Abstract
Cystathionine beta-synthase (CBS) deficiency is an inborn error of metabolism characterized by extremely elevated levels of plasma total homocysteine. The vast majority of CBS-deficient patients have missense mutations located in the CBS gene that result in the production of either misfolded or unstable protein. Here, we examine the effect of proteasome inhibitors on mutant CBS function using two different mouse models of CBS deficiency. These mice lack mouse CBS and express a missense mutant human CBS enzyme (either p.I278T or p.S466L) that has less than 5% of normal liver CBS activity, resulting in a 10–30 fold elevation in plasma homocysteine levels. We show that treatment of these mice with two different proteasome inhibitors can induce liver Hsp70, Hsp40, and Hsp27, increase levels of active CBS, and lower plasma homocysteine levels to within the normal range. However, response rates varied, with 100% (8/8) of the p.S466L animals showing correction, but only 38% (10/26) of the p.I278T animals. In total, our data shows that treatment with proteostasis modulators can restore significant enzymatic activity to mutant misfolded CBS enzymes and suggests that they may be useful in treating certain types of genetic diseases caused by missense mutations.
Keywords: genetic disease, missense mutation, chaperone, inborn error, aminoaciduria
Introduction
Missense mutations are genetic alterations that result in the production of proteins with single amino acid changes and are the most common alteration observed in several inborn errors of metabolism. For example, they make up 62.4% of the mutant alleles described in phenylketonuria patients, 65% of the alleles seen in ornithine transcarbamylase deficient patients, and 67% of the alleles seen in cystathionine beta-synthase (CBS, OMIM 613381) deficient patients. [Stenson, et al., 2003]. Most disease-causing missense mutations do not target key catalytic residues in proteins but rather cause problems in protein folding [DePristo, et al., 2005]. It is thought that missense mutations affect protein structure by preventing them from folding into their lowest-free energy native state, resulting in trapped misfolded protein intermediates that are either degraded or form large molecular weight aggregates [Bross, et al., 1999]. In theory, treatments that could reverse these protein folding defects and promote proper folding of missense mutant proteins would be of great utility in the treatment of a wide variety of genetic diseases.
Previously, it has been demonstrated that it is possible to restore significant enzymatic function to a large percentage of missense mutant human CBS alleles expressed in S. cerevisiae and E. coli by various genetic and pharmacological manipulations that alter the protein folding (i.e., chaperone) environment [Kopecka, et al., 2011; Majtan, et al., 2010; Mayfield, et al., 2012; Singh, et al., 2007; Singh, et al., 2010; Singh and Kruger, 2009]. CBS deficiency is the most common inborn error of sulfur metabolism and is the cause of classical homocystinuria, a condition characterized by very high levels of plasma total homocysteine (tHcy) [Mudd, et al., 1964]. CBS deficient patients suffer from various pathologies including arteriosclerosis, osteoporosis, mental retardation, and dislocated lenses [Mudd, et al., 1985]. Current treatment for CBS deficiency involves lowering tHcy levels with a combination of vitamins, protein restriction, and cysteine supplementation [Wilcken and Wilcken, 1997; Yap, et al., 2001; Yap, et al., 2000]. Although those treatments can be effective, the dietary regimes are difficult and compliance is often a problem. Thus, a drug that could stimulate residual CBS enzyme activity could be useful in treating this disease.
One class of pharmacologic agents used to stimulate mutant CBS enzyme activity is proteasome inhibitors [Singh, et al., 2010; Singh and Kruger, 2009]. The proteasome is the major intracellular machine that is responsible for the degradation of excess or misfolded proteins [Adams, et al., 1999]. Proteasome inhibitors both stop the degradation of misfolded mutant protein and induce the expression of molecular chaperone proteins, such as Hsp70, which can help stimulate proper protein folding [Hartl and Hayer-Hartl, 2002]. Clinically, proteasome inhibitors are used primarily to treat certain types of cancer, such as multiple myeloma [Orlowski and Kuhn, 2008].
The goal of the current study was to investigate the effects of proteasome inhibitors in two different humanized mouse models of CBS deficiency. Our results show that it is possible to restore significant enzymatic activity to mutant CBS alleles in vivo, resulting in significant reductions in plasma homocysteine levels. Our findings suggest that proteasome inhibitors may be useful in the treatment of certain types of genetic disease caused by missense mutations.
Material and Methods
All experimental animal procedures were carried out according to the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Fox Chase Cancer Center.
Mouse Models
All mice used in the studies are shown in Supp. Table 1. Tg-S466L Cbs−/−, Tg-I278T Cbs−/−, and Tg-hCBS Cbs−/− mice were generated as previously described [Gupta, et al., 2008; Wang, et al., 2005; Wang, et al., 2004]. All three mouse strains were backcrossed to C57BL6 three times before intercrosses were performed. These mice expressed human CBS under control of the zinc-inducible metallothionein promoter. All mice were maintained on LabDiet 5013 chow. Before being used for experiments, mice were put on water containing 25 μM ZnSO4 for 11 days to induce transgene expression. Only female animals were used for drug studies as they tend to have more consistent transgene induction [Wang, et al., 2004]. A description of all the mice used in the study is given in Supp. Table 1.
Drug Studies
Bortezomib (Velcade, Millennium Pharmaceuticals) was obtained from the pharmacy at Fox Chase Cancer Center. Female adult animals were implanted with an Alzet micro –osmotic pump (Model 1007D) filled with either 0.9% NaCl or bortezomib diluted in 0.9% NaCl. This pump has a mean pumping rate of 0.5 ml/hr. Mice were weighed, and the amount of bortezomib added to each pump was calculated to deliver a final dosage of 0.49 mg/kg/day. Each pump was implanted subcutaneously on the back of the animal. The amount of fluid left in the pump at the end of the experiment was measured to verify that pumping rates were accurate. After 3 or 4 days, the animals were sacrificed, and the liver, kidney and serum were harvested for analysis.
ONX0912 was obtained from Onyx Pharmaceuticals. ONX0912 was administered as a suspension in 1% (w/v) carboxymethylcellulose (CMC) in water. Solutions were made fresh each day. Each animal was dosed once per day by oral gavage. Animals were dosed for four days in a row. After the final dose, animals were sacrificed and tissue harvested 7 hours after last dose.
