Abstract
The current study clarifies the role of the Glycosaminoglycan (GAG)-binding domain of insulin-like growth factor binding protein-3 (IGFBP-3) in cell penetration. The cell penetration function of IGFBP-3 has been mapped to an 18-residue GAG-binding domain in the C-terminal region that mobilizes cellular uptake and nuclear localization of unrelated proteins. Uptake of KW-22, a 22-residue peptide that encompasses the 18-residue GAG-binding domain, and another IGFBP-3 peptide carrying a streptavidin protein cargo was investigated in Chinese hamster ovary (CHO) cells defective at several steps of biosynthesis of cell surface GAGs. The severity of GAG truncation was highly correlated to the impairment of uptake ranging from complete abrogation to only a partial reduction, suggesting that GAG-binding is required for uptake. The 18-residue GAG-binding domain consists of an 8-residue KK-8 basic sequence devoid of Arg and an adjacent 10-residue QR-10 sequence rich in Arg. Peptide mapping of uptake and GAG-binding activities within the KW-22 peptide showed that the 8-residue KK-8 basic peptide retained 80% of GAG-binding activity with no uptake activity while the 10-residue QR-10 peptide retained 53% of uptake activity and 18% of GAG-binding activity. This suggests that KK-8 carries out the majority of GAG-binding function while QR-10 carries out the majority of the cell entry function. To our knowledge, this is the first report of physical separation of the uptake and GAG-binding functions within a short cell penetrating peptide and may shed light on the general mechanism of uptake of Arg-rich CPPs and guide new design of Arg-rich CPP-assisted drug/gene delivery systems.
Keywords: IGFBP-3, C-terminus, GAG, Cellular Uptake, cell penetration
INTRODUCTION
Insulin-like growth factors (IGFs) are potent mitogens that play a role in cell survival, proliferation, and differentiation [1]. A family of IGF-binding proteins mediates IGF actions through specific and high affinity binding to IGFs [2,3]. IGFBP-3, the major member of the family in adult serum, serves as a carrier of IGFs and directly modulates the biological effects of IGFs by prolonging their half lives and regulating their bioavailability [4]. In addition to its role of carrying IGFs, IGFBP-3 itself can exert effects on cellular growth and apoptosis. Although their exact mechanisms are not known, these IGF-independent effects may involve specific binding to many extracellular matrix and cell surface molecules, conditional proteolysis, rapid entry of intact IGFBP-3 (or its fragments) into target cells, and nuclear transport within target cells [5,6].
Efforts have been focused on IGFBP-3 C-terminus that consists of several domains with distinct functions. IGFBP-3 binds to transferrin (Tf), which depends on a domain in the C-terminus. A two amino acid substitution in the domain dramatically disrupts Tf-binding affinity [7]. The bipartite nuclear localization sequence (NLS) overlaps with the Tf-binding domain [8,9]. Another highly basic heparin-binding domain overlaps with both the NLS and the Tf-binding domain. Since several GAGs bind to this highly basic domain, it is also known as GAG-binding domain [10,11]. A caveolin-scaffolding domain consensus sequence, FCWCVDKY, has also been identified near the very end of the C-terminus [8,12].
Nuclear import of IGFBP-3, the appearance of exogenously added IGFBP-3 in the nucleus of cultured mammalian cells, has been well described and demonstrated in a variety of cellular models [8,9,13–15]. The C-terminus of IGFBP-3 contains a region with strong sequence homology to NLS consensus motif [16] and nuclear transport is facilitated by importin-β factor [14], suggesting the use of the nuclear import mechanism from the cytosol. In contrast to the nuclear localization, the mechanism and pathways by which IGFBP-3 enters cells remain poorly understood. Chemical inhibition of receptor-mediated endocytosis did not affect nuclear uptake of IGFBP-3, suggesting that IGFBP-3 uses a receptor-independent entry pathway for translocation across the plasma membrane [9]. However, recent reports assert that blocking transferrin receptor-1-mediated endocytosis prevents IGFBP-3 uptake and inhibitors of caveolae formation also retard IGFBP-3 nuclear appearance [8,12]. To identify the sequences essential for IGFBP-3 internalization, several short peptides within the C-terminus were used as carriers of unrelated reporter proteins such as green fluorescent protein and streptavidin-horseradish peroxidase [17]. It was found that a 14-mer peptide present in the C-terminus of IGFBP-3 was sufficient for cellular uptake. Later, a longer 22-mer peptide expanded from the 14-mer was exploited as the carrier of therapeutic molecules selectively targeting cancer cells [18]. These two peptides encompass the Tf-binding domain, but not the complete caveolin-scaffolding domain. There is no solid evidence to support that these two peptides permeate into the cells by binding to transferrin or to proteins harboring the caveolin scaffolding docking sequence.
While all of these sequences/domains in the C-terminus of IGFBP-3 can conceivably be implicated in various steps of cellular entry, the GAG-binding domain appears more directly involved in cellular entry since it may mediate the first step of IGFBP-3 cellular internalization. Studies on the import mechanism of other positively charged basic peptides [19] into cells have shown that heparan sulfate proteoglycans are surface receptors for these basic peptides. It is not clear whether the GAG-binding domain in IGFBP-3 also uses cell surface GAGs as receptors to enter cells. In view of potential clinical application of IGFBP-3 and an increasing interest in using IGFBP-3 peptides for intracellular cargo delivery [18,20], we further characterized the role of the GAG-binding domain in cellular entry. We found that GAGs are indeed cell surface receptors for GAG-binding domain and within the GAG-binding domain the cell entry function and the GAG-binding function are physically separated.
