This long-distance acoustic behavior likely serves to attract the sexes and crepitation patterns and sounds are species-specific and tend to be similar among related taxa (Rentz & Weisman 1981; Otte 1984). We can only speculate as to why crepitation has been lost in Nebulatettix. In addition, visual cues associated with flight displays within the Chortophaga group appear to be on a trend towards reduced pigmentation of the disc of the hind wing. Shotwellia isleta, which is sister to the remaining species of the group, has among the strongest pigmentation of the disc of the hind wing (Fig. 25), and species further nested tend to having nearly pellucid flight wing discs like Nebulatettix pallidus or completely pellucid in the case of Chortophaga cubensis and Encoptolophus otomitus (Fig. 28). Muted pigmentation and lack of crepitation would appear to be less conspicuous than a similar flight with a distinctly pigmented flight wing and crepitation.
Key to the genera of the Chortophaga group
| 1. | Fastigium parallel-sided basally (Fig. 2); hind margin of pronotal shield distinctly rounded (Fig. 10); lateral carinae of entire pronotum absent (Fig. 10); anterior medial vein and posterior medial vein sector of hind wing with a vein (Fig. 25) | Shotwellia Gurney |
| – | Fastigium not parallel-sided basally (Figs 3–9); hind margin of pronotal shield pointed (Figs 11–18); lateral carinae of entire pronotum present (Figs 11–18); anterior medial vein and posterior medial vein sector of hind wing without a vein (Fig. 28) or with a very short vein arising laterally (Figs 26 and 27) | 2 |
| 2. | Cells of costal-subcostal sector of hind wing greatly enlarged (Fig. 28); if not greatly enlarged (e.g., E. fuliginosus (Fig. 27)) then dorsal surface of meta-femur with a small, narrow, and oblique dark macula (Figs 24 and 28) | Encoptolophus Scudder |
| – | Cells of costal-subcostal sector not greatly enlarged (Fig. 26); dorsal surface of metafemur variable, without a small, narrow and oblique dark macula (Figs 22 and 23) | 3 |
| 3. | Fastigium strongly triangulate (Fig. 9); distance between lateral carinae of prozona narrow (Fig. 13); length of pronotum from base to apex long, 1.9 × head length or greater … | Chortophaga Saussure |
| – | Fastigium not strongly triangulate, more elongate and narrow (Figs 6–8); distance between lateral carinae of prozona moderately broad (Figs 14–18); length of pronotum not as long, less than 1.9 × head length | 4 |
| 4. | Fastigium distinctly elongate and narrow, deep to markedly deep (Figs 6 and 8); frontal ridge distinctly projecting beyond front of compound eyes; occipital carinula typically distinct (Figs 20 and 21); lateral carinae continuous (Figs 14 and 15); if discontinuous then pronotum also heavily rugose (Ch. pacifica) | Chimarocephala Scudder |
| – | Fastigium moderately elongate and narrow, shallow to moderately deep (Fig. 7); frontal ridge not distinctly projecting beyond front of compound eyes; occipital carinula not distinct (Fig. 19); lateral carinae discontinuous (Figs 16–18) | Nebulatettix Gómez, Lightfoot & Miller, gen.n. |
Fig. 29.
One of two most parsimonious cladograms from combined parsimony analysis with morphological characters mapped using ‘fast’ optimization in WinClada. Filled hatchmarks indicate unique character state transformations, open hatchmarks indicate reversals or homoplasious transformations. Numbers above hatchmarks are character numbers. Numbers below hatchmarks are character state numbers (derived at that branch).
Fig. 30.
Two of two most parsimonious cladograms from combined parsimony analysis with morphological characters mapped using ‘fast’ optimization in WinClada. Filled hatchmarks indicate unique character state transformations, open hatchmarks indicate reversals or homoplasious transformations. Numbers above hatchmarks are character numbers. Numbers below hatchmarks are character state numbers (derived at that branch).
Fig. 31.
Strict consensus of two most parsimonious trees based on phenotypic and molecular characters. Numbers in bold above branches are bootstrap support values when greater than 50%. Numbers below branches are Bremer support values. Total Bremer support values are shown outside parentheses followed by partitioned Bremer support values for (morphology/COII/16S/12S).
Acknowledgments
Financial research support for R.A.G. came in part from the MARC program, funded by Award Number T34GM00851 from the National Institute of General Medical Sciences, without which this research would not be possible. We thank William C. Edelman for his supporting work of this project. We thank the molecular biology facility at the University of New Mexico for sequencing. We thank the following institutions for loaning specimens: Academy of Natural Sciences of Philadelphia, California Academy of Sciences, Cornell University, the Florida State Collection of Arthropods, Michigan State University, and the University of California Berkeley. We thank D. B. Weissman (Department of Entomology, CAS), John Capinera (Department of Entomology and Nematology, University of Florida) and A. Joern (Department of Ecology, University of Kansas) for providing much-needed DNA samples. Portions of this project were funded in part by USA National Science Foundation grant DEB-0845984 (K.B.M., PI).