CBS Western Blot, Enzyme Activity, and Serum tHcy Measures
Liver and kidney homogenates for Western blot and enzyme assays were made as previously described [Wang, et al., 2004]. CBS activity was analyzed in the presence of Adomet, as previously described, by employing a Biochrom 30 amino acid analyzer (Cambridge, UK) to measure cystathionine levels [Gupta, et al., 2008]. One unit of activity is defined as one nanomole of cystathionine formed per milligram of protein per hour. Serum tHcy was measured by Biochrom 30 amino acid analyzer, as previously described [Gupta, et al., 2009]. Native and denaturing gels and Western blotting were performed as previously described [Gupta, et al., 2008]. CBS was detected using a polyclonal anti-human primary antibody [Shan and Kruger, 1998] (1:10,000) and secondary anti-rabbit antibody (1:30,000) conjugated to horseradish peroxidase (Amersham Biosciences, UK). For chaperone proteins, the following antibodies were purchased from Cell Signaling Technology: HSPA4/Apg-2 (#3303); HSPA8 (#8444); HSP90 (#4874); HSP40 (#4868); HSP27 (#2442). Antibody from Stressgen (Hsp70/72, mAb C92F3A-5 #SPA-810) was used to detect Hsp70. A total of 5 micrograms of serum protein was used in the Western blotting for CBG. CBG antibody (rabbit anti-mouse) was gifted by Dr. Geoffrey L. Hammond, Child and Family Research Institute, University of British Columbia, Vancouver, Canada.
Immunoprecipitation
For each immunoprecipitation reaction, 10 μl of human polyclonal CBS anti-serum (Abnova #H00000875-A01) was conjugated to 50 μl of protein A/G agarose (Santa Cruz Biotechnology #sc2003) by incubation overnight at 4°C. To this was added 100 μg of liver cell extract prepared in lysis buffer containing 10 μM Tris pH 7.5 plus protease inhibitor cocktail (Complete mini, Roche). After four hour incubation at 4°C, samples were washed three times with lysis buffer, and material was eluted by boiling in 4X-SDS sample buffer (Novex, Life Technologies). Material was then analyzed by running on 7% SDS Tris-Acetate gels followed by Western blot.
Proteasome Activity Assay
Proteasome activity was quantified via a biochemical assay for chymotrypsin-like (CT-L) activity, utilizing a fluorogenic enzymatic substrate, Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (LLVY-AMC) as described [Kuhn, et al., 2007]. Mouse liver samples for LLVY-AMC testing were dissected from animals, rinsed in PBS, and snap-frozen. Tissue lysates were prepared by bead homogenization of liver in hypotonic lysis buffer (20mM Tris-HCl pH 8.0, 5mM EDTA), followed by centrifugation and collection of supernatant. Lysate was assayed at a final concentration 0.35mg/mL, in combination with 60μM LLVY-AMC. The production of fluorescent AMC was monitored kinetically on a fluorometer, and the detected signal at 30 minutes was used to calculate the nanograms of AMC generated per microgram of total protein assayed (CT-L activity), based on an AMC standard curve.
Microarray Assay
Microarray analysis was done using a mouse 44K oligoarray (G4122F, Agilent technologies) with Cy3 labeling as described previously [Gupta, et al., 2009]. A total of eight hybridizations were performed. Expression intensities were background corrected using the “normexp” method (R/Bioconductor package limma), and quantile-normalized. Differential expression was assessed using limma. A list of differentially regulated genes for responders and non-responders was selected on the basis of log (base 2) fold-change ≥ 1 or ≤ −1 and a p-value less than 0.05. This list was then used for functional pathway analysis employing the WebGestalt data mining system (http://genereg.ornl.gov/webgestalt).
Statistical Analysis
For comparison between two groups, we used a non-parametric two-sided Mann-Whitney U-test using Graphpad Prism software. A P-value less than 0.05 is considered significant.
Results
Proteasome inhibitor ONX 0912 can rescue p.S466L
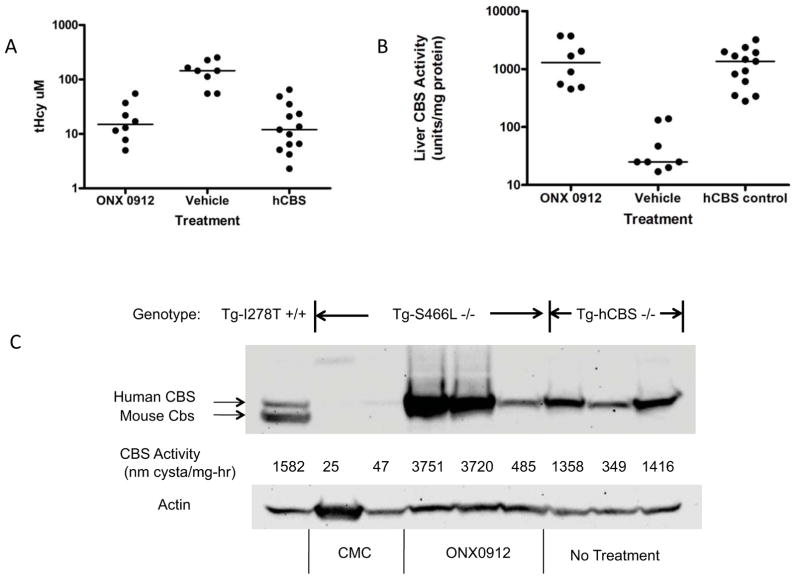
Previously, our lab created three humanized mouse models for CBS deficiency [Gupta, et al., 2008; Wang, et al., 2005; Wang, et al., 2004]. These mice have an insertionally-inactivated mouse Cbs allele and a zinc-inducible transgene that expresses either a wild-type (Tg-hCBS Cbs−/−) human CBS or one of two patient-derived missense-containing human CBS alleles (Tg-I278T Cbs−/− or Tg-S466L Cbs−/−). Like human patients with the p.S466L mutation, Tg-S466L mice have greatly elevated tHcy levels [Gaustadnes, et al., 2000; Gupta, et al., 2008]. The reason for this elevation is that the mutant protein is degraded, resulting in little or no detectable CBS activity in the liver. However, heterologous expression studies in E. coli and S. cerevisiae have shown that when human p.S466L CBS is not degraded, does not respond to AdoMet as an allosteric activator, but is highly enzymatically active [Gupta, et al., 2008; Maclean, et al., 2002]. To see if we could rescue p.S466L CBS function in vivo, we treated a group of Tg-S466L Cbs−/− mice with the proteasome inhibitor ONX 0912. ONX 0912 is an oral proteasome inhibitor developed by Onyx Pharmaceuticals and is currently in human clinical trials as an anti-cancer agent [Zhou, et al., 2009]. As a control, we also treated animals with carboxymethylcellulose (CMC), the vehicle used to make up the ONX 0912 suspension. Vehicle and drug dissolved in vehicle were delivered by oral gavage once daily for four consecutive days, after which the animals were sacrificed and tissues were collected for analysis. ONX 0912 treated animals had on average an 85% drop in median homocysteine levels compared to vehicle treated animals (Fig. 1A and Supp. Table 1; 145 vs. 15 μM, P<0.0003). Consistent with this data, we found that the median liver CBS enzyme activity increased from 25 to 1290 units (P<0.003), a level that is nearly identical to that observed in untreated control Tg-hCBS Cbs−/− animals (Fig. 1B). Increased enzyme activity was observed in the kidney (29 vs. 105 units, P<0.04). Western analysis of liver extracts show that vehicle-treated animals have barely detectable levels of CBS protein, while treated animals have levels at or exceeding those found in Tg-hCBS Cbs−/− animals (Fig. 1C). We also examined if the S466L enzyme in the ONX 0192 treated livers were responsive to AdoMet. In control Tg-hCBS Cbs−/− extracts we observed a 450% increase in CBS activity upon AdoMet addition, while in Tg-S466L Cbs−/− mice treated with ONX 0912, we saw only a 30% increase in activity (n=3, P<0.0007). These results demonstrate that treatment with ONX 0912 stabilizes p.S466L protein, and this stable protein is enzymatically competent to lower plasma homocysteine levels in vivo even though AdoMet responsiveness is greatly diminished.