MATERIALS AND METHODS
Reagents
All peptides were synthesized and characterized by CHI Scientific, Inc. (Maynard, MA). At least 85% of molecules in a peptide population were full-length. Chondroitin sulfates were purchased from Sigma-Aldrich (St. Louis, MO), heparan sulfate from Celsus Laboratories (Sharonville, OH), 7-Aminoactinomycin D (7-AAD) staining solution from BD Biosciences (San Jose, CA). Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12), fetal bovine serum (FBS), penicillin/streptomycin were purchased from Life Technologies (Carlsbad, CA)
Cell Lines and Culture
The wild type CHO K1 cell line and its derivatives deficient in GAG biosynthesis were graciously provided by Jeffrey D. Esko (University of California at San Diego, CA). Mutant CHO K1 derivative cell lines include pgs A-745, pgs C-605, pgs D-677, and pgsF-17. All cell lines were cultured in DMEM/F12 supplemented with 10% FBS, 1% penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37 °C.
Cellular Uptake of FITC-labeled Peptides
Wild-type CHO and mutant cells were seeded in 6 or 24-well plates. After 48 hours, cell cultures at ~75% confluence were washed with PBS and incubated for 2 h at 37°C with medium containing 20 μM FITC-labeled peptides. For GAG supplement experiments, FITC-labeled peptides premixed with 0.6 mg/ml of chondroitin sulfate A, B, C or heparan sulfate in media were added to the cultures. After 2 h incubation, cells were washed once with PBS, trypsinized for 5 min at 37°C, washed three times by centrifugation, and resuspended in PBS. The total cell-associated fluorescence was quantified using flow cytometry.
Flow Cytometric Analysis of Peptide Uptake
Cells following the uptake step described above were counted and resuspended in PBS at a final density of 2×106 cells/ml. 7-AAD was added to a final concentration of 0.5 ng/μl 10 min prior to flow cytometry analysis. Cells stained with 7-AAD were excluded as dead cells from analysis. Fluorescence analysis was performed using a 488-nm argon ion laser line in a Cytomics™ FC 500 flow cytometer from Beckman coulter (Fullerton, CA). A minimum of 5,000 events per sample was collected from the 7-AAD-negative population.
Confocal Microscopy for Internalized Peptides
Cells were treated as described above for cellular uptake and attached to chamber cover glasses pre-coated with poly-D-lysine. Internalized peptides were visualized using an oil immersion objective on an inverted Leica TCS SP5 AOBS laser-scanning confocal microscope in XYZ mode using the sequential imaging acquisition to avoid color spill from one channel to another. A middle section of about 0.40 micron thickness of the Z-stacks was presented as each reported image.
Cellular Uptake of Peptide-horseradish peroxidase (HRP) complex
Biotinylated KG-28 peptide (Table 1) and streptavidin-horseradish peroxidase (SA-HRP) at the molar ratio of 50:1 were co-incubated in PBS for 30 min at room temperature to form a KG-28/SA-HRP complex. Free peptide was separated from the complex by filtration using centricon-30 filter from Millipore (Billerica, MA). The enzymatic activity of the complex was determined before use. Cells were seeded in 6-well plates and incubated for 48 hours to about 75% confluence. Complexes were added to fresh media at 240 ng/ml and incubated with the cells for 30 min at 37°C. Afterwards, the cells were washed twice with PBS plus 1% calf serum. Cell extracts were made using M-PER Mammalian Protein Extraction Reagent (Thermo Scientific, Rockford, IL) and assayed for peroxidase activity using tetramethylbenzidine (TMB) liquid substrate system (Sigma, T0440). For each uptake experiment, SA-HRP alone was added to the media as a negative control. 96-well plates loaded with assayed cell lysates were read at 405 nm on a GENios Pro plate reader (Tecan, Switzerland).
Table 1.
Primary structures of the peptides used in this study.
| Name | AA in IGFBP-3 | Sequence |
|---|---|---|
| KG-28 | 215–242 | Biotin-Ahx-KKGFYKKKQCRPSKGRKRGFCWAVDKYG-NH2 |
| KW-22 | 215–236 | FITC-Ahx-KKGFYKKKQCRPSKGRKRGFCW-NH2 |
| QW-14 | 223–236 | FITC-Ahx-QCRPSKGRKRGFCW–NH2 |
| KK-8 | 215–222 | FITC-Ahx-KKGFYKKK–NH2 |
| QW-14m | 223–236 | FITC-Ahx-QGRPSKGRKRGFCW–NH2 |
| QR-10 | 223–232 | FITC-Ahx-QCRPSKGRKR–NH2 |
GAG-binding
FITC-labeled peptides (20μM) were incubated with 0.6 mg/ml of chondroitin sulfates A, B, C, and heparan sulfate, respectively, in PBS for one hour at 37°C. The bound and unbound peptides were separated by filtration using 10 KD Microcon from Millipore (Billerica, MA) and the fluorescence of the bond peptides placed in 96-well plates were read at the excitation/emission of 485/535 nm in the plate reader.
Statistical Analysis
Data are expressed as mean of triplicate experiments ± S.D. Statistical analyses were performed using unpaired, two-tailed Student’s t test. Differences are considered statistically significant when the p values are <0.05.