References
- Baker RH, DeSalle R. Multiple sources of character information and the phylogeny of Hawaiian Drosophila. Systematic Biology. 1997;46:654–673. doi: 10.1093/sysbio/46.4.654. [DOI] [PubMed] [Google Scholar]
- Bruner L. Orthoptera. The Acrididae. Biologia Centrali-Americana. 1905;2:105–176. [Google Scholar]
- Brust ML, Hoback WW, Wright RJ. A review of the genus Chortophaga (Orthoptera: Acrididae) among Nebraska populations: questioning the validity of Chortophaga australior Rehn & Hebard. Journals of Orthoptera Research. 2008;17:101–105. [Google Scholar]
- Chapco W, Martel RKB, Kuperus WR. Molecular phylogeny of North American band-winged grasshoppers (Orthoptera: Acrididae) Annals of the Entomological Society of America. 1997;90:555–562. [Google Scholar]
- Chapco W, Contreras D. Subfamilies Acridinae, Gomphocerinae and Oedipodinae are “Fuzzy Sets”: A Proposal for a Common African Origin. Journal of Orthoptera Research. 2011;20:173–190. [Google Scholar]
- Dirsh VM. The Phallic Complex in Acridoidea (Orthoptera) in relation to the taxonomy. Transactions of the Royal Entomological Society of London. 1956;108:223–356. [Google Scholar]
- Eades DC, Otte D, Cigliano MM, Braun H. [accessed September 2011];Orthoptera Species File Online, Version 2.0/4.0. 2011 Available online at http://Orthoptera.SpeciesFile.org.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughputs. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries M, Chapco W, Contreras M. A molecular phylogenetic analysis of the Oedipodinae and their intercontinental relationships. Journal of Orthoptera Research. 2007;16:115–125. [Google Scholar]
- Goloboff P. NONA. Published by the author; Tucumán, Argentina: 1995. [Google Scholar]
- Hebard M. The Dermaptera and Orthoptera of Illinois. Bulletin of the Illinois State Natural History Survey. 1934;20:125–279. [Google Scholar]
- Miller KB, Alarie Y, Wolfe GW, Whiting MF. Association of insect life stages using DNA sequences: The larvae of Philodytes umbrinus (Motschulsky) (Coleoptera: Dytiscidae) Systematic Entomology. 2005;30:499–509. [Google Scholar]
- Miller KB, Bergsten J, Whiting MF. Phylogeny and classification of the diving beetle tribe Cybistrini (Coleoptera: Dytiscidae) Zoologica Scripta. 2007;36:41–59. [Google Scholar]
- Nixon KC. WinClada. Published by the author; Ithaca, NY: 2002. [Google Scholar]
- Otte D. A comparative study of communicative behavior in grasshoppers. Miscellaneous Publications of the University of Michigan Museum of Zoology. 1970;141:1–168. [Google Scholar]
- Otte D. The North American Grasshoppers, Volume 1 (Acrididae: Gomphocerinae and Acridinae) Harvard University Press; Cambridge, MA: 1981. p. 208. [Google Scholar]
- Otte D. The North American Grasshoppers, Volume 2, (Acrididae: Oedipodinae) Harvard University Press; Cambridge, MA: 1984. p. 366. [Google Scholar]
- Rehn JAG, Hebard M. Fixation of single type (lectotype) specimens of species of American Orthoptera. I. Proceedings of the Academy of Natural Sciences of Philadelphia. 1912;64:235–276. [Google Scholar]
- Rentz DCF. Two new grasshoppers from California (Orthoptera, Acrididae, Oedipodinae) Acrida. 1977;6:141–151. [Google Scholar]
- Rentz DCF, Weissman DB. Faunal Affinities, systematics, and bionomics of the Orthoptera of the California Channel Islands. University of California Publications in Entomology. 1981;94:1–240. [Google Scholar]
- Roberts HR. A comparative study of the subfamilies of the Acrididae (Orthoptera) primarily on the basis of their phallic structures. Proceedings of the Academy of Natural Sciences of Philadelphia. 1941;XCII:201–246. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America. 1994;87:651–701. [Google Scholar]
- Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proceedings of the National Academy of Sciences. 2008;105:13486–13491. doi: 10.1073/pnas.0803076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson MD. TreeRot, Version 2. Boston University; Boston, MA: 1999. [Google Scholar]
- Strohecker HF, Middlekauff WW, Rentz DC. The grasshoppers of California. Bulletin of the California Insect Survey. 1968;10:1–177. [Google Scholar]
- Svenson GJ, Whiting MF. Phylogeny of Mantodea based on molecular data: evolution of a charismatic predator. Systematic Entomology. 2004;29:359–370. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Weissman DB, Rentz DCF. Cytological, morphological, and crepetational characteristics of the Trimerotropine (Aerochoreutes, Circotettix, and Trimerotropis) grasshoppers (Orthoptera: Oedipodinae) Transactions of the American Entomological Society. 1980;106:253–272. [Google Scholar]
- Whiting MF. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoologica Scripta. 2002;31:93–104. [Google Scholar]