Figure 1.
ONX 0912 rescue of S466L CBS. A: Graph of tHcy in Tg-S466L Cbs−/− treated with ONX 0912 or vehicle. The column labeled “hCBS” shows values from either untreated or vehicle treated Tg-hCBS Cbs−/− animals. For each column, the dots show individual animals. The line indicates the median. B: Graph of CBS activity in liver extracts from ONX 0912, vehicle, and Tg-hCBS Cbs−/− control mice. C: Western blot of liver extracts in Tg-S466L Cbs−/− mice treated with either vehicle (CMC; carboxymethyl cellulose) or ONX 0912. Also included are extracts from untreated Tg-hCBS Cbs−/− animals (right three lanes), and a control extract from a Tg-I278T Cbs+/− animal. For each lane, CBS activity is also indicated. CMC indicates animals treated with vehicle only.
Heterogeneous response of p.I278T to bortezomib and ONX 0912
We next examined the effect of proteasome inhibitors on mice expressing p.I278T CBS. The p.I278T mutation is the most commonly observed mutation in CBS-deficient patients, and has minimal residual enzyme activity when expressed in human fibroblasts, E. coli, or S. cerevisiae [Chen, et al., 2006; Kluijtmans, et al., 1999]. Previously, our lab has shown that in Tg-I278T Cbs−/− mouse liver, the steady-state level of the protein is somewhat reduced compared to wild-type CBS, and the remaining protein has minimal enzyme activity [Wang, et al., 2005].
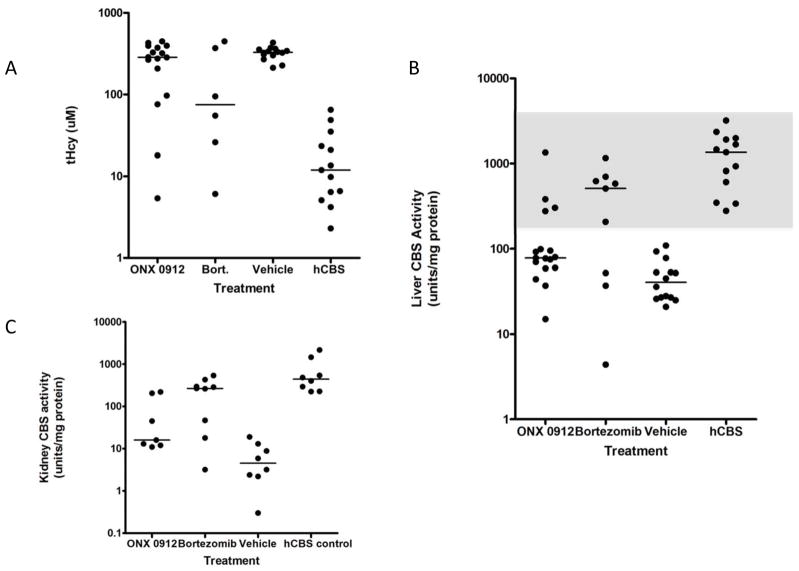
We treated Tg-I278T Cbs−/− mice with two different proteasome inhibitors, ONX 0912 and bortezomib. In humans, bortezomib is used to clinically to treat myeloma and certain lymphomas, and is usually given intravenously. For the studies described here, bortezomib (n=9) was given at a dose of 0.49 mg/kg/day through the use of a surgically implanted osmotic pump, while ONX 0912 (n=17) was given by oral gavage at 40 mg/kg/day. Based on the standard conversion between human and mouse doses [Reagan-Shaw, et al., 2008], both drugs are being given at near the level used in cancer patients. Control animals consisted of seven animals implanted with a saline containing pump, and seven animals gavaged with the ONX 0192 vehicle, CMC. In bortezomib-treated animals, the median liver CBS activity increased from 40.5 to 513 units (P<0.03), median kidney CBS activity increased from 4.55 to 267 units (P<0.0017) and median tHcy decreased from 329 to 75 μM (P<0.20) compared to control animals (Fig. 2). For ONX 0912 treated animals, median liver CBS activity increased from 40.5 to 78 units (P<0.008), median kidney CBS increased from 4.5 to 16 units (P<0.01), and median tHcy decreased from 329 μM to 286 μM (P<0.12).
Figure 2.
ONX 0912 and bortezomib rescue of I278T CBS. A: Graph of tHcy in Tg-I278T Cbs−/− mice treated with ONX 0912, bortezomib, or vehicle (Saline and CMC combined). The column labeled “hCBS” shows values from Tg-hCBS Cbs−/− animals. For each column, the dots show individual animals. Median values are also shown. B: Graph of CBS activity in liver extracts from Tg-I278T Cbs−/− mice treated with ONX 0912, bortezomib, or vehicle. Gray region shows a box identifying responders. C: Graph of CBS activity in kidney extracts from Tg-I278T Cbs−/− mice treated with ONX 0912, bortezomib, or vehicle.