RESULTS
Cellular Uptake of IGFBP-3-derived Peptide KW-22 Depends on Cell Surface GAGs
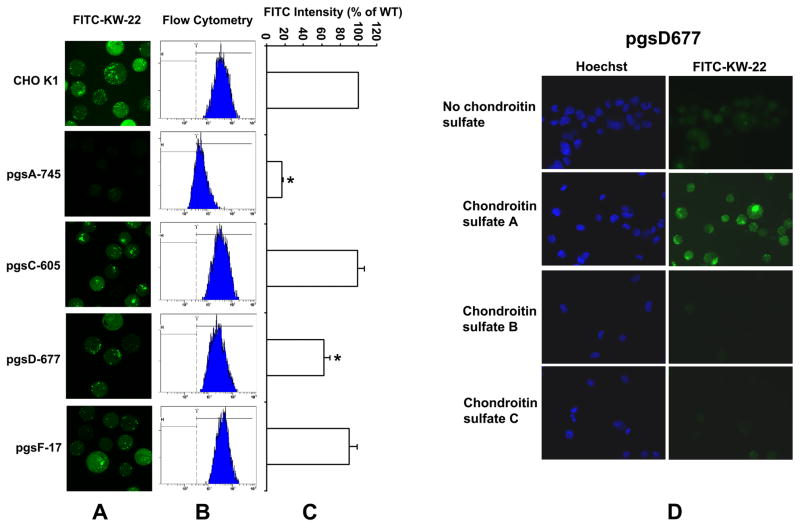
Most polycationic macromolecules and cationic peptides enter cells initially through electrostatic interaction with cell-surface heparan sulfate molecules, followed by endocytosis of the resulting complexes [21]. It is also known that cells internalize surface heparan sulfate proteoglycans through an endocytic pathway and may internalize ligands that bind to their GAG chains [22,23]. The existence of the GAG-binding domain in the IGFBP-3 C-terminal region suggests that cell surface GAGs may be receptors for IGFBP-3 and its fragments. To test this hypothesis, wild-type CHO K1 cells and several mutant cell lines derived from CHO K1 were used to examine the cellular internalization of KW-22, a 22-mer peptide from IGFBP-3 C-terminal region that encompasses the GAG-binding domain (Fig. 1). These mutant cell lines are defective at different steps of GAG biosynthesis, resulting in mutant GAGs with varying degrees of loss of glycosylation.
Figure 1.

IGFBP-3 C-terminal region sequence, showing several putative functional domains located 22 residues to the very C-terminus of the protein. They include the nuclear localization sequence (NLS) domain, the transferring-binding region, the glycosaminoglycan-binding domain (GAG-binding domain) and the caveolin scaffolding docking domain [8]. The first three domains are overlapping. Shown also is the double point mutation (K228E, R230G) in IGFBP-3 that impaired transferring binding activity [7]. The boxed sequence is the QR-10 uptake sequence uniquely identified in this study
Mutant pgs A-745 cell line produces less than 1% of GAGs produced by the wild-type (wt) CHO K1 cell line, since it has a defect in xylosyltransferase - the first sugar transferase in GAG synthesis [24]. Mutant cell line pgs C-605 is defective in the sulfate transporter of cell surface, but can produce heparan sulfate and chondroitin sulfate chains via endogenous formation of sulfate from sulfur containing amino acids [25]. Mutant cell line pgs D-677 produces chondroitin sulfate but is defective in the synthesis of heparan sulfate as a result of lacking N-acetylglucosaminyl transferase and glucuronyltransferase activities [26]. Mutant cell line pgs F-17 does not perform 2-O-sulfation of heparan sulfate, because it lacks sulfotransferase activity [27] (Fig. 2). All mutant cell lines were derivatives of the wt cell line; therefore, they are all isogenic.
Figure 2.
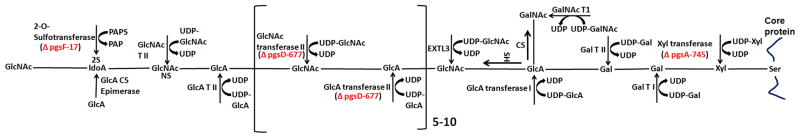
Glycosaminoglycan (GAG) biosynthesis pathway, showing several mutant cells defective at different steps of the pathway. pgs A-745 cell line lacks xylosyltransferase and does not produce detectable levels of GAGs. pgs D-677 cell line is defective for N-acetylglucosaminyl (GlcNAc)- and glucuronyl (GlcA)-transferase activities required for heparan sulfate synthesis. pgs F-17 cell line lacks sulfotransferase activity required for 2-O-sulfation of heparan sulfate. pgs C-605 cell line defective in the sulfate transporter on cell surface is not shown in this figure. Xyl, xylose; UDP, uridine-diphosphate; Gal, galactose; Gal T I, galactose transferase I; Gal T II, galactose transferase II; GalNAc, N-acetylgalactosamine; GalNAc T I, N-Acetylgalactosamine transferase I; CS, chondroitin sulfate; HS, heparan sulfate; GlcA, glucuronic acid; GlcNAc, N-Acetylglucosamine; EXTL3, exostosin-like 3; GlcA T II, glucuronic acid transferase II; GlcNAc T II, N-Acetylglucosamine transferase II; IdoA, Iduronic acid; PAPS, 3′-phosphoadenyl-5′-phospho sulfate; PAP, 3′-phosphoadenyl-5′-phosphate.
Fluorescence-labeled KW-22 peptide (FITC-KW-22) (Table 1) was added to the culture media of wild type and mutant cells, and cellular fluorescence was analyzed 2 h later by confocal microscopy and flow cytometry. In confocal microscopy (Fig. 3A), both punctate and diffuse types of fluorescence were seen within wild-type (wt) and other isogenic partially GAG-defective cells, except for severely GAG-defective A-745 cells, suggesting both endosomal and cytosolic localization of internalized peptide. Flow cytometry quantitatively showed the difference in uptake (Fig. 3B&C). Compared to wt CHO K1 cells, fluorescence in A-745 cells was reduced to less than 20% of the wt level, consistent with the confocal images suggesting that GAG is required for cell penetration of IGFBP-3-derived peptides. Fluorescence in D-677 cells was reduced to ~62% of wt level. Since pgs D-677 cells are defective in the synthesis of heparan sulfate, the result suggests that heparan sulfate in GAG partially contributes to the uptake of FITC-KW-22 peptide (~ 38% of total uptake), with the remainder mainly contributed by other negatively charged components of GAG such as compensatory over-expression of chondroitin sulfates [26]. To test this hypothesis, we included chondroitin sulfate A, B or C, in the KW-22 uptake reactions by pgs D-677 cells (Fig. 3D). The results showed that while chondroitin sulfate B and C inhibited uptake in pgs D-677 as expected, exogenous choidroitin sulfate A actually increased uptake. Interpretation of the effects of chondroitin sulfate A, B and C needs further studies but the results are suggestive of a role of chondroitin sulfate GAGs in KW-22 uptake. Fluorescence level of pgs C-605 cells is comparable with that of the wt cells, probably because of endogenous sulfate production compensatory to the cell surface sulfate transport defect in this mutant cell line [25]. Similarly, uptake in F-17 cells was comparable to that in wt cells since F-17 cells are defective only for 2-O-sulfation of heparan sulfate, resulting in a minor loss of negative charge at the last step of GAG synthesis [27]. Taken together, Fig. 3 shows for the first time that the cellular uptake of IGFBP-3 C-terminal GAG-binding peptides indeed relies on the existence of cell surface GAGs.
Figure 3.
GAG-dependent uptake of FITC-labeled KW-22 peptide in wild-type CHO K1 and its GAG-defective isogenic cell lines. Cells were incubated with DMEM/F12 medium containing 20 μM FITC-KW-22 for 2h at 37°C and washed before study. (A) confocal images, (B) cellular fluorescence intensity distribution in flow cytometry and (C) uptake amount relative to that in wild-type cell line CHO K1. * indicates statistically significance between the wild-type cell line and a mutant cell line in uptake.
IGFBP-3 Peptide-mediated Protein Uptake
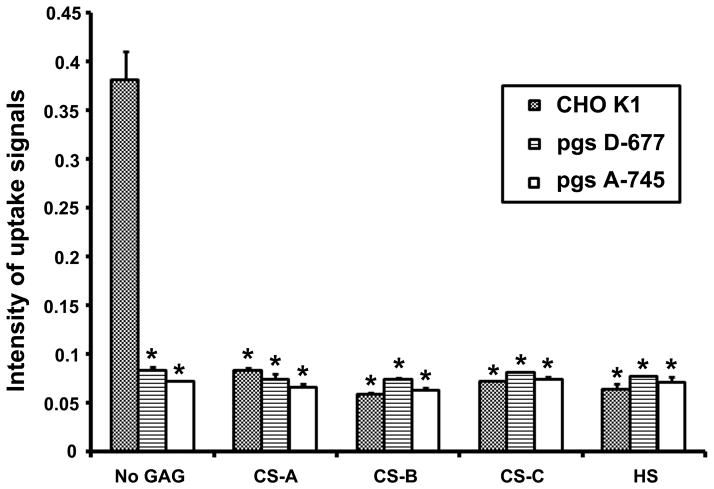
Previous work indicates that the uptake of an 18-mer peptide (position 215–232, 4 amino acids shorter than peptide KW-22, table 1) fused to GST protein is inhibited competitively by the presence of soluble heparin [28]. To further examine if the uptake of protein-conjugated peptides differs from unconjugated peptides and absolutely depends on the presence of cell surface heparan sulfate or other GAG receptors, we use a 28-mer peptide (KG-28, position 215–242) in this experiment that includes several previously described functional domains and exhibits strong activity to bring unrelated proteins into cells [17]. In addition, we also use heparan sulfate instead of heparin because the later is synthesized only from mast cells and differs from cell surface heparan sulfate in molecular size and sulfate density. When wild type CHO K1 cells were incubated with 240 ng/ml complex of biotinylated peptide KG-28 and streptavidin-horseradish peroxidase (SA-HRP) for 30 min, the complex was quickly taken up into cells, and no uptake of SA-HRP alone was detected (Figure 4). However, the uptake of KG28-SA-HRP complex was significantly arrested in mutant cells lacking surface GAGs (pgs A-745) or heperan sulfates (pgs D-677). Moreover, uptake of KG-28-SA-HRP was inhibited or remained at baseline levels in wt and mutant cells in the presence of soluble chondroitin sulfates-A, -B, -C, or heperan sulfates (Figure 4). These observations are in agreement with the previous report that heparin inhibits the uptake of IGFBP-3 and -5 derived peptides fused to unrelated GST protein [28]. In addition, all three chondroitin sulfates inhibited the uptake of KG-28-SA-HRP complex into CHO K1 cells to the same degree as heparan sulfate did, suggesting heparan sulfate is not the unique GAG receptor for cellular uptake of IGFBP-3. The partial reduction of FITC-KW-22 uptake in the heparan sulfate-deficient pgs D-677 cells also suggests the involvement of other GAG species (Fig. 3).
Figure 4.
GAG-dependent uptake of KG-28 peptide complexed with streptavidin-horseradish peroxidase (SA-HRP). Uptake into wild-type CHO K1, HS-defective pgs D-677 and GAG-defective pgs A-745 cells in the absence and presence of CS-A, B, C, or HS was described in Materials and Methods with SA-HRP as the negative control. * indicates statistically significance between uptake in wild-type cells in the absence of a GAG and presence of a GAG and between uptake in wild-type cells in the absence of a GAG and uptake in any one of the two mutant cell line regardless of presence or absence of a GAG (n=3, p< 0.05).