For both drugs, we noticed a high degree of heterogeneity in tHcy, liver CBS activity, and kidney CBS activity compared to the vehicle treated animal group. A proportion of animals in each of the drug treated groups had tHcy, liver CBS, and Kidney CBS activities outside the range defined by the vehicle treated animals, and within the range of those observed in the Tg-hCBS Cbs−/− control group. Using a liver CBS activity of 195 as our cut-point (half-way between the highest vehicle treated animal, and the lowest hCBS control animal; see gray box Fig. 2B), we designated each animal as either a responder or non-responder. Using this criteria, we found that 66% (6/9) of the bortezomib animals were responders, but only 24% (4/19) of the ONX 0912 animals were responders. This difference in response rates is significant (P<0.05, fishers exact). As expected, responsive animals had significantly lower median tHcy (40.5 vs. 349, P<0.0003) and higher kidney CBS activity (277 vs. 14.5, P<0.0003) compared to non-responsive animals, showing that all three measures are highly correlated.
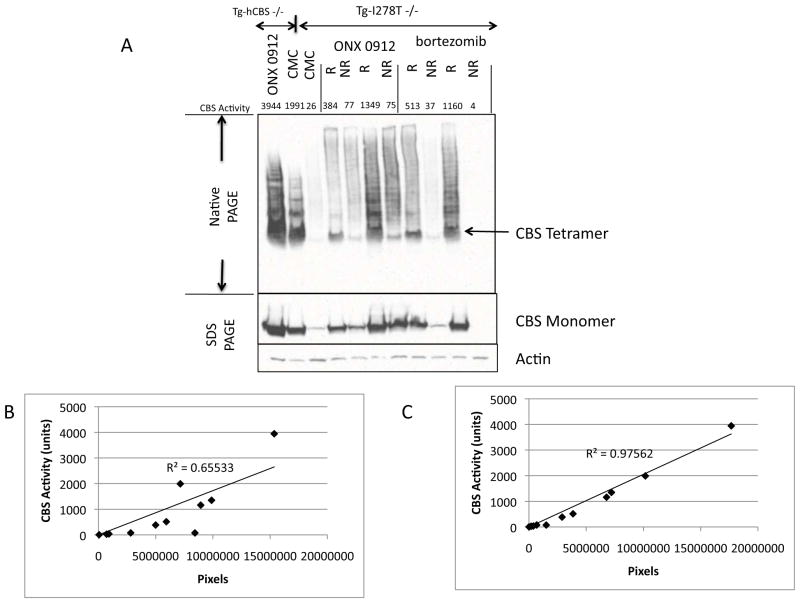
To understand the cause of this heterogeneity, we first examined steady-state CBS levels in untreated, treated non-responding, and treated responding animals. We found that treated animals generally had higher levels of liver p.I278T CBS than untreated animals, but that the amount of CBS protein present on denaturing gels did not correlate very well (R2=0.66) with the amount of CBS activity in the same extract (Fig. 3). Much better correlation (R2=0.97) was observed when activity was compared to the amount of tetrameric CBS as determined by native gels, suggesting that the differences in response are due the percentage of CBS that is correctly folded.
Figure 3.
Steady state CBS levels in ONX 0912- and bortezomib-treated mice. A: Native and denaturing gel Western blot analyses showing CBS levels in ONX 0912- and bortezomib-treated mice. Liver CBS enzyme activity level for the extract is shown above. Controls on left two lanes are Tg-hCBS Cbs−/− mice treated with ONX 0912 or Vehicle (CMC). R and NR stand for responder and non-responder. In Native gels, bands above the tetramer represent higher molecular weight CBS containing complexes. B: Plot of amount of CBS protein as determined by densitometry of the denaturing gel vs. CBS activity. C: Plot of amount of CBS protein as determined by densitometry of the native gel vs. CBS activity.
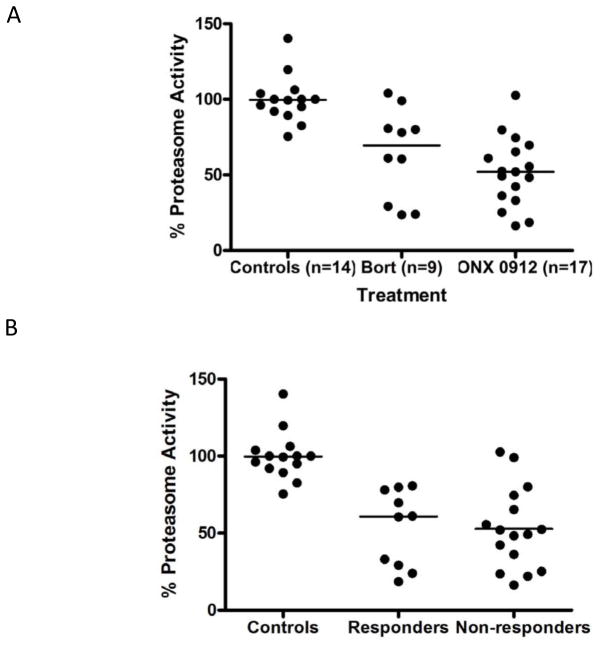
To see if differences in bortezomib and ONX 0912 pharmacokinetics might play a role in response, liver proteasome function was measured in both treated and untreated Tg-I278T animals. As expected, the median proteasome activity was significantly reduced in both bortezomb (P<0.004) and ONX 0912 (P<0.001) treated animals compared to controls (Fig. 4A), but they were not different from each other. When sorted by response status, there was also no statistically significant difference in proteasome activity (Fig. 4B). These findings indicate that differences in the level of proteasome inhibition are not the primary cause of the heterogeneity in CBS response to proteasome inhibitors.
Figure 4.
Inhibition of proteasome activity. A: Proteasome activity measured in liver extracts from mice treated with the indicated drugs. Activity is expressed as a % of the average of the control (vehicle-treated) animals. Line indicates median. B: Proteasome activity in Responders and Non-responders.