Identification of IGFBP-3 C-terminal Core Sequence Essential for Cell Internalization
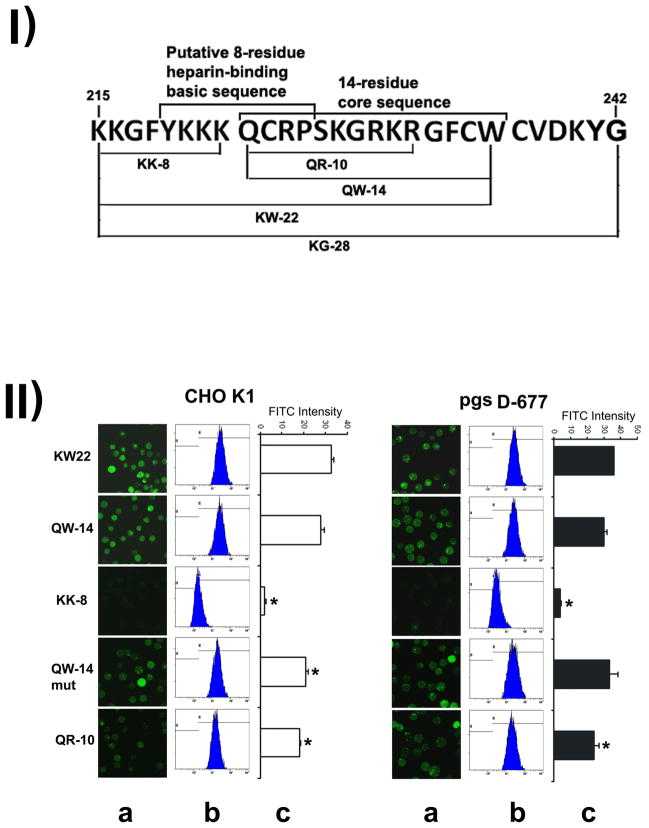
The region encompassing residue 215–232 in the C-terminus of IGFBP-3 was initially characterized as a GAG-binding domain important for IGFBP-3 internalization [10,11] (Fig. 1). Within this domain, a nuclear localization sequence [14] is obvious (Fig. 1). However, later it was shown by some investigators that the uptake of IGFBP-3 and various IGFBP-3 peptides fused to unrelated proteins is mediated by a shorter uptake core sequence (a 14-mer, residue 223–236) that spans from the middle of the GAG-binding domain (215–232) to residue 236 [17] (Fig. 1). Within the 14-mer, the uptake function of IGFBP-3 can be largely mapped to a still shorter 12-mer, a deduced Cys-Cys loop (224–235) [17] (Fig. 1). This observation indicates that the uptake core sequence (223–236) within the GAG-binding domain exists beyond the putative heparin-binding basic sequence YKKKQCRP (219–226; XBBBXXBX, B=basic amino acid, and X=non-basic amino acid) [29,30].
We hypothesized that the so-called GAG-binding domain encompasses both the GAG-binding core sequence and the uptake core sequence. To test this hypothesis, we analyzed several wild type and mutant peptides (Fig. 5-I) within the so-called GAG-binding domain for internalization into wt CHO K1 and heperan sulfate-defective pgsD-677 cells.
Figure 5.
Mapping of uptake function to the 10-residue QR-10 sequence within the 22-residue KW-22 peptide. Mapping was based on the uptake into CHO K1 and HS-deficient pgsD-677 cells. Fig. 5-I shows the sequences and relative location of several peptides (cf. table 1) described in Fig. 5-II. The previously proposed 8-residue heparin-binding basic sequence partially overlaps with the previously identified QW-14 sequence. Fig. 5-II shows the mapping of the uptake function to the 10-residue QR-10 sequence. Experimental conditions were the same as described for Fig. 3. (A) confocal images, (B) cellular fluorescence intensity distribution in flow cytometry and (C) absolute fluorescence levels of each treatment on the same scale. Note that in (C) the fluorescence level is presented as the absolute value, as opposed to the relative value in Fig. 3C. * indicates statistically significance between uptake in wild-type and uptake in a mutant cell line (n=3, p< 0.05).
The FITC-labeled QW-14, identical to the previously identified uptake core sequence (223–236) [17], exhibits robust uptake in both wt CHO K1 and heparan sulfate deficient pgs D-677 cell lines (Fig. 5-IIB and C). However, the upstream 8-mer sequence (KK-8) that contains 5 basic lysine residues was not able to permeate into the cells at all. Previously it was suggested that the 12-mer Cys-Cys loop in the peptide QW-14 may be important for its uptake activity [18], but in our study the replacement of the first cysteine in position 224 of the loop with glycine (QW-14mut) did not weaken the uptake much in both wt CHO K1 and the heparan sulfate-deficient pgs D-677 cells (Fig. 5-IIB and C). Moreover, a shorter 10-mer QR-10 peptide with complete deletion of caveolin-scaffolding domain consensus sequence, including the second Cys of the putative Cys-Cys loop, still remains about 64% uptake ability in CHO K1 and 80% in pgsD-677 cells (Figure 5B), suggesting that QR-10 retains a significant portion of KW-22’s cell entry function. The result also suggests that the postulated caveolin-scaffolding domain and the Cys-Cys loop either do not exist or are not important for cell penetration. Note that the 64% of QR-10 uptake level in CHO K1 and the 80% of QR-10 uptake level in pgs D-677 cannot be directly compared because they are referenced to respective KW-22 uptake values in two different cell types.
IGFBP-3 C-terminal Peptides Differentially Bind to Various Types of Glycosaminoglycans
To examine whether the extracellular uptake of IGFBP-3 C-terminal peptides correlates with their binding ability to GAGs, FITC-labeled peptides were added to various GAG species. As shown in Figure 6, all peptides show relatively strong binding to chondroitin sulfate B and heparan sulfate, but the binding affinity markedly decreases with the progressive shortening of the peptides. Exceptionally, the 8-mer basic KK-8 peptide show stronger binding to most GAG species than the 14-mer QW-14 and the 10-mer QR-10 peptides though no uptake was observed in cell culture (Figure 5B).
Figure 6.
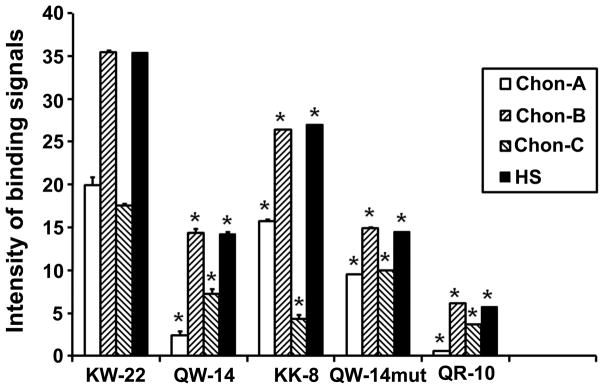
Mapping of GAG-binding function to the 8-residue KK-8 sequence within the 22-residue KW-22 peptide. FITC-labeled peptides (20μM) were incubated with 0.6 mM of CS -A, -B, -C, and HS, respectively, for one hour at 37°C. The bound peptides were separated from unbound peptides using Microcon (10KD) filter units and the bound fluorescence was read at 485/535 nm. * indicates that the difference between KW-22 and any one of other four shorter peptides in solution binding to a GAG (N=3, p<0.05).
DISCUSSION
There is now considerable evidence to support that IGFBP-3 is not only the binding protein of IGFs, it has direct IGF-independent effects on cellular functions. It is well noted that IGFBP-3 interacts with multiple extracellular components including transferrin, heparin, fibronectin, and collagen [3,7,26]. IGFBP-3 has also been proposed to bind cell surface receptors [31], but the intracellular effects may be mostly mediated by internalization and nucleus localization of IGFBP-3. It has been demonstrated that endogenous IGFBP-3 must be secreted before re-uptake and targeting to the nucleus [8]. However, the mechanisms and pathways by which IGFBP-3 is internalized into the cell are still poorly understood. Based on documented observations [17,28], and the characterization of GAG-binding domain present in the C-terminal region of IGFBP-3 [11], we recognized that the cell surface GAGs may be important role players for the protein transduction pathway.
In our study, IGFBP-3 C-terminal peptides readily entered cells as CPPs in a GAG-dependent manner, extending previously published studies demonstrating their cell penetrating property [16,18,28]. GAGs exist in animals as a part of heavily glycosylated proteoglycans, the other component being the core proteins. The core protein in a proteoglycan (PG) probably does not play a significant role in CPP binding and internalization. It is known that GAG, not the core protein, determines the binding specificity by GAG-binding proteins [32], which may be extended to GAG-CPP interaction. The cellular uptake of FITC-labeled C-terminal peptides of IGFBP-3 is almost eliminated in the pgsA-745 cell line, which express only 1% of wt GAGs, indicating that their presence is required. The loss of uptake in pgs A-745 was also observed in the case of widely used TAT CPP [33]. As described therein, the internalization of the TAT CPP mainly depends on the presence of cell surface heparan sulfate (HS), and the uptake is almost equally affected in HS-deficient pgs D-677 cells [26] and GAG-deficient pgs A-745 cells. In contrast, the uptake of IGFBP-3 CPPs is seriously impaired in pgsA-745 cells but only drops off approximately 40% in pgs D-677 cells, suggesting other GAG species, probably chondroitin sulfates (CS), are effectively involved in the internalization process. Two of the three chondroitin sulfates (CSs) did inhibited KW-22 uptake in pgs D-677 (Fig. 3D), supporting this hypothesis. The differential loss of uptake in pgs D-677 by the TAT CPP and IGFBP-3 CPPs could be due to a combination of the differential dependence of uptake on GAG negative charge and, to a less extent, on binding specificity (GAG structural feature). The loss of terminal sulfation on the 2-O-induronic acid monosaccharide of GAG in pgs F-17 [27] results in a minor loss of total negative charge and a subtle structural change in the GAG of a PG. In this study, uptake of IGFBP-3 KW-22 peptide was barely affected in pgs F-17, which is in sharp contrast to the complete loss of binding of bFGF protein to GAG on pgs F-17 cells [27]. This suggests that, unlike a protein, a smaller Arg-rich CPP like KW-22 relies more on GAG charge than on GAG structural feature for its interaction with GAG. This is not surprising. Compared to protein-protein interaction, carbohydrate-protein interaction is weak with monosaccharide-protein binding Kd at mM to μM range [34] and low specificity [35]. A survey of the known structures of carbohydrate-lectin complexes suggests that the underlying mechanism is a shallow binding pocket on lectin proteins and a limited variety of chemical groups on the sugars [36]. To achieve high affinity binding to carbohydrates, nature has evolved a strategy of multivalency binding (avidity) for lectins, enzymes, receptors and other carbohydrate-binding proteins [37,38]. Arg-rich CPPs, including most IGFBP-3 peptides in this study, do not exist in multimers as many carbohydrate-binding proteins do and have much less sequence complexity. They are thus expected to exhibit lower GAG-binding specificity. However, they possess much higher positive charge density that certainly contributes to binding affinity to many highly negatively charged GAGs in PGs. In this regard, it remains to be clarified how different types of sulfated GAG in different PGs, tyrosine-sulfated glycoproteins and the less negatively charged sialoglycoproteins affect cell entry of Arg-rich CPPs and their cargo-conjugates.
Previous work shows that the uptake of the 18-mer peptide Pep3 (Figure 1, position 215–232) of IGFBP-3 conjugated to GST protein is competitively inhibited by the presence of soluble heparin [28], indicating that peptide-GST complex may use heparan sulfate (HS) receptors for internalization. In our experiment, the uptake of SA-HRP conjugated peptide KG-28 that contains the Pep3 region and more functional domains was abolished in HS-deficient pgs A-745 and D-677 cells, and competitively blocked by soluble HS and condroitin sulfates, indicating the importance of GAGs in IGFBP-3 entry into cells. However, the uptake of unconjugated KW-22 or shorter peptides is only partially inhibited in cells lacking HS, and even enhanced in the pgs D-677 cells with the addition of exogenic chondroitin sulfate A (Figure 3D). These results indicate that unconjugated peptides can penetrate into cells using additional pathways differing from those followed by peptides conjugated to proteins, similar to the case of TAT peptide [33].