Induction of chaperone proteins
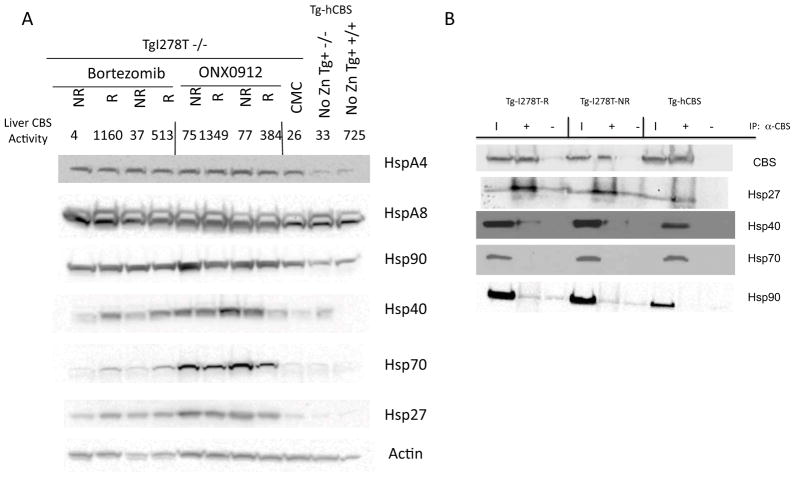
Since Hsp70 induction was required for rescue of p.I278T function in S. cerevisiae [Singh and Kruger, 2009], we wondered if the heterogeneity in response might be related to differential induction of various liver chaperone proteins. Therefore, we measured the steady state levels of a variety of liver chaperones in control, responsive, and non-response mice (Fig. 5A). The chaperones examined included HspA4, HspA8, Hsp90, Hsp40, Hsp70, and Hsp27. Treatment with either bortezomib or ONX 0912 increased expression of all of the Hsp’s over the CMC treated control with the exception of HspA8, a known constitutively-expressed Hsp70 family member [Green and Liem, 1989]. We did observe a slight induction of HspA4 by CMC treatment alone, but an even higher induction occurred when the drugs were given. Hsp90 was only slightly induced by either drug treatment, and no differences were observed between responders and non-responders. Hsp27, Hsp70, and Hsp40 were all robustly induced by ONX 0912, and to a lesser extent by bortezomib. In the bortezomib-treated animals, responders seemed to have higher steady state levels of Hsp70 and Hsp40 compared to non-responders, but in ONX 0912-treated animals, this relationship was reversed. Thus, differential induction of Hsp70 and Hsp40 by itself cannot explain the difference between responsive and non-responsive animals.
Figure 5.
Effect of ONX 0912 and bortezomib on chaperone proteins. A: Western analysis of chaperone proteins in liver from bortezomib, ONX 0912, and control mice. Liver CBS activity for each extract is also shown. Mice are classified as either R (responder) or NR (non-responder) based on CBS activity level. Control mice include a Tg-I278T Cbs−/− mice treated with vehicle (CMC), a Tg-hCBS CBS −/− mouse that was uninduced for CBS expression (No Zn Tg+ −/−), and a mouse that expresses endogenous levels of mouse CBS (No Zn Tg+ +/+). B: Immunoprecipitation experiment examining interaction of Hsp90, Hsp70, Hsp40 and Hsp27 with CBS. Extracts from a Tg-I278T Cbs−/− responder (Tg-I278T-R), Tg-I278T Cbs−/− non-responder (Tg-I278T-NR) and a Tg-hCBS Cbs−/− mouse, all treated with ONX 0912, were used. Lane “I” shows input extract; lane “+” shows material brought down with a-CBS antibody conjugated to beads; lane “−“ shows material brought down by beads alone.
We also examined the interaction of various chaperone proteins with I278T and WT CBS protein by immunoprecipitation (Fig. 5B). In these experiments, we immunoprecipitated CBS from three different liver extracts; I278T responder (R), I278T nonreponder (NR), and hCBS. We were able to pull down easily detectable levels of Hsp27 and Hsp40 with p.I278T CBS, but only weak interaction with Hsp27 and no interaction with Hsp40 was detected with WT hCBS. We were unable to detect any Hsp70 or Hsp90 with either WT or p.I278T CBS. However, we did not observe any consistent differences in extracts from the I278T responder vs. non-responder, suggesting that differences in chaperone interaction cannot explain the increase in ezyme activity seen in responders.
Microarray analysis
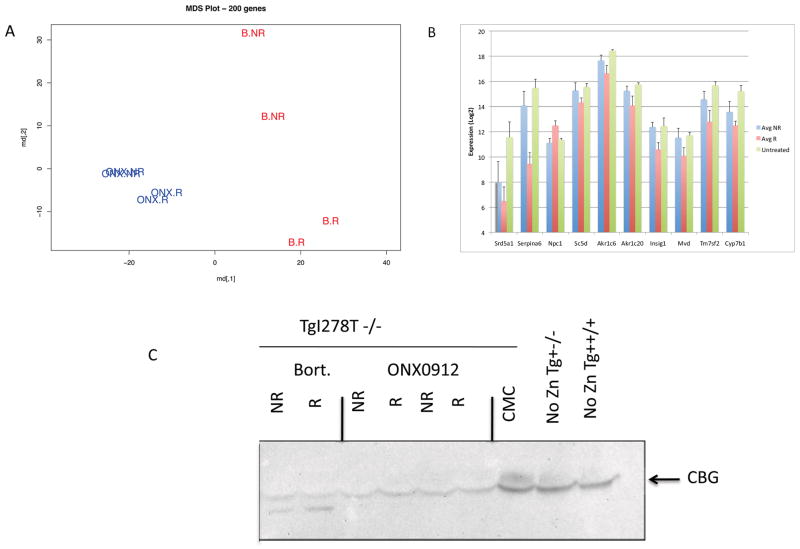
To identify the factors that might contribute to the response heterogeneity of I278T, we turned to microarray expression analysis. We performed expression analysis on livers from 8 animals, four treated with bortezomib and four treated with ONX 0912. We examined two responsive and two non-responsive animals in each group. Multidimensional scaling analysis (MDS) revealed that the largest separation occurred between animals treated with bortezomib and animals treated with ONX 0912, indicating that there are differences in the biological effects of these two compounds (Fig. 6A). We also observed separation within each group between responders and non-responders, although this difference was more pronounced in the bortezomib treated animals. To identify genes that might distinguish between responders and non-responders, we searched for genes that were either up or down-regulated in both bortezomib and ONX 0912 responders. We identified 178 known transcripts that were at least 2-fold up or down regulated with P<0.05 (Supp. Table 2). We then looked for enrichment in gene ontology classes using the Webgestalt Gene Analysis Toolkit 2.0 [Zhang, et al., 2005]. We observed significant enrichment in several categories (see Supp. Table 3), but the most significant P-values were observed in oxido reduction genes (n=23), lipid biosynthetic processes (n=13), steroid metabolic processes (n=10), and steroid biosynthetic process (n=8). As the steroid metabolic process genes were present as a subset in all the groups, we chose to focus on this group for further analysis. Of the 10 genes in this category, 9 were down-regulated in responders relative to non-responders (Fig. 6B). We did not observe significant differences between non-responsive and untreated animals, suggesting that down-regulation of these transcripts is in some way related to the response mechanism. The largest fold-change occurred in the SERPINA6 gene, whose known function is to act as the major corticosteroid binding protein (CBG) in the serum [Henley and Lightman, 2011]. However, we did not observe elevation in CBG levels in serum (Fig. 6C), suggesting that the regulation of CBG may have a large post-transcriptional component.