It is also noteworthy that the previously identified 14-mer sequence is not the minimal functional domain for penetrating into cells. In our experiments, a shorter 10-mer sequence without caveolin scaffolding docking domain still remains good penetrating activity. Also, a mutation of cysteine in the 14-mer sequence hardly loses its ability, indicating that the postulated 12-mer Cys-Cys loop is not necessary for this activity. Thus, the essential nuclear localization sequence “KGRKR” possibly is the core functional domain for cell penetration. Nevertheless, it remains to examine if peptides shorter than the 14-mer sequence can be internalized into cells when they are conjugated to macromolecules. It is also noteworthy that the uptake efficiency of these shorter peptides not only depends on their positive charge for binding to GAGs, the Arg-rich sequence appears to be important for the internalization activity. The 8-mer “basic domain” peptide KK-8 (KKGFYKKK) is devoid of Arg, binds to chondroitin sulfates and heparan sulfate with an affinity comparable to KW-22 (Fig. 6), but no uptake was observed when it was incubated with cells.
In summary, our experiments provide genetic and biochemical evidence to further substantiate the key role of cell surface glycosaminoglycans in mediating extracellular uptake of IGFBP-3 C-terminal peptides. Our data also suggests that GAG-binding and cell penetration functions could be mainly assigned to two adjacent 8-mer and 10-mer sequences in IGFBP-3 C-terminal region, which could shed light on Arg-rich CPP mechanism and design. As IGFBP-3 is a moderately abundant human protein in circulation, these peptides have enormous potential for applications in drug delivery as an alternative to the HIV-Tat derived cell penetrating peptide. Previous study indicates that IGFBP-3-derived peptides can selectively deliver therapeutic molecules into cancer cells, but the mechanism remains poorly understood [18]. There is some evidence that abnormal expression of glycosaminoglycans is present in cancer and found to correlate with clinical prognosis in several malignant neoplasms [39,40]. Based on our results, it is possible that the aberrant expression of GAGs on cell surface may contribute to selectivity of IGFBP-3 C-terminal peptides targeting cancer cells. Further study is necessary to explore the potential of these peptides for applications in clinical therapy as drug targeting moiety.
Acknowledgments
This work was supported by the NIH Grant AI 51214. We thank Dr. Jeffrey D. Esko for providing us wild type and mutant CHO cell lines. We also want to thank Dr. C. Gardner for assistance in confocal microscopy, Ms. Theresa Choi and Joan Dubois for performing some of FACS experiments at the Environmental and Occupational Health Science Institute/Cancer Institute of New Jersey Core Facility.
Footnotes
CONFLICT OF INTEREST
There are no conflicts of interests of the authors.
References
- 1.Laviola L, Natalicchio A, Giorgino F. The IGF-I signaling pathway. Curr Pharm Des. 2007;13:663–669. doi: 10.2174/138161207780249146. [DOI] [PubMed] [Google Scholar]
- 2.Clemmons DR. Use of mutagenesis to probe IGF-binding protein structure/function relationships. Endocr Rev. 2001;22:800–817. doi: 10.1210/edrv.22.6.0449. [DOI] [PubMed] [Google Scholar]
- 3.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 4.Duan C, Xu Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen Comp Endocrinol. 2005;142:44–52. doi: 10.1016/j.ygcen.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 5.Jogie-Brahim S, Feldman D, Oh Y. Unraveling IGFBP-3 Actions in Human Disease. Endocr Rev. 2009;30:417–437. doi: 10.1210/er.2008-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada PM, Lee KW. Perspectives in mammalian IGFBP-3 biology: local vs. systemic action. Am J Physiol Cell Physiol. 2009;296:C954–976. doi: 10.1152/ajpcell.00598.2008. [DOI] [PubMed] [Google Scholar]
- 7.Weinzimer SA, Gibson TB, Collett-Solberg PF, Khare A, Liu B, Cohen P. Transferrin is an insulin-like growth factor-binding protein-3 binding protein. J Clin Endocrinol Metab. 2001;86:1806–1813. doi: 10.1210/jcem.86.4.7380. [DOI] [PubMed] [Google Scholar]
- 8.Lee KW, Liu B, Ma L, Li H, Bang P, Koeffler HP, Cohen P. Cellular internalization of insulin-like growth factor binding protein-3: distinct endocytic pathways facilitate re-uptake and nuclear localization. J Biol Chem. 2004;279:469–476. doi: 10.1074/jbc.M307316200. [DOI] [PubMed] [Google Scholar]
- 9.Schedlich LJ, Young TF, Firth SM, Baxter RC. Insulin-like growth factor-binding protein (IGFBP)-3 and IGFBP-5 share a common nuclear transport pathway in T47D human breast carcinoma cells. J Biol Chem. 1998;273:18347–18352. doi: 10.1074/jbc.273.29.18347. [DOI] [PubMed] [Google Scholar]
- 10.Booth BA, Boes M, Andress DL, Dake BL, Kiefer MC, Maack C, Linhardt RJ, Bar K, Caldwell EE, Weiler J, Bar RS. IGFBP-3 and IGFBP-5 association with endothelial cells: role of C-terminal heparin binding domain. Growth Regul. 1995;5:1–17. [PubMed] [Google Scholar]
- 11.Fowlkes JL, Serra DM. Characterization of glycosaminoglycan-binding domains present in insulin-like growth factor-binding protein-3. J Biol Chem. 1996;271:14676–14679. doi: 10.1074/jbc.271.25.14676. [DOI] [PubMed] [Google Scholar]
- 12.Couet J, Li S, Okamoto T, Ikezu T, Lisanti MP. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J Biol Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- 13.Jaques G, Noll K, Wegmann B, Witten S, Kogan E, Radulescu RT, Havemann K. Nuclear localization of insulin-like growth factor binding protein 3 in a lung cancer cell line. Endocrinology. 1997;138:1767–1770. doi: 10.1210/endo.138.4.5177. [DOI] [PubMed] [Google Scholar]
- 14.Schedlich LJ, Le Page SL, Firth SM, Briggs LJ, Jans DA, Baxter RC. Nuclear import of insulin-like growth factor-binding protein-3 and -5 is mediated by the importin beta subunit. J Biol Chem. 2000;275:23462–23470. doi: 10.1074/jbc.M002208200. [DOI] [PubMed] [Google Scholar]
- 15.Wraight CJ, Liepe IJ, White PJ, Hibbs AR, Werther GA. Intranuclear localization of insulin-like growth factor binding protein-3 (IGFBP-3) during cell division in human keratinocytes. J Invest Dermatol. 1998;111:239–242. doi: 10.1046/j.1523-1747.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- 16.Radulescu RT. Nuclear localization signal in insulin-like growth factor-binding protein type 3. Trends Biochem Sci. 1994;19:278. doi: 10.1016/0968-0004(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 17.Singh B, Charkowicz D, Mascarenhas D. Insulin-like growth factor-independent effects mediated by a C-terminal metal-binding domain of insulin-like growth factor binding protein-3. J Biol Chem. 2004;279:477–487. doi: 10.1074/jbc.M307322200. [DOI] [PubMed] [Google Scholar]
- 18.Huq A, Singh B, Meeker T, Mascarenhas D. The metal-binding domain of IGFBP-3 selectively delivers therapeutic molecules into cancer cells. Anticancer Drugs. 2009;20:21–31. doi: 10.1097/CAD.0b013e3283144610. [DOI] [PubMed] [Google Scholar]
- 19.Mai JC, Shen H, Watkins SC, Cheng T, Robbins PD. Efficiency of protein transduction is cell type-dependent and is enhanced by dextran sulfate. J Biol Chem. 2002;277:30208–30218. doi: 10.1074/jbc.M204202200. [DOI] [PubMed] [Google Scholar]
- 20.Seligson DB, Yu H, Tze S, Said J, Pantuck AJ, Cohen P, Lee KW. IGFBP-3 nuclear localization predicts human prostate cancer recurrence. Horm Cancer. 2013;4:12–23. doi: 10.1007/s12672-012-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belting M. Heparan sulfate proteoglycan as a plasma membrane carrier. Trends Biochem Sci. 2003;28:145–151. doi: 10.1016/S0968-0004(03)00031-8. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs SM, Raines RT. Pathway for polyarginine entry into mammalian cells. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanagishita M, Hascall VC. Cell surface heparan sulfate proteoglycans. J Biol Chem. 1992;267:9451–9454. [PubMed] [Google Scholar]
- 24.Esko JD, Stewart TE, Taylor WH. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esko JD, Elgavish A, Prasthofer T, Taylor WH, Weinke JL. Sulfate transport-deficient mutants of Chinese hamster ovary cells. Sulfation of glycosaminoglycans dependent on cysteine. J Biol Chem. 1986;261:15725–15733. [PubMed] [Google Scholar]
- 26.Lidholt K, Weinke JL, Kiser CS, Lugemwa FN, Bame KJ, Cheifetz S, Massague J, Lindahl U, Esko JD. A single mutation affects both N-acetylglucosaminyl transferase and glucuronosyltransferase activities in a Chinese hamster ovary cell mutant defective in heparan sulfate biosynthesis. Proc Natl Acad Sci U S A. 1992;89:2267–2271. doi: 10.1073/pnas.89.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai X, Esko JD. An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J Biol Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- 28.Goda N, Tenno T, Inomata K, Shirakawa M, Tanaka T, Hiroaki H. Intracellular protein delivery activity of peptides derived from insulin-like growth factor binding proteins 3 and 5. Exp Cell Res. 2008;314:2352–2361. doi: 10.1016/j.yexcr.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 30.Fowlkes JL, Serra DM, Rosenberg CK, Thrailkill KM. Insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) functions as an IGF-reversible inhibitor of IGFBP-4 proteolysis. J Biol Chem. 1995;270:27481–27488. doi: 10.1074/jbc.270.46.27481. [DOI] [PubMed] [Google Scholar]
- 31.Oh Y, Muller HL, Pham H, Rosenfeld RG. Demonstration of receptors for insulin-like growth factor binding protein-3 on Hs578T human breast cancer cells. J Biol Chem. 1993;268:26045–26048. [PubMed] [Google Scholar]
- 32.Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interaction with proteins. Chem Biol Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 33.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J Biol Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 34.Zeng X, Andrade CA, Oliveira MD, Sun XL. Carbohydrate-protein interactions and their biosensing applications. Anal Bioanal Chem. 2012;402:3161–3176. doi: 10.1007/s00216-011-5594-y. [DOI] [PubMed] [Google Scholar]
- 35.Lee RT, Hsu TL, Huang SK, Hsieh SL, Wong CH, Lee YC. Survey of immune-related, mannose/fucose-binding C-type lectin receptors reveals widely divergent sugar-binding specificities. Glycobiology. 2011;21:512–520. doi: 10.1093/glycob/cwq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis WI, Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu Rev Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 37.Sansone F, Casnati A. Multivalent glycocalixarenes for recognition of biological macromolecules: glycocalyx mimics capable of multitasking. Chem Soc Rev. 2013;42:4623–4639. doi: 10.1039/c2cs35437c. [DOI] [PubMed] [Google Scholar]
- 38.Mammen M, Choi S, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphateglycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 40.Yip GW, Smollich M, Gotte M. Therapeutic value of glycosaminoglycans in cancer. Mol Cance Ther. 2006;5:2139–2148. doi: 10.1158/1535-7163.MCT-06-0082. [DOI] [PubMed] [Google Scholar]