Figure 6.
Microarray analysis of ONX 0912- and bortezomib-treated responders and non-responders. A: Multi-dimensional scaling plot of the samples. B: Expression data of the nine genes identified as part of the steroid metabolism family (GO:0008202). Shown is expression in NR, R, and untreated animals. Expression levels are shown using a log2 scale. C: Western blot analysis of serum serpin A6 (CBG) levels. Drug treatment and response status of mice are as 27 indicated. Controls on right include vehicle-treated mouse (CMC), Tg-I278T Cbs−/− uninduced mouse (very high tHcy), and Tg-I278T Cbs+/+ (normal tHcy).
Discussion
In this paper, we demonstrate that treatment with proteasome inhibitors can restore significant levels of enzymatic activity to missense mutant human CBS proteins expressed in mice. Rescue of the p.S466L allele occurred in 100% of the animals treated, while rescue of the p.I278T mutation was more variable, with 23.5–66% of the treated animals showing response depending on the drug being used. When response did occur, it was robust, with liver CBS activity and tHcy restored to the normal range. These findings suggest that this class of drugs may be useful in the treatment of CBS deficiency and other inborn errors of metabolism caused by missense alterations that effect protein folding.
The most consistent and robust rescue was observed in animals expressing p.S466L CBS. In mammalian liver, this mutation appears to cause the protein to become unstable and degraded, explaining why it causes CBS deficiency [Gupta, et al., 2008]. However, when the protein is expressed in non-liver systems including E. coli, S. cerevisiae, or Chinese hamster fibroblasts the protein is stable and folds into a conformation that has levels of enzymatic activity approaching that of the AdoMet stimulated wild-type enzyme [Gupta, et al., 2008; Janosik, et al., 2001a; Maclean, et al., 2002]. A hypothesis to explain these findings is that the mutation slows down the kinetics of protein folding, but if the protein is given enough time, it will fold into a native conformation. In fact, it has been recently demonstrated that recombinant human S466L CBS (as well as several other mutant CBS proteins) have reduced thermostability stability relative to wild-type CBS [Pey, et al., 2013]. We speculate that in heterologous expression systems, there is sufficient time for this folding to occur, but in mammalian cells, the partially folded protein is targeted for degradation before it can complete the folding process. It seems likely that inhibition of the proteasome allows the p.S466L to find its native conformation before it gets degraded. This behavior is not unique to p.S466L CBS. There are multiple examples of unique missense mutations present in patients with various inborn errors of metabolism that have significant enzyme activity when expressed in heterologous systems [Carter, et al., 1998; Eiken, et al., 1996; Kim, et al., 1997; Shan, et al., 1999]. It seems likely that at least some of these mutations may have stability defects that only manifest themselves when expressed in the appropriate mammalian or human tissue.
The p.I278T mutant form of the enzyme was more difficult to reactivate. This mutant, unlike p.S466L, appears largely stable and inactive when expressed in either E. coli or S. cerevisiae, but tends to form large aggregates[Chen, et al., 2006; Janosik, et al., 2001b; Singh, et al., 2010]. Thus, in heterologous systems the protein appears to misfold into a stable inactive form. In yeast, it is possible to reverse this misfolding by various strategies, including growing cells in the presence of chemical chaperones, genetic manipulation (e.g., deletion of the small heat shock protein Hsp26), or stressing cells with treatments that induce Hsp70 [Singh, et al., 2007; Singh, et al., 2010; Singh and Kruger, 2009]. In yeast, mutational analysis showed definitively that Hsp70 (SSA2) is required for rescue [Singh and Kruger, 2009]. In the studies described here, we used two different proteasome inhibitors to induce proper folding of p.I278T in vivo. In both cases, we observed heterogenous response, with some animals responding and others displaying minimal detectable changes in tHcy levels and CBS activity. Bortezomib showed a higher rate of response (66%) than ONX 0912 (23.5%). This difference in response rates does not appear to be due to differences in effectiveness of the two drugs on proteasome inhibition, as there was no significant difference in the amount of proteasome inhibition between the two drugs, and there was no difference in proteasome inhibition between responsive and non-responsive animals. A possible explanation for the difference in effectiveness of these drugs is off-target effects. Recent studies indicate that bortezomib has more off-target effects than Carfilzomib (a highly related molecule to ONX-0912), so it is possible that these bortezomib-specific off-target effects are contributing to this increased response rate [Arastu-Kapur, et al., 2011]. However, given the observation that 1) all responsive animals showed robust induction of Hsp70; 2) responders were able to be identified with two structurally different proteasome inhibitors; and 3) p.S466L was efficiently rescued by the more specific proteasome inhibitor ONX-0912, it seems unlikely that the entire mechanism of action of these drugs involves off-target effects.
That being said, the reason for the heterogeneous response is still not understood. It seems unlikely that the differences are related to genetics or aging, as all mice treated were of the same inbred B6 background, and we tended to use age- and sex-matched siblings as controls. We also failed to observe consistent differences in the levels of chaperone proteins between responsive and non-responsive animals. However, we were able to find differences in gene expression between responders and non-responders. Strikingly, mRNA levels of several genes involved in steroid hormone metabolism were found at lower levels in the responders vs. the non-responder animals. These changes are not secondary effects mediated by lower tHcy levels, as previous microarray studies failed to identify them as differentially regulated by tHcy [Gupta, et al., 2009]. How exactly this steroid hormone gene signature change relates to the stochastic variability in drug response remains to be determined.
Our findings in mice show that proteasome inhibitors can restore substantial enzyme activity to some mutant proteins in vivo, and potentially could be used clinically to treat patients with certain missense mutations in CBS or other genetic diseases. Bortezomib was first approved by the FDA in 2003 for the treatment of multiple myeloma and is also used in the treatment of mantle cell lymphoma. The major noted toxicity is peripheral neuropathy, but patients can be on the drug for several years [Gilardini, et al., 2008]. In fact, there is a report of an individual receiving the drug for over seven years with no reported harmful side effects [Richardson, et al., 2012], suggesting that very long-term treatment is possible. In 2012, a second drug in this class, carfilzomib, received FDA approval for the same indication. While bortezomib is a boronate-based inhibitor, carfilzomib and ONX 0912 (the drug used in the current studies) are peptide epxoyketones. Evidence suggests that this chemistry reduces off-target effects of these drugs [Arastu-Kapur, et al., 2011]. Consistent with this idea, carfilzomib does not exhibit a strong peripheral neuropathy effect like bortezomib [Siegel, et al., 2012; Vij, et al., 2012]. It is interesting to note that the liver microarray studies performed here indicate very large differences in the gene expression profiles of animals treated with bortezomib vs. ONX 0912, suggesting that the drugs have very different mechanisms of action. A major unknown in using these drugs for non-cancer related diseases is the proper schedule of dosing.
In summary, the results here show for the first time that proteasome inhibitors can be used to restore function to disease causing missense alleles in mice. Our data provides strong pre-clinical evidence that this class of drugs should be investigated as potentially being useful in treating inborn errors of metabolism and other genetic diseases caused by missense mutations.
Supplementary Material
Supplemental Table 3: Functional Gene Ontology Enrichment Analysis of Responders vs. Non-responders
Supplemental Table 1: All mice used in the study
Supplemental Table 2: Genes differentially expressed in Responders vs. Non-responders
Acknowledgments
We acknowledge the contribution of the FCCC Genomics Facility, Laboratory Animal Facility, Biostatistics Facility in carrying out these studies. We also thank Yue-Sheng Li and Tony Lerro for technical assistance. We also thank the Pharmacy at FCCC for providing the bortezomib.
Grant Sponsors: This work was supported in part by grants from the Hempling Foundation for Homocystinuria Research, the NIH CA06927, R01GM098772 (W.D.K.), Onyx Pharmaceuticals, and an appropriation from the Commonwealth of Pennsylvania.
Footnotes
Disclosure Statement: Partial funding for this work was provided to Dr. Kruger’s laboratory from Onyx Pharmaceuticals.
References
- Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 1999;59:2615–2622. [PubMed] [Google Scholar]
- Arastu-Kapur S, Anderl JL, Kraus M, Parlati F, Shenk KD, Lee SJ, Muchamuel T, Bennett MK, Driessen C, Ball AJ, Kirk CJ. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: a link to clinical adverse events. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2734–2743. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- Bross P, Corydon TJ, Andresen BS, Jorgensen MM, Bolund L, Gregersen N. Protein misfolding and degradation in genetic diseases. Hum Mutat. 1999;14:186–198. doi: 10.1002/(SICI)1098-1004(1999)14:3<186::AID-HUMU2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Carter KC, Byck S, Waters PJ, Richards B, Nowacki PM, Laframboise R, Lambert M, Treacy E, Scriver CR. Mutation at the phenylalanine hydroxylase gene (PAH) and its use to document population genetic variation: the Quebec experience. European journal of human genetics : EJHG. 1998;6:61–70. doi: 10.1038/sj.ejhg.5200153. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang L, Fazlieva R, Kruger WD. Contrasting behaviors of mutant cystathionine beta-synthase enzymes associated with pyridoxine response. Hum Mutat. 2006;27:474–482. doi: 10.1002/humu.20320. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- Eiken HG, Knappskog PM, Apold J, Flatmark T. PKU mutation G46S is associated with increased aggregation and degradation of the phenylalanine hydroxylase enzyme. Hum Mutat. 1996;7:228–238. doi: 10.1002/(SICI)1098-1004(1996)7:3<228::AID-HUMU7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Gaustadnes M, Rudiger N, Rasmussen K, Ingerslev J. Intermediate and severe hyperhomocysteinemia with thrombosis: a study of genetic determinants. Thromb Haemost. 2000;83:554–558. [PubMed] [Google Scholar]
- Gilardini A, Marmiroli P, Cavaletti G. Proteasome inhibition: a promising strategy for treating cancer, but what about neurotoxicity? Curr Med Chem. 2008;15:3025–3035. doi: 10.2174/092986708786848622. [DOI] [PubMed] [Google Scholar]
- Green LA, Liem RK. Beta-internexin is a microtubule-associated protein identical to the 70-kDa heat-shock cognate protein and the clathrin uncoating ATPase. The Journal of biological chemistry. 1989;264:15210–15215. [PubMed] [Google Scholar]
- Gupta S, Kühnisch J, Mustafa A, Lhotak S, Schlachterman A, Slifker MJ, Klein-Szanto A, High KA, Austin RC, Kruger WD. Mouse models of cystathionine β-synthase deficiency reveal significant threshold effects of hyperhomocysteinemia. FASEB J. 2009;23:883–893. doi: 10.1096/fj.08-120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Wang L, Hua X, Krijt J, Kozich V, Kruger WD. Cystathionine β-synthase p.S466L mutation causes hyperhomocysteinemia in mice. Hum Mutat. 2008;29:1048–1054. doi: 10.1002/humu.20773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Henley DE, Lightman SL. New insights into corticosteroid-binding globulin and glucocorticoid delivery. Neuroscience. 2011;180:1–8. doi: 10.1016/j.neuroscience.2011.02.053. [DOI] [PubMed] [Google Scholar]
- Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry (Mosc) 2001a;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- Janosik M, Oliveriusova J, Janosikova B, Sokolova J, Kraus E, Kraus JP, Kozich V. Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am J Hum Genet. 2001b;68:1506–1513. doi: 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CE, Gallagher PM, Guttormsen AB, Refsum H, Ueland PM, Ose L, Folling I, Whitehead AS, Tsai MY, Kruger WD. Functional modeling of vitamin responsiveness in yeast: a common pyridoxine-responsive cystathionine beta-synthase mutation in homocystinuria. Hum Mol Genet. 1997;6:2213–2221. doi: 10.1093/hmg/6.13.2213. [DOI] [PubMed] [Google Scholar]
- Kluijtmans LA, Boers GH, Kraus JP, van den Heuvel LP, Cruysberg JR, Trijbels FJ, Blom HJ. The molecular basis of cystathionine beta-synthase deficiency in Dutch patients with homocystinuria: effect of CBS genotype on biochemical and clinical phenotype and on response to treatment. Am J Hum Genet. 1999;65:59–67. doi: 10.1086/302439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecka J, Krijt J, Rakova K, Kozich V. Restoring assembly and activity of cystathionine beta-synthase mutants by ligands and chemical chaperones. J Inherit Metab Dis. 2011;34:39–48. doi: 10.1007/s10545-010-9087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FW, Chanan-Khan AA, Orlowski RZ. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean KN, Gaustadnes M, Oliveriusova J, Janosik M, Kraus E, Kozich V, Kery V, Skovby F, Rudiger N, Ingerslev J, Stabler SP, Allen RH, et al. High homocysteine and thrombosis without connective tissue disorders are associated with a novel class of cystathionine beta-synthase (CBS) mutations. Hum Mutat. 2002;19:641–655. doi: 10.1002/humu.10089. [DOI] [PubMed] [Google Scholar]
- Majtan T, Liu L, Carpenter JF, Kraus JP. Rescue of cystathionine beta-synthase (CBS) mutants with chemical chaperones: purification and characterization of eight CBS mutant enzymes. J Biol Chem. 2010;285:15866–15873. doi: 10.1074/jbc.M110.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield JA, Davies MW, Dimster-Denk D, Pleskac N, McCarthy S, Boydston EA, Fink L, Lin XX, Narain AS, Meighan M, Rine J. Surrogate genetics and metabolic profiling for characterization of human disease alleles. Genetics. 2012;190:1309–1323. doi: 10.1534/genetics.111.137471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd SH, Finkelstein JD, Irreverre F, Laster L. Homocystinuria: an enzyme defect. Science. 1964;143:1443–1444. doi: 10.1126/science.143.3613.1443. [DOI] [PubMed] [Google Scholar]
- Mudd SH, Skovby F, Levy HL, Pettigrew KD, Wilcken B, Pyeritz RE, Andria G, Boers GH, Bromberg IL, Cerone R, et al. The natural history of homocystinuria due to cystathionine beta-synthase deficiency. Am J Hum Genet. 1985;37:1–31. [PMC free article] [PubMed] [Google Scholar]
- Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- Pey AL, Majtan T, Sanchez-Ruiz JM, Kraus JP. Human cystathionine beta-synthase (CBS) contains two classes of binding sites for S-adenosylmethionine (SAM): complex regulation of CBS activity and stability by SAM. Biochem J. 2013;449:109–121. doi: 10.1042/BJ20120731. [DOI] [PubMed] [Google Scholar]
- Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- Richardson PG, Laubach JP, Schlossman RL, Ghobrial IM, Redman KC, McKenney M, Warren D, Noonan K, Lunde L, Doss D, Colson K, Hideshima T, et al. The potential benefits of participating in early-phase clinical trials in multiple myeloma: long-term remission in a patient with relapsed multiple myeloma treated with 90 cycles of lenalidomide and bortezomib. Eur J Haematol. 2012;88:446–449. doi: 10.1111/j.1600-0609.2012.01765.x. [DOI] [PubMed] [Google Scholar]
- Shan X, Kruger WD. Correction of disease-causing CBS mutations in yeast. Nat Genet. 1998;19:91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- Shan X, Wang L, Hoffmaster R, Kruger WD. Functional characterization of human methylenetetrahydrofolate reductase in Saccharomyces cerevisiae. J Biol Chem. 1999;274:32613–32618. doi: 10.1074/jbc.274.46.32613. [DOI] [PubMed] [Google Scholar]
- Siegel DS, Martin T, Wang M, Vij R, Jakubowiak AJ, Lonial S, Trudel S, Kukreti V, Bahlis N, Alsina M, Chanan-Khan A, Buadi F, et al. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood. 2012 doi: 10.1182/blood-2012-05-425934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LR, Chen X, Kozich V, Kruger WD. Chemical chaperone rescue of mutant human cystathionine β-synthase. Mol Genet Metab. 2007;91:335–342. doi: 10.1016/j.ymgme.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LR, Gupta S, Honig NH, Kraus JP, Kruger WD. Activation of mutant enzyme function in vivo by proteasome inhibitors and treatments that induce Hsp70. PLoS Genet. 2010;6:e1000807. doi: 10.1371/journal.pgen.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LR, Kruger WD. Functional rescue of mutant human cystathionine β-synthase by manipulation of Hsp26 and Hsp70 levels in Saccharomyces cerevisiae. J Biol Chem. 2009;284:4238–4245. doi: 10.1074/jbc.M806387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Vij R, Siegel DS, Jagannath S, Jakubowiak AJ, Stewart AK, McDonagh K, Bahlis N, Belch A, Kunkel LA, Wear S, Wong AF, Wang M. An open-label, single-arm, phase 2 study of single-agent carfilzomib in patients with relapsed and/or refractory multiple myeloma who have been previously treated with bortezomib. Br J Haematol. 2012;158:739–748. doi: 10.1111/j.1365-2141.2012.09232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen X, Tang B, Hua X, Klein-Szanto A, Kruger WD. Expression of mutant human cystathionine beta-synthase rescues neonatal lethality but not homocystinuria in a mouse model. Hum Mol Genet. 2005;14:2201–2208. doi: 10.1093/hmg/ddi224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jhee KH, Hua X, DiBello PM, Jacobsen DW, Kruger WD. Modulation of cystathionine beta-synthase level regulates total serum homocysteine in mice. Circ Res. 2004;94:1318–1324. doi: 10.1161/01.RES.0000129182.46440.4a. [DOI] [PubMed] [Google Scholar]
- Wilcken DE, Wilcken B. The natural history of vascular disease in homocystinuria and the effects of treatment. J Inherit Metab Dis. 1997;20:295–300. doi: 10.1023/a:1005373209964. [DOI] [PubMed] [Google Scholar]
- Yap S, Boers GH, Wilcken B, Wilcken DE, Brenton DP, Lee PJ, Walter JH, Howard PM, Naughten ER. Vascular outcome in patients with homocystinuria due to cystathionine beta-synthase deficiency treated chronically: a multicenter observational study. Arterioscler Thromb Vasc Biol. 2001;21:2080–2085. doi: 10.1161/hq1201.100225. [DOI] [PubMed] [Google Scholar]
- Yap S, Naughten ER, Wilcken B, Wilcken DE, Boers GH. Vascular complications of severe hyperhomocysteinemia in patients with homocystinuria due to cystathionine beta-synthase deficiency: effects of homocysteine-lowering therapy [In Process Citation] Semin Thromb Hemost. 2000;26:335–340. doi: 10.1055/s-2000-8100. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HJ, Aujay MA, Bennett MK, Dajee M, Demo SD, Fang Y, Ho MN, Jiang J, Kirk CJ, Laidig GJ, Lewis ER, Lu Y, et al. Design and synthesis of an orally bioavailable and selective peptide epoxyketone proteasome inhibitor (PR-047) J Med Chem. 2009;52:3028–3038. doi: 10.1021/jm801329v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 3: Functional Gene Ontology Enrichment Analysis of Responders vs. Non-responders
Supplemental Table 1: All mice used in the study
Supplemental Table 2: Genes differentially expressed in Responders vs. Non-responders